Abstract
Named entity recognition in online medical consultation aims to address the challenge of identifying various types of medical entities within complex and unstructured social text in the context of online medical consultations. This can provide important data support for constructing more powerful online medical consultation knowledge graphs and improving virtual intelligent health assistants. A dataset of 26 medical entity types for named entity recognition for online medical consultations is first constructed. Then, a novel approach for deep named entity recognition in the medical field based on the fusion context mechanism is proposed. This approach captures enhanced local and global contextual semantic representations of online medical consultation text while simultaneously modeling high- and low-order feature interactions between local and global contexts, thereby effectively improving the sequence labeling performance. The experimental results show that the proposed approach can effectively identify 26 medical entity types with an average F1 score of 85.47%, outperforming the state-of-the-art (SOTA) method. The practical significance of this study lies in improving the efficiency and performance of domain-specific knowledge extraction in online medical consultation, supporting the development of virtual intelligent health assistants based on large language models and enabling real-time intelligent medical decision-making, thereby helping patients and their caregivers access common medical information more promptly.
1. Introduction
With the rapid development and advancements in mobile internet technology and improvements in health awareness, an increasing number of patients and their caregivers tend to obtain preliminary health advice through online medical consultation before going to the hospital [1]. Although online medical consultation can effectively alleviate social problems such as the uneven distribution of and shortages in medical resources and high medical expenses [2], the timeliness of information acquisition for online medical consultations cannot be guaranteed due to the lack of doctors who can provide online medical consultation services 24/7.
Virtual intelligent health assistants constructed by fully using the massive quantity of real doctor–patient question-and-answering data accumulated in online medical consultation [3] will provide timely, inexpensive, and accurate health services. However, the text of doctor–patient questions and answers in online medical consultation is mostly unstructured, which hinders medical knowledge discovery and clinical information extraction [4]. This further affects in-depth healthcare-related applications, such as medical history analysis and disease early warning. Medical named entity recognition can convert unstructured text into structured text effectively, providing important data support for constructing online medical consultation knowledge graphs and virtual intelligent health assistants.
Current medical named entity recognition mainly uses electronic medical records [5], medical literature [6], and clinical records [7] as data sources; online medical consultation texts are rarely used as a data source. As a result, there are no publicly available high-quality annotated datasets. This may be because online medical consultation in the context of social media is facing more serious problems related to nonstandard expression. This includes internet slang, lexical inflections, and spelling errors [4]. This problem may lead to a serious out-of-vocabulary (OOV) problem and seriously affect the performance of medical named entity recognition. In addition, the current research on medical named entity recognition for online medical consultation faces severe challenges, such as the scarcity of entity types and the complex semantics of the context [1,4].
To address the above problems, a large online medical consultation named entity recognition dataset containing 26 medical entity types is constructed. On this basis, a novel deep medical named entity recognition approach based on the fusion context mechanism named CBERT-SCNN-Local_Context-BiLSTM-Global_Context-DeepFM-Fusion_Context-MCRF is proposed. First, by comprehensively considering character embedding, glyph embedding, pinyin embedding, and position embedding in ChineseBERT [8], the OOV issue is mitigated, enhancing the model’s ability to generalize. Second, this approach combines a stacked convolutional neural network (CNN) and a local context mechanism to capture the local contextual semantic representation of the online medical consultation text. Third, this approach combines bidirectional long short-term memory (BiLSTM) and the global context mechanism to capture the global contextual semantic representation of online medical consultation text. Then, a deep factorization machine is combined with a fusion context mechanism to capture the high- and low-order feature interaction relationships between the local and global contextual semantic representations. Finally, the semantic representation of the fusion context obtained through the above steps is input to the masked conditional random field (MCRF) to determine the final medical entity tag sequence.
From the perspective of theoretical and practical significance, the novel contributions of this study are as follows:
- An online medical consultation named entity identification dataset for up to 26 medical entity types is constructed, which far exceeds those of the existing studies;
- This study provides important data support for constructing more powerful online medical consultation knowledge graphs and further virtual intelligent health assistants;
- A fusion context mechanism that can fuse the local and global contextual semantic representations is developed;
- This study investigates the performance of the state-of-the-art (SOTA) large language model (LLM) ChatGPT-4o in fine-grained named entity recognition tasks within a specific domain and discusses its applicability to the research problem.
2. Related Work
Named entity recognition constitutes an essential foundational task in natural language processing for building domain-specific knowledge graphs and developing virtual intelligent question-answering systems. Early named entity recognition methods were mainly based on complex human predefined rules and domain-specific dictionary matching. The extraction efficiency of specific information is higher; however, the generalization ability is poor. Adapting to the massive new vocabulary that is emerging in the mobile internet era and the high intellectual costs of domain-specific experts remain challenging [9]. Zeng et al. developed many regular expression-based matching rules to extract core medical concepts from discharge summaries [10]. Proux et al. formulated a series of rules with different properties to detect named entities in biological texts [11].
With the initial progress in computer software and hardware technology, machine learning-based named entity recognition methods represented by CRF and hidden Markov methods have gradually begun to increase. The shortcomings of rule- and dictionary-based approaches have been somewhat mitigated. However, in the modeling stage, eliminating the reliance on manual work remains challenging. Before training the machine learning model, manual feature engineering is needed; that is, a series of features that can effectively characterize the data characteristics of a specific dataset must be extracted. Settles combined a variety of traditional and novel features to identify five types of entities in biomedical abstracts using CRF [12]. Bikel et al. employed a modified version of the conventional hidden Markov model to detect nonrecursive entities within texts [13].
In the past ten years, breakthroughs have been made in computer software and hardware technology, and computing power has greatly increased. In particular, NVIDIA’s ongoing advancements in CUDA technology have significantly facilitated the emergence of deep learning-driven named entity recognition techniques based on neural networks. The novel end-to-end learning method has completely removed the dependence on manual labor in the modeling stage; the model is capable of autonomously adapting and extracting relevant features based on the contextual information. Strubell et al. explored the feasibility of an iterated dilated CNN for named entity recognition and proposed the ID-CNN-CRF model based on this approach [14]. Huang et al. applied the combination of BiLSTM and CRF to the natural language processing sequence annotation task for the first time, and the performance of entity recognition was improved by effectively using past and future input features [15]. On the basis of the study by Huang et al., Lample et al. further introduced a character-based word representation model [16] and a pretrained distributional representation model [17] to capture spelling and morphological information, effectively improving the entity recognition performance [18]. Building on this foundation, Li and Guo developed a combination of CNN, BiLSTM, and CRF, proposing the CNN-BiLSTM-CRF model, which effectively enhances entity recognition performance [19]. Based on the study of Li and Guo, Shang and Ran proposed the fusion multi-feature-CNN-BiLSTM-CRF model. By designing a novel word embedding algorithm that effectively integrates pinyin, radical, and word meaning features at the model’s word embedding layer, they successfully addressed the limitations of existing word embedding methods, leading to further improvements in entity recognition performance [6]. Tehseen et al. proposed the W2V-ELMo-CNN-BiLSTM-CRF model to enhance the performance of Shahmukhi named entity recognition [20]. Liu et al. proposed a word segmentation method based on dynamically updating a custom dictionary, and on this basis, they developed the BiLSTM-CRF-Loaded model, which effectively improved the performance of named entity recognition in Traditional Chinese Medicine chest discomfort cases [21]. Yu et al. proposed a document-level named entity recognition method that can simultaneously capture both word-level and sentence-level global contextual information, effectively mitigating the inherent ambiguity issues at the sentence level [22]. Zha et al. proposed a contrastive learning enhanced prototypical network that effectively captures boundary information and hidden relationships between entities across sequences for few-shot named entity recognition, achieving a strong performance [23]. Yang et al. proposed an uncertainty-aware contrastive learning-based semi-supervised named entity recognition method, which effectively improved the utilization of unlabeled data [24].
Table 1 presents a comparison of representative studies in the named entity recognition task in terms of methods, performance, and disadvantages.

Table 1.
Comparisons of existing named entity recognition methods.
Although the above studies have effectively promoted the technical progress of the named entity recognition task, they still have certain limitations: (1) they cannot effectively address the OOV problem, (2) they struggle to fully capture the complex contextual semantic information of the data, and (3) classic CRF may occasionally generate illegal tag sequences.
3. The Proposed Approach
This section provides a detailed explanation of our medical named entity recognition approach designed for online medical consultations. The overall model framework is shown in Figure 1. The framework comprises five core components, including the pretrained model, the stacked CNN with a local context mechanism, BiLSTM with a global context mechanism, the deep factorization machine with a fusion context mechanism, and MCRF. The framework operation process is divided into five main stages:
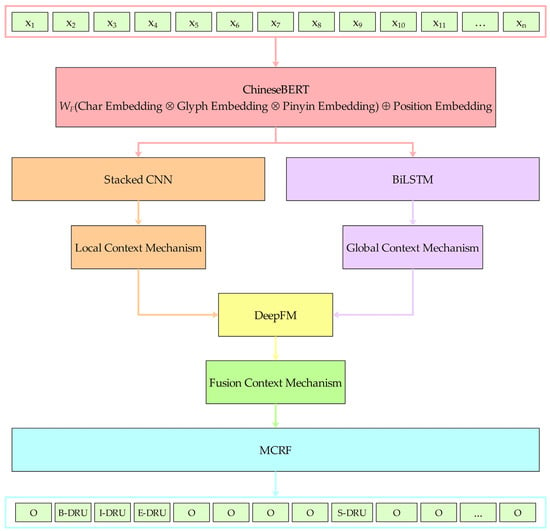
Figure 1.
Framework for a medical named entity recognition model utilizing a fusion context mechanism. denotes the weight matrix of the fully connected fusion layer; and , respectively, denote vector concatenation and addition.
- A data sample sentence is randomly selected from the experimental dataset. In the pretrained model component, ChineseBERT maps this sentence into a real-valued matrix. The matrix’s row count matches the sentence’s character count, while its column count corresponds to the final hidden layer dimension of ChineseBERT. Then, the real-valued matrix is input into the stacked CNN with the local context mechanism component and BiLSTM with the global context mechanism component to model the sentence’s contextual semantic representation from both local and global perspectives simultaneously.
- In the stacked CNN with the local context mechanism component, the real-valued matrix first learns local semantic features from simple to complex layer by layer through a three-layer stacked CNN to improve the local understanding ability of the input sentence. Then, the learned local semantic features are input to the multihead self-attention mechanism in the local context mechanism module. Each head within the multihead self-attention module independently acquires representations from distinct segments of the input sentence. These distinct segments are represented by concatenation and linear transformation. The representation comprehensively considers the information from different perspectives to capture the local contextual semantic representation of the input sentence. Subsequently, the local contextual semantic representation is input to the deep factorization machine with the fusion context mechanism component.
- In BiLSTM with the global context mechanism component, the real-valued matrix first learns global semantic features through BiLSTM’s capture of long-distance dependency to improve the global understanding ability of the input sentence. Then, the learned global semantic features are input into the global context mechanism module to further learn the global contextual semantic representation of the input sentence. Subsequently, the global contextual semantic representation is input to the deep factorization machine with the fusion context mechanism component.
- In the deep factorization machine with the fusion context mechanism component, the factorization machine module of DeepFM is used to capture the first-order and second-order low-order feature interaction relationships, and DeepFM’s deep module is used to capture high-order feature interaction relationships to fully fuse the local and global contextual semantic representations to obtain the fusion contextual semantic representation. Subsequently, the fusion contextual semantic representation is input to the MCRF component.
- In the MCRF component, with the help of the Viterbi algorithm and the masked transition matrix, the MCRF decodes the fusion contextual semantic representation into the tag sequence path with the highest probability score, and the occasional generation of illegal tag sequences is effectively prevented.
For better intuitive understanding, we also present the workflow of the proposed medical named entity recognition framework in pseudocode format in Algorithm 1.
| Algorithm 1 Our proposed medical named entity recognition framework |
| Input: The given data sample sentence X = {x1, x2, …, xn} Output: The predicted label sequence S 1: for i = 1 to n do 2: zi = ChineseBERT(xi) 3: li = SCNN(zi)//SCNN denotes the stacked convolutional neural network 4: hi = BiLSTM(zi)//BiLSTM denotes the bidirectional long short-term memory 5: LCi = LCM(li)//LCM denotes the local context mechanism 6: GCi = GCM(hi)//GCM denotes the global context mechanism 7: end for 8: LC = {LC1, LC2, …, LCn}//LC denotes the local context 9: GC = {GC1, GC2, …, GCn}//GC denotes the global context 10: FC = LC‖GC//FC denotes the fusion context 11: J = DeepFM(FC)//DeepFM denotes the deep factorization machine 12: S = MCRF(J)//MCRF denotes the masked conditional random field |
3.1. Pretrained Model Component
In this study, the pretrained model component utilizes a variant of bidirectional encoder representations from transformers (BERT), ChineseBERT, which comprehensively considers the glyph and pinyin information of Chinese characters [8]. As shown in Figure 2, ChineseBERT first splices character, glyph, and pinyin embedding into one, which is subsequently mapped into a fusion embedding through a fully connected fusion layer. Subsequently, the fusion and position embedding are added and input to the BERT layer, thus obtaining richer syntactic and semantic information for language understanding [8]. Specifically, given a data sample sentence , represents the number of characters contained in the sentence, which can be mapped by ChineseBERT to a representation containing context information as follows:
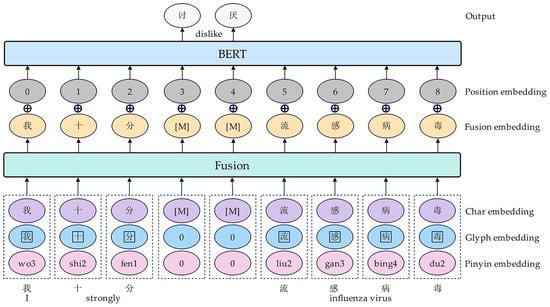
Figure 2.
Architecture diagram of the pretrained model component.
3.2. Stacked CNN with a Local Context Mechanism Component
As shown in Figure 3, in this study, we first use a three-layer stacked CNN to learn local semantic features . Subsequently, the local context mechanism module employs the multihead self-attention mechanism to capture the local context semantic representation [6,25,26]:
where denotes the rectified linear unit (ReLU) activation function, KP represents the convolution kernel parameters, WS represents the convolution window size, represents the dimension of the key vector in each head, m represents the number of heads, and , , , and are learnable parameter matrices.
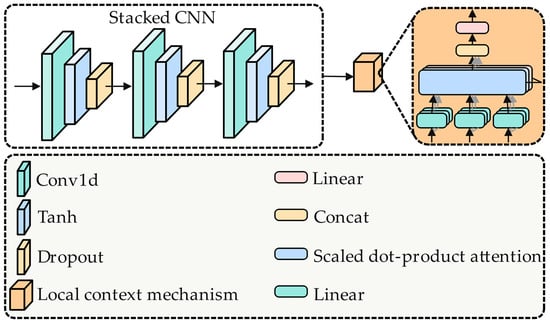
Figure 3.
Architecture diagram of stacked CNN with local context mechanism component.
3.3. BiLSTM with Global Context Mechanism Component
As shown in Figure 4, in the BiLSTM with the global context mechanism component, BiLSTM models the sentence structure by capturing the forward and backward long-distance dependency [20]. Taking time step t as an example, the hidden semantic representation of Z based-BiLSTM models X is as follows [25,27]:
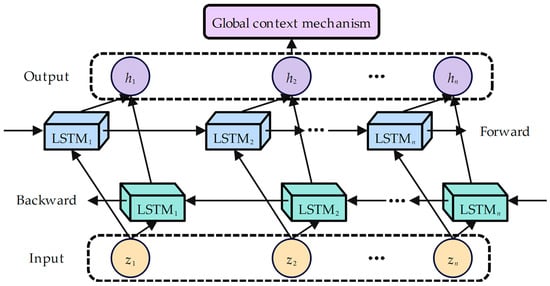
Figure 4.
Architecture diagram of BiLSTM with global context mechanism component.
Thus, the output of BiLSTM can be obtained as . The global context mechanism was proposed by Xu et al., who found that the representation of the entire sentence found in the forward last unit and the backward first unit of the BiLSTM can effectively complement the single-sentence representation for each unit [25]. Taking time step t as an example, the modeling process is as follows:
where and are learnable parameter matrices, represents the sigmoid function, represents the weight of the current sentence’s hidden semantic representation , represents the weight assigned to the entire sentence representation , represents the element-wise product, and represents the weighted combination of the entire sentence and current unit . Thus, the global context semantic representation GC can be obtained:
3.4. Deep Factorization Machine with Fusion Context Mechanism Component
In terms of the deep factorization machine with the fusion context mechanism component, in the present study, we introduce a comprehensive end-to-end learning framework, DeepFM, into the recommender system field, which is capable of concurrently grasping both low- and high-order feature interactions [28]. As shown in Figure 5, by improving its input and output design, a fusion context mechanism suitable for information fusion field is developed, marking the first application of DeepFM for model fusion. This approach effectively fuses local and global context semantic representations, resulting in a fused context semantic representation. There are two main parts of DeepFM: the factorization machine module FM and the deep module DNN. The factorization machine module is used to capture low-order (first- and second-order) feature interaction relationships, while the deep module specifically addresses the capture of high-order feature interaction relationships. The process unfolds as follows [28]:
where , , k represents the factorization dimension, represents the concatenation of the local context semantic representation and the global context semantic representation , represents the fusion context semantic representation, denotes the count of hidden layers within the deep module, and represents the ReLU activation function.
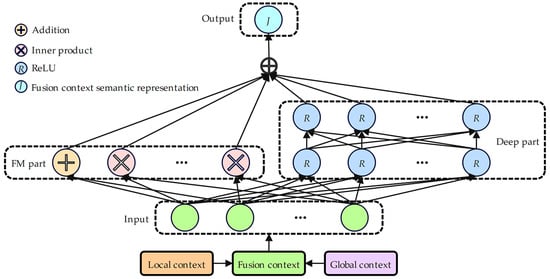
Figure 5.
Architecture diagram of deep factorization machine with fusion context mechanism component.
3.5. MCRF Component
As shown in Figure 6, in this study, we introduce an MCRF, which can effectively address the occasional illegal tag sequence generated by the traditional conditional random field. This method is both easy to implement and achieves performance improvements over traditional CRF with little additional cost [29]. For a given input data sample sentence , its predicted tag sequence path can be expressed as ; the scoring process is as follows:
where represents a tag sequence path, is the transfer score matrix, represents the i-th logit generated by the encoder network of the model, indicates the -th entry of , represents the score from label transferring to label in the transfer score matrix , represents the set of all possible tag sequence paths, represents the set of all illegal tag sequence paths, represents the set of all legal tag sequence paths, represents the set of all training samples, represents a test sample sentence, and represents the minimum loss value.
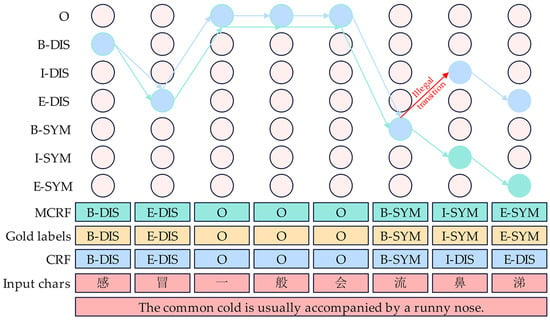
Figure 6.
An example of the MCRF component workflow.
4. Research Design and Analysis of Results
4.1. Data Collection and Annotation Approach
The dataset used in this study is based on the HaoDF collected in our previous work [30]. HaoDF (https://www.haodf.com/) is one of China’s leading and most popular online medical consultation platforms. Since its establishment in 2006, it has provided online medical consultation services to over 84 million patients and their caregivers as of July 2023. Based on the medical entity recognition dataset, substantial updates have been made: (1) the number of supported medical entity types has expanded to 26 types [6,30,31,32,33], which is far more than those of the existing studies, and (2) some outdated data samples have been updated to include data samples that can reflect the latest medical developments, such as interactive question-and-answer pairs between doctors and patients related to the COVID-19 pandemic. The BIOES-Y tagging strategy was used for the dataset, where B, I, O, E, S, and Y represent the abbreviation of beginning tag, inside tag, outside tag, end tag, single-character tag, and entity type, respectively. To balance data annotation efficiency and reliability, this study employed a two-stage annotation approach combining algorithmic auto-annotation with expert verification. First, a bidirectional maximum matching algorithm combined with a medical dictionary was used for large-scale, rapid automatic annotation. Then, three medical experts were invited to verify the annotation results. To ensure consistency and eliminate subjective bias among annotators, a voting mechanism was implemented to determine the final entity boundaries and types in cases of disagreement. The complete types and samples of medical entities are shown in Table 2. Table 3 presents the actual annotated samples from the experimental dataset.

Table 2.
Medical entity types and examples.

Table 3.
Experimental dataset samples. Entities are displayed in both bold and italics.
4.2. Experimental Setup
For this study, the experimental dataset was segmented into training, validation, and test sets in a 3:1:1 ratio. The detailed statistics of the experimental datasets are shown in Table 4. Table 5 details the software and hardware settings used in the experiments. Table 6 outlines the key parameter settings of the proposed approach, aiming to guarantee the reliability and reproducibility of the experimental outcomes, where max sequence length represents the maximum input sequence length supported by the model, learning rate indicates the learning rate at which the model converges to the optimal solution, optimizer represents the optimizer used in the model to minimize the objective function, and Adam epsilon is usually added to the denominator to prevent division by zero errors during model calculation; the hidden dropout ratio is the regularization strategy employed to curb model overfitting, warmup proportion represents the proportion of the warmup period in the total training steps in the learning rate warmup strategy, and weight decay serves as a regularization strategy designed to mitigate model overfitting. Max epochs represent the number of epochs for the entire training set to completely pass through and return once. The batch size denotes the quantity of data samples processed per iteration in training, while BiLSTM size specifies the dimension of the hidden layer of the long short-term memory (LSTM) unit in one direction.

Table 4.
Statistics of the experimental dataset.

Table 5.
Experimental setup.

Table 6.
Parameter settings of the proposed approach.
4.3. Evaluation Metrics
To assess model performance, this study employed three standard evaluation metrics common in named entity recognition: precision (P), recall (R), and the F1 score (F1) [34]. To verify the reliability and stability of the experimental results, all experiments were performed for five complete rounds. The average experimental result of the three evaluation indicators was used as the final experimental result, and the standard deviation of the average experimental result was provided.
4.4. Experimental Results and Discussion
4.4.1. Evaluative Comparison Across Methods
As illustrated by the first nine rows in Table 7, to ensure a balanced comparison with previous research, in this study, we established nine baseline methods based on the literature to validate the efficacy of the proposed approach. As Rows 10 and 11 of Table 7 show, to evaluate the performance of the SOTA LLM on the constructed dataset in this study, we established two baseline methods based on zero-shot prompting: ChatGPT-4o_26_tags and ChatGPT-4o_5_tags. ChatGPT-4o_26_tags represents the overall performance of ChatGPT-4o (https://chatgpt.com/, accessed on 3 February 2025) across all 26 entity categories in the constructed dataset, while ChatGPT-4o_5_tags reflects its overall performance on the five common entity categories (i.e., body, symptom, check, disease, and treatment) consistent with the categories used in the literature [35]. As Row 12 of Table 7 shows, to prove the novelty of the approach proposed in this study, the latest SOTA method [25] was also introduced for comparison. Notably, for a fair comparison, we replaced the BERT models in all baseline methods with CBERT, which demonstrates a superior performance and is consistent with the proposed approach in this study. From Table 7, the following conclusions can be drawn:

Table 7.
Comparative analysis of our approach against baseline models. Here, P, R, and F1 denote precision, recall, and F1 scores, respectively, which are three performance evaluation metrics for models. Optimal results are highlighted in bold. CBERT denotes the ChineseBERT pretrained model, SCNN denotes the stacked convolutional neural network, and MCRF represents the masked conditional random field.
- According to Rows 1–3 in Table 7, the F1 scores of CBERT, CBERT-CRF, and CBERT-MCRF were 80.33%, 82.10%, and 83.42%, respectively. The experimental results show that the model performance can be effectively improved by adding the decoding layer constraint on the CBERT basis. Better performance is obtained because MCRF provides a stronger constraint relationship than CRF and effectively solves the problem of CRF occasionally generating illegal tag sequences.
- According to Rows 3, 5, and 7 in Table 7, the F1 scores of CBERT-MCRF, CBERT-SCNN-MCRF, and CBERT-BiLSTM-MCRF were 83.42%, 84.06%, and 84.37%, respectively. The data indicate that both the SCNN and BiLSTM neural networks can enhance the model’s capability to learn features, with BiLSTM demonstrating a more robust feature learning capacity compared to SCNN. This is because SCNN is more suitable for spatial feature extraction scenarios (such as video processing and image segmentation), while BiLSTM is more suitable for processing sequential data (such as text).
- As Rows 5, 7, and 8 in Table 7 show, the F1 scores of CBERT-SCNN-MCRF, CBERT-BiLSTM-MCRF, and CBERT-SCNN-BiLSTM-MCRF were 84.06%, 84.37%, and 85.17%, respectively. The data indicate that the combination of SCNN and BiLSTM in parallel can complement the advantages of the two methods, which can fully utilize the advantage of SCNN in local feature extraction and fully exploit the advantage of BiLSTM in global feature extraction, thus improving the model’s performance further.
- As Rows 8 and 9 in Table 7 show, the F1 scores of CBERT-SCNN-BiLSTM-MCRF and CBERT-SCNN-Local_Context-BiLSTM-Global_Context-MCRF were 85.17% and 85.24%, respectively. The experimental results show that adding the local context mechanism module after SCNN and the global context mechanism module after BiLSTM can notably enhance model performance, thereby confirming the efficacy of the local context mechanism module in learning local contextual semantic representation and the global context mechanism module in learning global contextual semantic representation.
- As Rows 9 and 13 in Table 7 show, the F1 scores of CBERT-SCNN-Local_Context-BiLSTM-Global_Context-MCRF and CBERT-SCNN-Local_Context-BiLSTM-Global_Context-DeepFM-Fusion_Context-MCRF were 85.24% and 85.47%, respectively. The experimental results show that adding a deep factorization machine with fusion context mechanism component DeepFM-Fusion_Context to the model can better fuse the local contextual semantic representation learned by the stacked CNN with the local context mechanism component, and the global contextual semantic representation learned by BiLSTM with the global context mechanism component, further improving the performance. This is mainly because the factorization machine module in the deep factorization machine with the fusion context mechanism component can capture the low-order (first- and second-order) feature interactions between the local and global contextual semantic representations, while the deep module can capture the high-order feature interactions between the local and global contextual semantic representations to finally obtain the fusion contextual semantic representation.
- As Rows 10 and 11 in Table 7 show, the F1 scores of ChatGPT-4o_26_tags and ChatGPT-4o_5_tags were 42.14% and 54.29%, respectively. The experimental results demonstrate that, under the premise of zero-shot prompting, although large language models (LLMs) have achieved remarkable success in general domains, their performance in specific domains remains suboptimal, with significant gaps compared to traditional methods. Through an in-depth analysis of the prediction results, we found that ChatGPT-4o struggles to recognize fine-grained entity categories that are uncommon in the specific domain. Even for the five common entity categories, ChatGPT-4o often generates hallucinations or makes incorrect judgments in its responses. This may be attributed to the fact that the training corpus of ChatGPT-4o is more focused on general domain generalization, lacking specialized optimization for specific domains. This finding highlights the necessity of training domain-specific entity recognition models for fine-grained named entity recognition tasks in specialized domains, as LLMs under zero-shot prompting are currently inadequate for such tasks.
- As Rows 12 and 13 in Table 7 show, the F1 scores of CBERT-BiLSTM-context (SOTA) and CBERT-SCNN-Local_Context-BiLSTM-Global_Context-DeepFM-Fusion_Context-MCRF were 84.48% and 85.47%, respectively. The data indicate that our proposed approach outperforms the current SOTA method by a significant margin of 0.99%. This is because the SOTA method only considers the extraction of global contextual semantic representation, neglecting the extraction of local contextual semantic representation. It also fails to incorporate a fusion context mechanism that can capture high-order and low-order feature interaction relationships. Furthermore, it does not use a decoding layer constraint, MCRF, to enhance the validity of the predicted tag sequence.
4.4.2. Ablation Study
As Table 8 shows, ablation experiments were conducted using the experimental dataset. From Table 8, the following conclusions can be drawn:

Table 8.
Ablation study conducted on the experimental dataset.
- As Rows 1 and 2 in Table 8 show, after the approach proposed in this study removed the deep factorization machine with the fusion context mechanism component DeepFM-Fusion_Context, the model’s F1 score dropped by 0.23%, indicating that this component is effective in fusing the local and global context semantic representations.
- As Rows 1 and 3 of Table 8 show, after removing the deep factorization machine with the fusion context mechanism component DeepFM-Fusion_Context from the approach proposed in this study, the local context mechanism module Local_Context and the global context mechanism module Global_Context were further removed and the model’s F1 score dropped by 0.30%, indicating the effectiveness of the Local_Context module and the Global_Context module in improving model performance.
- As Rows 1 and 4 in Table 8 show, after removing the deep factorization machine with the fusion context mechanism component DeepFM-Fusion_Context, the local context mechanism module Local_Context, and the global context mechanism module Global_Context from the approach proposed in this study, on this basis, further removing the stacked CNN (SCNN) module decreased the F1 score of the model by 1.10%, indicating that the local semantic features extracted by this module are important in improving model performance.
- As Rows 1 and 5 of Table 8 show, after removing the deep factorization machine with the fusion context mechanism component DeepFM-Fusion_Context, the local context mechanism module Local_Context, and the global context mechanism module Global_Context from the approach proposed in this study, on this basis, the BiLSTM module was further removed and then the model’s F1 score dropped by 1.41%, indicating that the global semantic features extracted by this module are critical for improving model performance.
- As Rows 1 and 6 of Table 8 show, after the components of the deep factorization machine with the fusion context mechanism DeepFM-Fusion_Context, the stacked CNN with local context mechanism SCNN-Local_Context, and the BiLSTM with global context mechanism BiLSTM-Global_Context were simultaneously removed from the proposed approach, and the model’s F1 score dropped by 2.05%, indicating that these three components synergistically enhance the effectiveness of medical named entity recognition within online medical consultation.
4.4.3. Performance of the Proposed Approach on Different Medical Entity Types
Table 9 illustrates the varied performance of the proposed approach across different medical entity types. Based on the data presented in Table 9, several conclusions can be drawn:

Table 9.
Experimental results of the proposed approach for different medical entity types.
- As Rows 1, 11, and 22 in Table 9 show, the performance indicators for entity types AMO, DRT, and REA not only have low mean values but also very high standard deviations. This is due to the scarcity of these three types of medical entities in the experimental dataset, which prevented the model from adequately learning effective features.
- As Rows 5, 7, 8, 13, 17, 19, and 20 in Table 9 show, the F1 scores of the entity types CON, DEG, DEP, DUR, FRE, PAS, and POS were all above 90%, indicating that the model has a strong ability to identify these entity types. This arises from the relatively small number of unique entities within these types, facilitating the model’s ability to learn their intrinsic feature patterns.
- As Rows 10, 14, 15, 16, 18, 23, and 25 in Table 9 show, the F1 scores of the entity types DRD, EFF, EQU, FOG, MIC, SIG, and SYM were all below 80%. The reason for the weak identification ability of the model to the entity types is that they are easier to express freely in the context of online medical consultation, and the pronounced inconsistency in expression significantly hampers the model’s ability to thoroughly learn the intrinsic feature patterns.
4.4.4. Effects of Hyperparameters
As Table 10 shows, in this research, we further investigated how the learning rate, a crucial hyperparameter, impacts the performance of the proposed method. The learning rate influences the model’s training efficiency, convergence, stability, and generalization capabilities, making it vital for achieving an effective and efficient model. Table 10 shows that the proposed approach performs best when the learning rate is set to 2 × 10−05. A higher learning rate will make it difficult for the model to converge effectively, and a lower learning rate will make it difficult for the model to jump out of the local minima.

Table 10.
Model performance under different learning rates. Optimal results are highlighted in bold.
Figure 7 illustrates how the number of epochs impacts model performance during a single experimental cycle. It shows that the F1 score of the proposed approach increases rapidly as the number of epochs increases; it then starts to fluctuate and increase after the third epoch, and then gradually stabilizes. This indicates that the proposed approach can quickly learn the intrinsic feature patterns of the data samples, achieving near-optimal performance at the fastest speed, thereby demonstrating the effectiveness of the approach developed in this study. Moreover, by integrating an early stopping strategy, the model training can be completed more rapidly, reducing computational resource consumption.
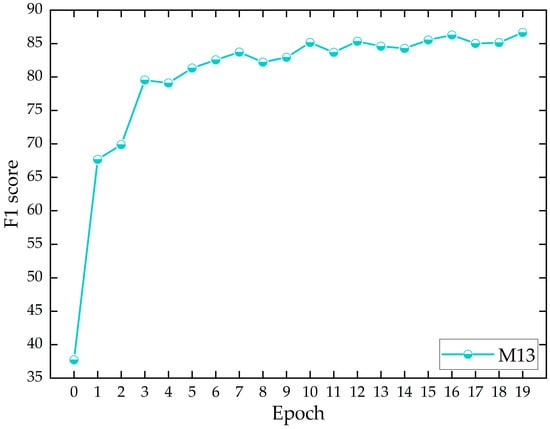
Figure 7.
The effect of the number of epochs on the F1 score during a single experimental cycle. M13 represents our proposed approach.
4.4.5. Research Implications and Limitations
With the increasing maturity of large language models (LLMs) represented by ChatGPT-4o and DeepSeek-V3, an increasing number of patients and their caregivers are turning to LLMs for basic health advice, while more physicians are exploring the use of LLMs to assist in medical decision-making. This trend holds the potential to reduce healthcare costs and improve medical efficiency. However, LLMs exhibit a phenomenon known as hallucination, which can result in generated content lacking factual consistency, posing significant challenges for their deeper clinical applications. Integrating medical knowledge graphs to provide additional domain-specific knowledge can offer more precise factual support for LLM reasoning, effectively mitigating hallucination issues and reducing clinical application risks. A key prerequisite for constructing high-quality medical knowledge graphs is high-performance medical named entity recognition technology, underscoring the significant practical value of this research.
Compared to electronic medical records, online medical consultation texts in social environments are more complex and less standardized. The high cost of obtaining high-quality annotated data continues to limit the size of experimental datasets, which, in turn, restricts the types and quantity of supported medical entities. The diversity of medical entity types determines the granularity of the extracted medical knowledge, while the quantity of entities within the same type determines their statistical significance. An insufficient number of entities within a given type can lead to significant misclassification, thereby reducing the overall recognition performance of the model.
5. Conclusions and Future Work
5.1. Conclusions
In this research, we proposed an innovative deep named entity recognition approach for the medical field based on a fusion context mechanism for online medical consultations and developed an experimental dataset comprising as many as 26 different medical entity types. The OOV problem was effectively alleviated by introducing ChineseBERT; the local contextual semantic representation of the data samples was effectively learned by the design of the stacked CNN with a local context mechanism component; the global contextual semantic representation of the data samples was effectively learned by designing a BiLSTM with a global context mechanism component; the local and global contextual semantic representations were effectively fused using a deep factorization machine with a fusion context mechanism component; and the occasional generation of illegal tag sequences was effectively prevented by introducing the MCRF component. Each of these can effectively improve the entity recognition performance, resulting in the proposed method outperforming previous SOTA research by 0.99% in terms of the average F1 score. This study also provides important data support for the main step of knowledge extraction in constructing domain-specific knowledge graphs. Additionally, the proposed approach in this study is based on Chinese characters, making it easily applicable to other character-level Chinese NER datasets in various domains, such as OntoNotes 4.0 [39] in the news domain and Weibo [40] in the social media domain. However, when applying this approach to NER datasets in other languages, the ChineseBERT framework needs to be replaced with a BERT model or its variants pretrained in the target language.
5.2. Future Work
Although the approach proposed in this study is effective, there is still room for further optimization: (1) both the variety of entity types and the quantity of each entity within the experimental dataset require further expansion; (2) a pretrained model that contains richer domain-specific information should be introduced; (3) a stronger data sample semantic representation extraction scheme can be designed; and (4) a fusion context mechanism with a stronger information fusion ability can be designed.
Author Contributions
Conceptualization, Z.H.; methodology, Z.H.; data curation, W.L.; writing—original draft, Z.H.; writing—review and editing, W.L. and H.Y.; visualization, W.L.; supervision, H.Y.; funding acquisition, Z.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant numbers 62201576 and U2433205) and the Supporting Fund of the National Natural Science Foundation of China (grant number 3122023PT10).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author due to privacy, legal, or ethical reasons.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wen, G.; Chen, H.; Li, H.; Hu, Y.; Li, Y.; Wang, C. Cross Domains Adversarial Learning for Chinese Named Entity Recognition for Online Medical Consultation. J. Biomed. Inform. 2020, 112, 103608. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, T.; Chen, Y.; Zhang, H. Knowledge sharing in online health communities: A social exchange theory perspective. Inform. Manag. 2016, 53, 643–653. [Google Scholar] [CrossRef]
- Guo, Q.; Cao, S.; Yi, Z. A medical question answering system using large language models and knowledge graphs. Int. J. Intell. Syst. 2022, 37, 8548–8564. [Google Scholar] [CrossRef]
- Yang, H.; Gao, H. Toward sustainable virtualized healthcare: Extracting medical entities from Chinese online health consultations using deep neural networks. Sustainability 2018, 10, 3292. [Google Scholar] [CrossRef]
- Sudat, S.E.; Robinson, S.C.; Mudiganti, S.; Mani, A.; Pressman, A.R. Mind the Clinical-Analytic Gap: Electronic Health Records and COVID-19 Pandemic Response. J. Biomed. Inform. 2021, 116, 103715. [Google Scholar] [CrossRef]
- Shang, F.; Ran, C. An Entity Recognition Model Based on Deep Learning Fusion of Text Feature. Inform. Process. Manag. 2022, 59, 102841. [Google Scholar] [CrossRef]
- Lybarger, K.; Ostendorf, M.; Thompson, M.; Yetisgen, M. Extracting COVID-19 Diagnoses and Symptoms from Clinical Text: A New Annotated Corpus and Neural Event Extraction Framework. J. Biomed. Inform. 2021, 117, 103761. [Google Scholar] [CrossRef]
- Sun, Z.; Li, X.; Sun, X.; Meng, Y.; Ao, X.; He, Q.; Wu, F.; Li, J. Chinesebert: Chinese pretraining enhanced by glyph and pinyin information. arXiv 2021, arXiv:2106.16038. [Google Scholar]
- Ji, J.; Chen, B.; Jiang, H. Fully-Connected LSTM-CRF on Medical Concept Extraction. Int. J. Mach. Learn. Cyb. 2020, 11, 1971–1979. [Google Scholar] [CrossRef]
- Zeng, Q.T.; Goryachev, S.; Weiss, S.; Sordo, M.; Murphy, S.N.; Lazarus, R. Extracting Principal Diagnosis, Co-Morbidity and Smoking Status for Asthma Research: Evaluation of a Natural Language Processing System. BMC Med. Inform. Decis. Mak. 2006, 6, 30. [Google Scholar] [CrossRef]
- Proux, D.; Rechenmann, F.; Julliard, L.; Pillet, V.; Jacq, B. Detecting Gene Symbols and Names in Biological Texts A First Step Toward Pertinent Information Extraction. Genome Inform. 1998, 9, 72–80. [Google Scholar]
- Settles, B. Biomedical named entity recognition using conditional random fields and rich feature sets. In Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications, Geneva, Switzerland, 28–29 August 2004; pp. 104–107. [Google Scholar]
- Bikel, D.M.; Miller, S.; Schwartz, R.; Weischedel, R. Nymble: A high-performance learning name-finder. In Proceedings of the Fifth Conference on Applied Natural Language Processing, Washington, DC, USA, 31 March–3 April 1997; pp. 194–201. [Google Scholar]
- Strubell, E.; Verga, P.; Belanger, D.; McCallum, A. Fast and accurate entity recognition with iterated dilated convolutions. arXiv 2017, arXiv:1702.02098. [Google Scholar]
- Huang, Z.; Xu, W.; Yu, K. Bidirectional LSTM-CRF models for sequence tagging. arXiv 2015, arXiv:1508.01991. [Google Scholar]
- Ling, W.; Luís, T.; Marujo, L.; Astudillo, R.F.; Amir, S.; Dyer, C.; Black, A.W.; Trancoso, I. Finding function in form: Compositional character models for open vocabulary word representation. arXiv 2015, arXiv:1508.02096. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.; Dean, J. Distributed representations of words and phrases and their compositionality. In Proceedings of the 26th International Conference on Neural Information Processing Systems, Sydney, NSW, Australia, 12–15 December 2013; pp. 3111–3119. [Google Scholar]
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural Architectures for Named Entity Recognition. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, San Diego, CA, USA, 12–17 June 2016; pp. 260–270. [Google Scholar]
- Li, L.; Guo, Y. Biomedical named entity recognition with CNN-BLSTM-CRF. J. Chin. Inform. Process. 2018, 32, 116–122. [Google Scholar]
- Tehseen, A.; Ehsan, T.; Liaqat, H.B.; Kong, X.; Ali, A.; Al-Fuqaha, A. Shahmukhi named entity recognition by using contextualized word embeddings. Expert Syst. Appl. 2023, 229, 120489. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, L.; Ren, G.; Zou, B. Research on named entity recognition of Traditional Chinese Medicine chest discomfort cases incorporating domain vocabulary features. Comput. Biol. Med. 2023, 166, 107466. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, Z.; Wei, W.; Zhang, R.; Mao, X.-L.; Feng, S.; Wang, F.; He, Z.; Jiang, S. Exploiting global contextual information for document-level named entity recognition. Knowl.-Based Syst. 2024, 284, 111266. [Google Scholar] [CrossRef]
- Zha, E.; Zeng, D.; Lin, M.; Shen, Y. Ceptner: Contrastive learning enhanced prototypical network for two-stage few-shot named entity recognition. Knowl.-Based Syst. 2024, 295, 111730. [Google Scholar] [CrossRef]
- Yang, K.; Yang, Z.; Zhao, S.; Yang, Z.; Zhang, S.; Chen, H. Uncertainty-Aware Contrastive Learning for semi-supervised named entity recognition. Knowl.-Based Syst. 2024, 296, 111762. [Google Scholar] [CrossRef]
- Xu, C.; Shen, K.; Sun, H. Supplementary features of BiLSTM for enhanced sequence labeling. arXiv 2023, arXiv:2305.19928. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; pp. 1–11. [Google Scholar]
- Li, R.; Xiao, Q.; Yang, J.; Ren, H.; Chen, Y. Few-Shot Relation Extraction via the Entity Feature Enhancement and Attention-Based Prototypical Network. Int. J. Intell. Syst. 2023, 2023, 1186977. [Google Scholar] [CrossRef]
- Guo, H.; Tang, R.; Ye, Y.; Li, Z.; He, X. DeepFM: A factorization-machine based neural network for CTR prediction. arXiv 2017, arXiv:1703.04247. [Google Scholar]
- Wei, T.; Qi, J.; He, S.; Sun, S. Masked Conditional Random Fields for Sequence Labeling. arXiv 2021, arXiv:2103.10682. [Google Scholar]
- Hu, Z.; Ma, X. A novel neural network model fusion approach for improving medical named entity recognition in online health expert question-answering services. Expert Syst. Appl. 2023, 223, 119880. [Google Scholar] [CrossRef]
- Chen, W.; Li, Z.; Fang, H.; Yao, Q.; Zhong, C.; Hao, J.; Zhang, Q.; Huang, X.; Peng, J.; Wei, Z. A benchmark for automatic medical consultation system: Frameworks, tasks and datasets. Bioinformatics 2023, 39, btac817. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liang, W. Medical name entity recognition based on lexical enhancement and global pointer. Int. J. of Adv. Comput. Sc. 2023, 14, 592–600. [Google Scholar] [CrossRef]
- Saad, S.; Zikun, H. Leveraging Transfer Learning and Label Optimization for Enhanced Traditional Chinese Medicine Ner Performance. Asia-Pac. J. Inf. Technol. Multimed. 2024, 13, 47. [Google Scholar] [CrossRef]
- Zhang, F.; Ma, L.; Wang, J.; Cheng, J. An MRC and adaptive positive-unlabeled learning framework for incompletely labeled named entity recognition. Int. J. Intell. Syst. 2022, 37, 9580–9597. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhao, Q.; Li, J.; Ge, Y.; Ding, X.; Gu, T.; Zou, J.; Lv, S.; Wang, S.; Yang, J.-J. Comparative Analysis of Large Language Models in Chinese Medical Named Entity Recognition. Bioengineering 2024, 11, 982. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, Y.; Mu, J.; Li, W.; Jiao, S.; Wang, Z.; Lv, P.; Zhu, Y. Chinese mineral named entity recognition based on BERT model. Expert Syst. Appl. 2022, 206, 117727. [Google Scholar] [CrossRef]
- Yang, B.; Zhou, C.; Li, S.; Wang, Y. A Chinese named entity recognition method for landslide geological disasters based on deep learning. Eng. Appl. Artif. Intel. 2025, 139, 109537. [Google Scholar] [CrossRef]
- Ke, J.; Wang, W.; Chen, X.; Gou, J.; Gao, Y.; Jin, S. Medical entity recognition and knowledge map relationship analysis of Chinese EMRs based on improved BiLSTM-CRF. Comput. Electr. Eng. 2023, 108, 108709. [Google Scholar] [CrossRef]
- Weischedel, R.; Pradhan, S.; Ramshaw, L.; Palmer, M.; Xue, N.; Marcus, M.; Taylor, A.; Greenberg, C.; Hovy, E.; Belvin, R. Ontonotes release 4.0. LDC2011T03, Philadelphia, Penn.: Linguistic Data Consortium. LDC2011T03 Philadel-Phia Penn. Linguist. Data Consort. 2011, 17, 1–53. [Google Scholar]
- Peng, N.; Dredze, M. Named entity recognition for chinese social media with jointly trained embeddings. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, 17–21 September 2015; pp. 548–554. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).