Integrating Biological Domain Knowledge with Machine Learning for Identifying Colorectal-Cancer-Associated Microbial Enzymes in Metagenomic Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. CRC-Associated Metagenomic Dataset
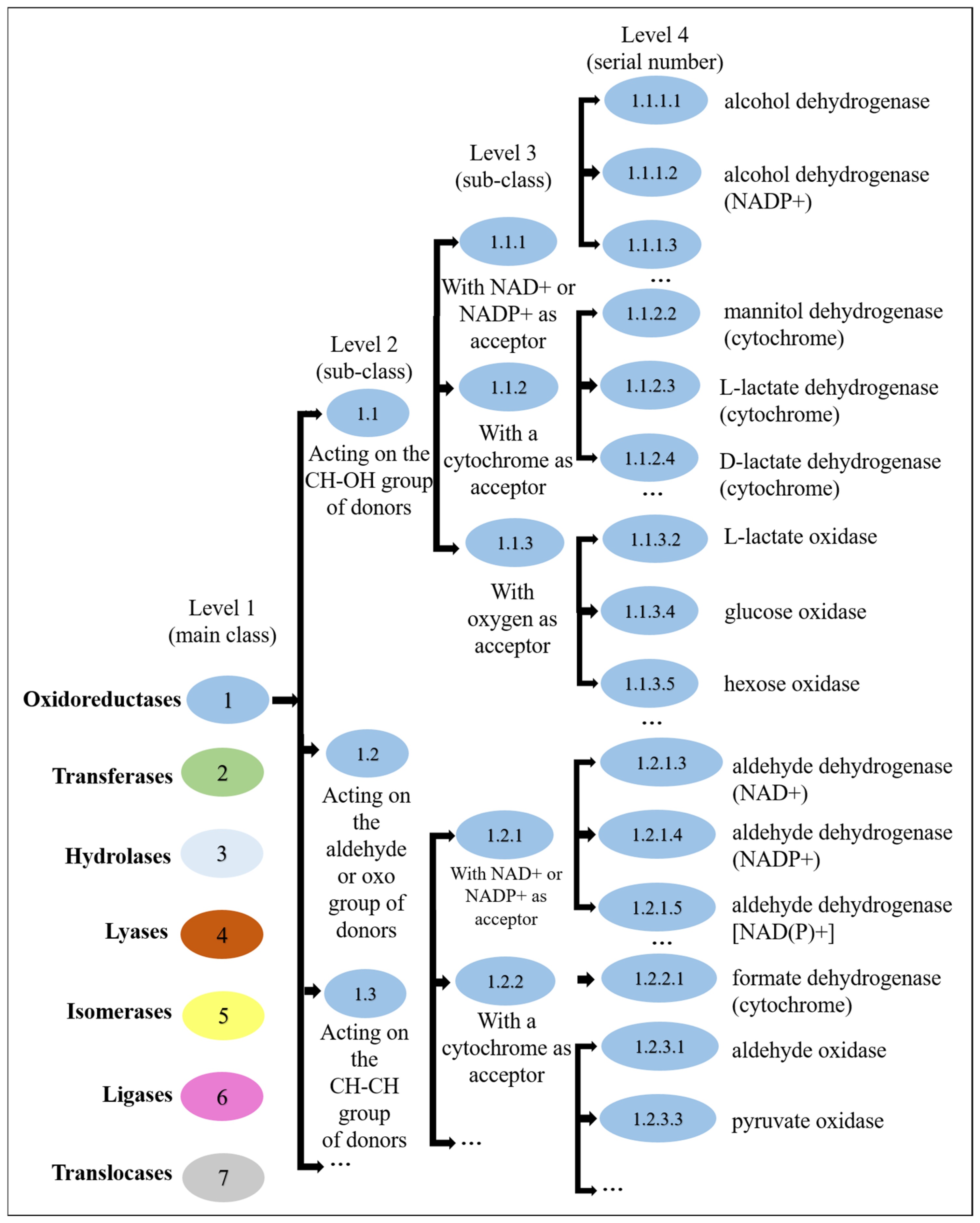
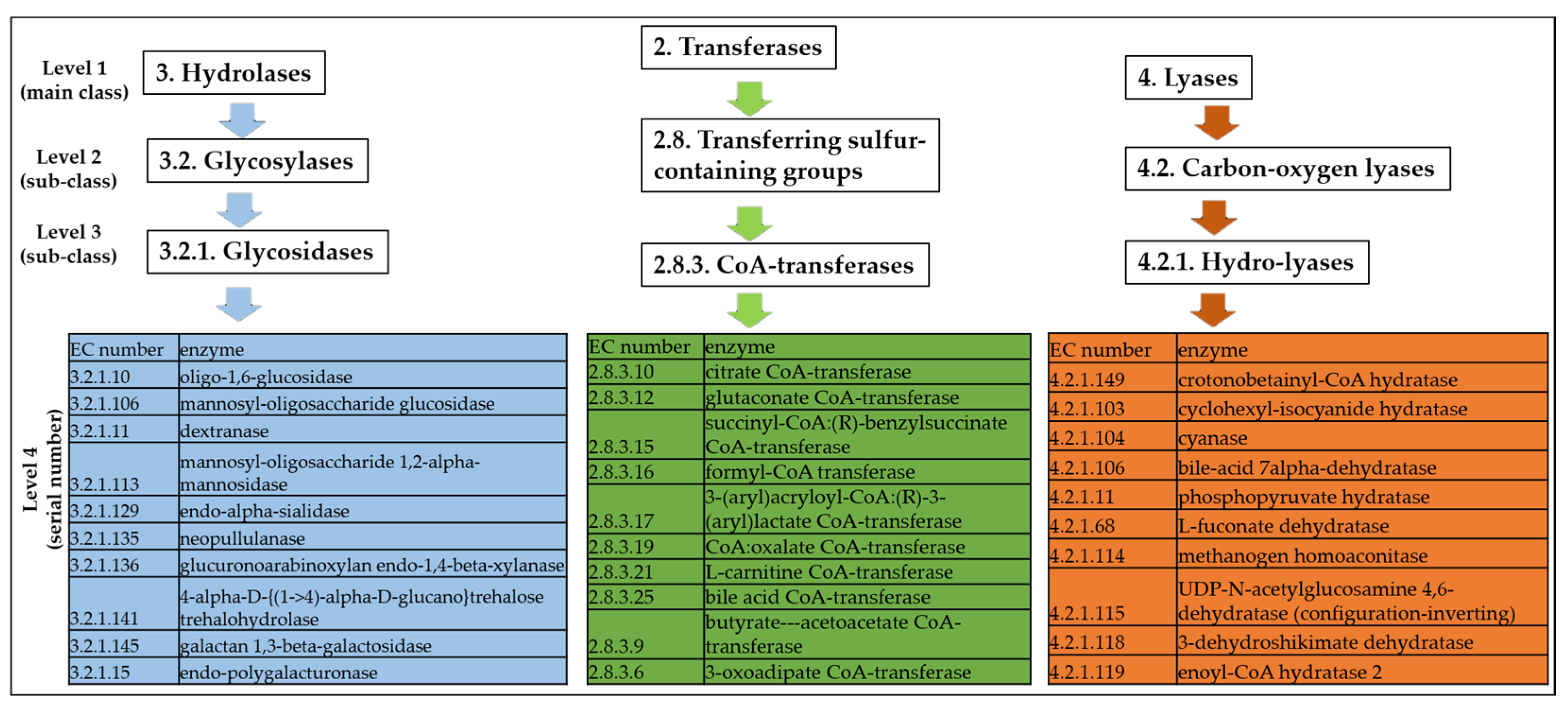
2.1.2. Enzyme Commission Dataset
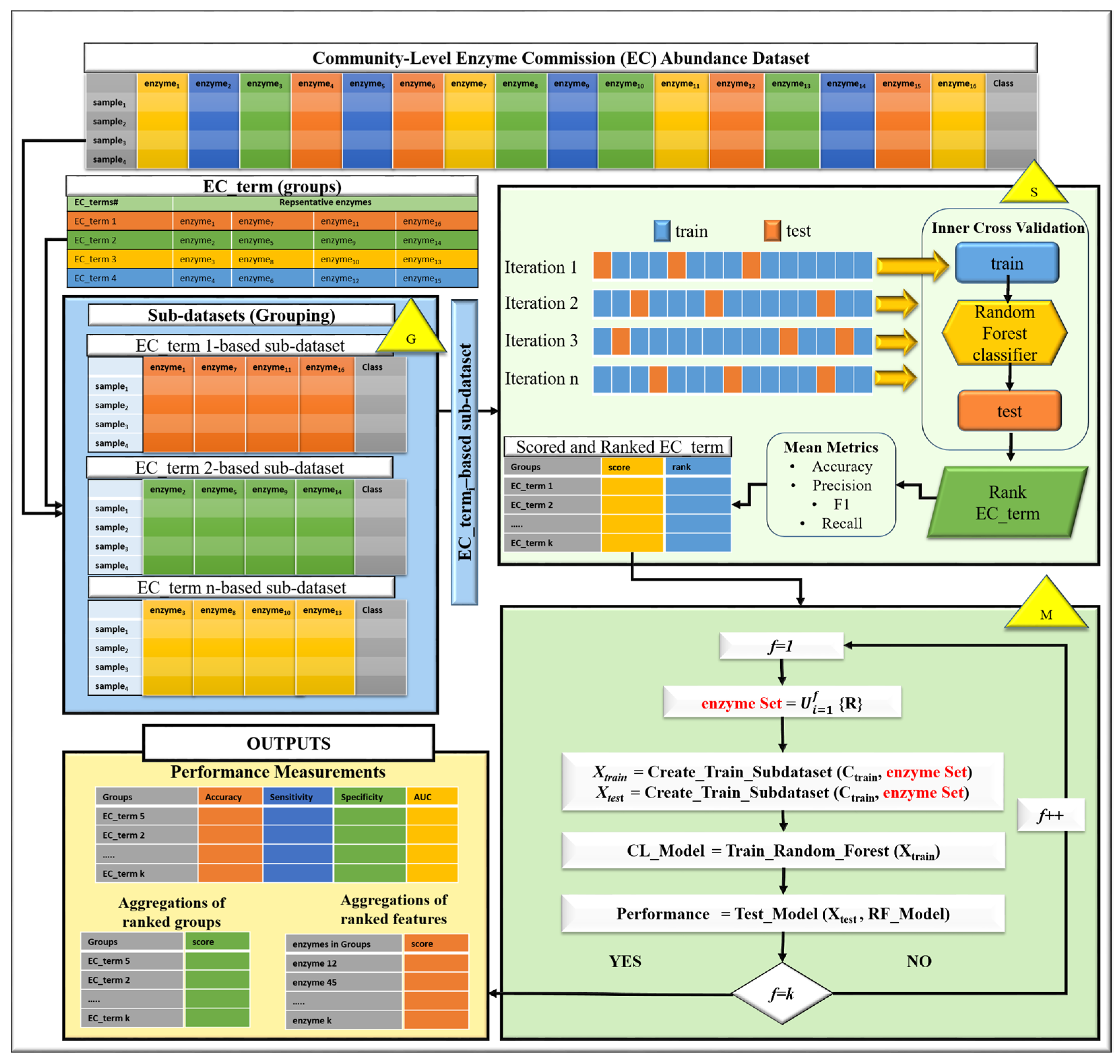
2.2. Our Proposed Method: EC-Nomenclature-Based G-S-M Approach
2.3. Implementation of the EC-Nomenclature-Based G-S-M Model
2.4. Comparative Evaluation with Traditional Feature Selection Methods and Classifiers
2.5. Performance Evaluation Metrics
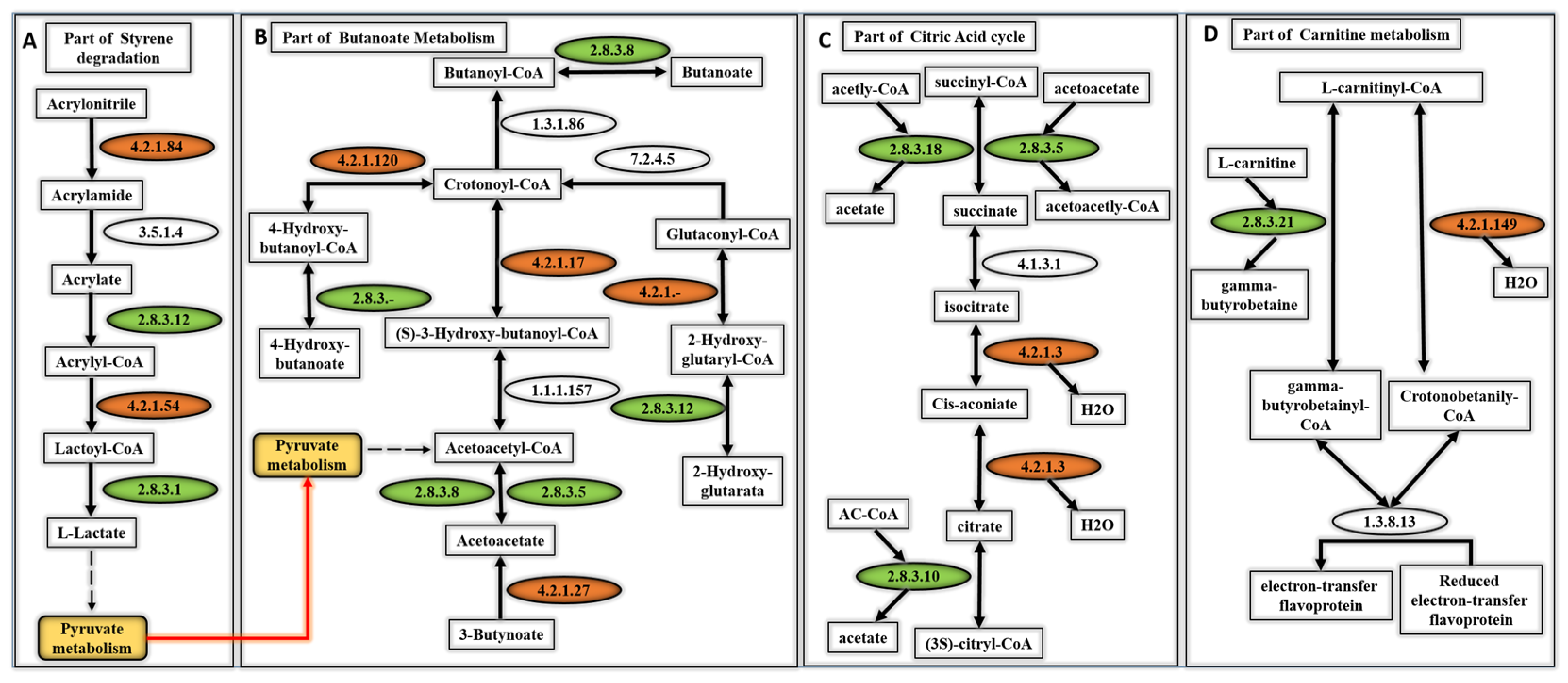
2.6. Molecular/Metabolic Pathways
3. Results
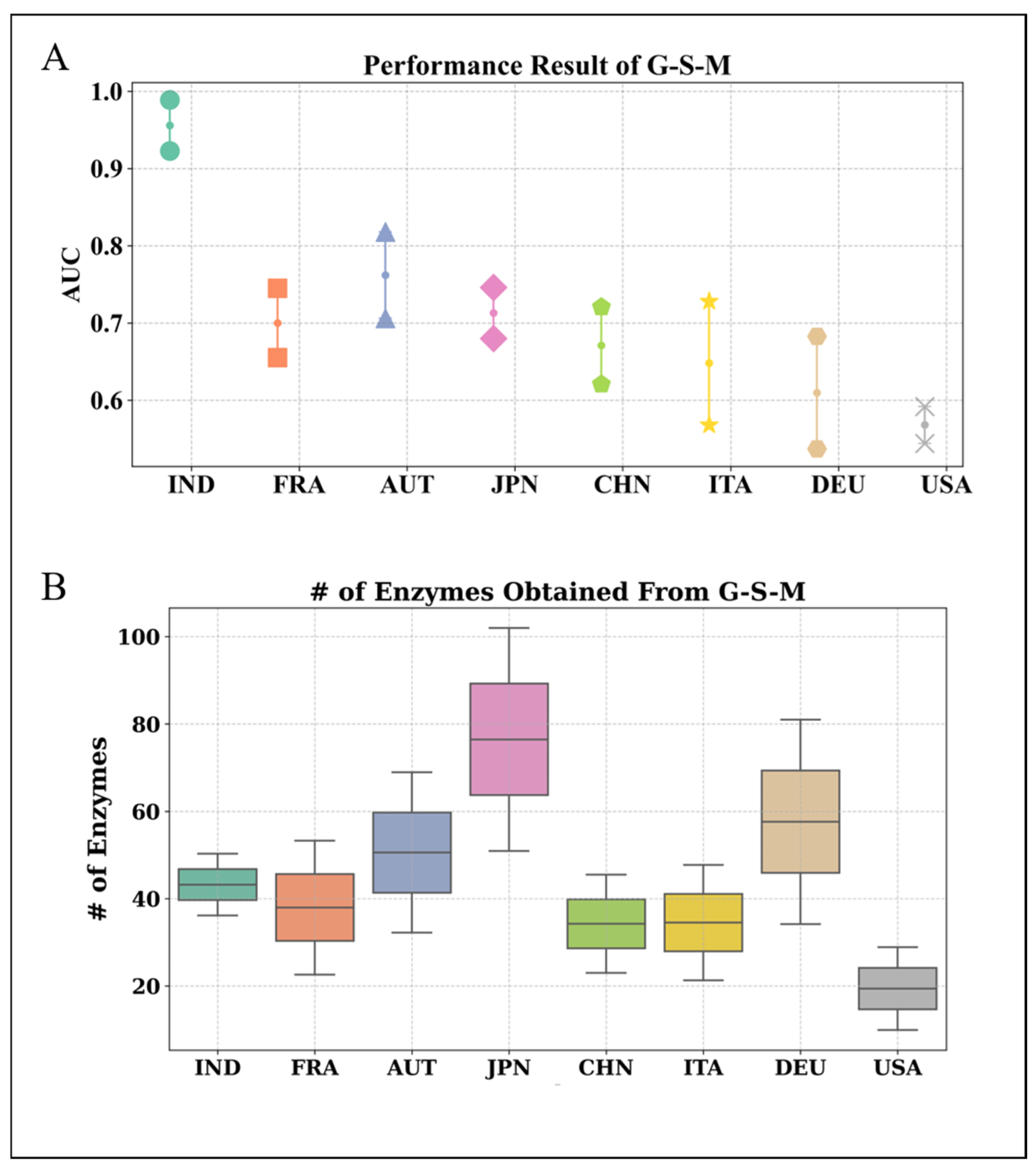
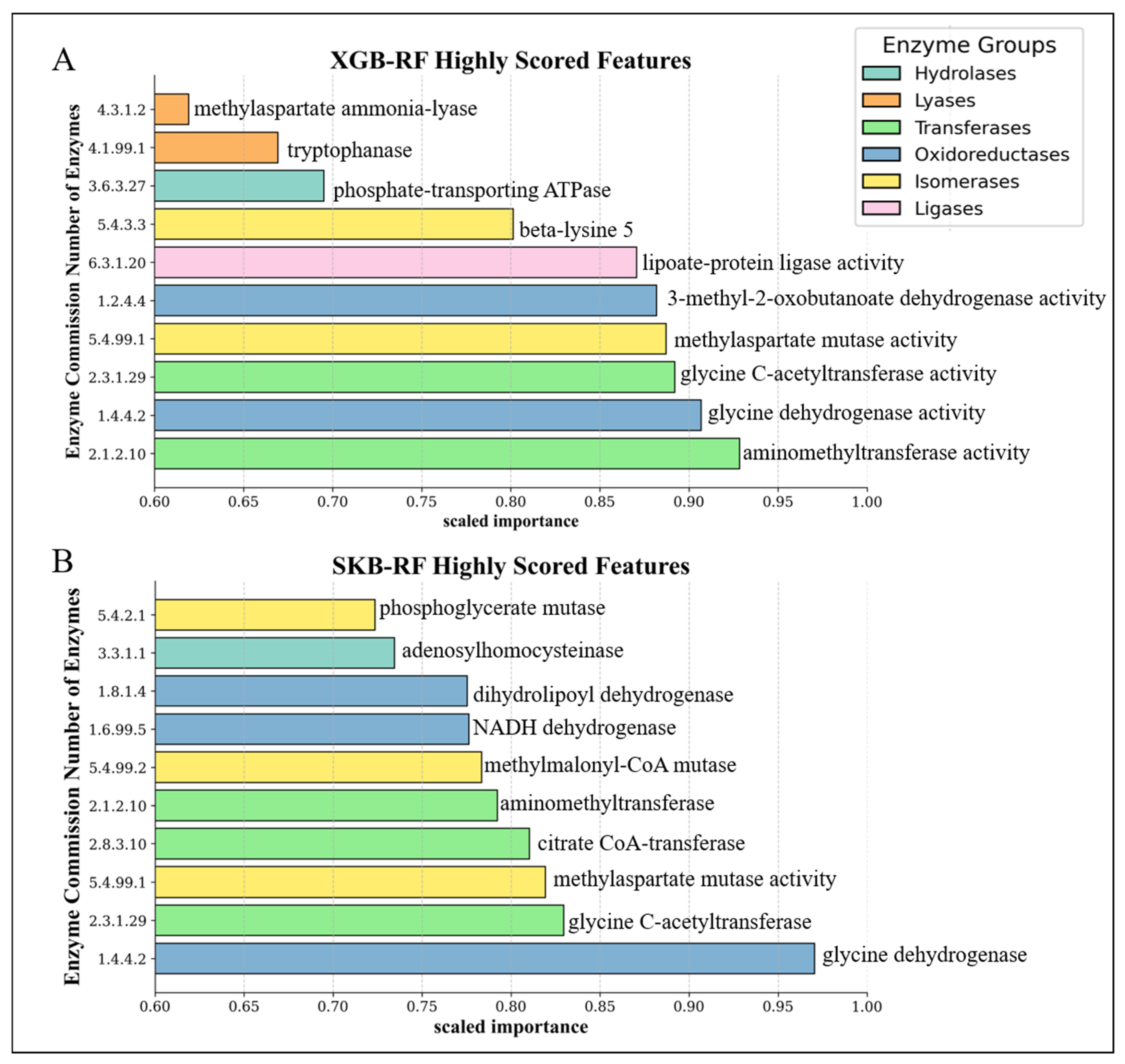
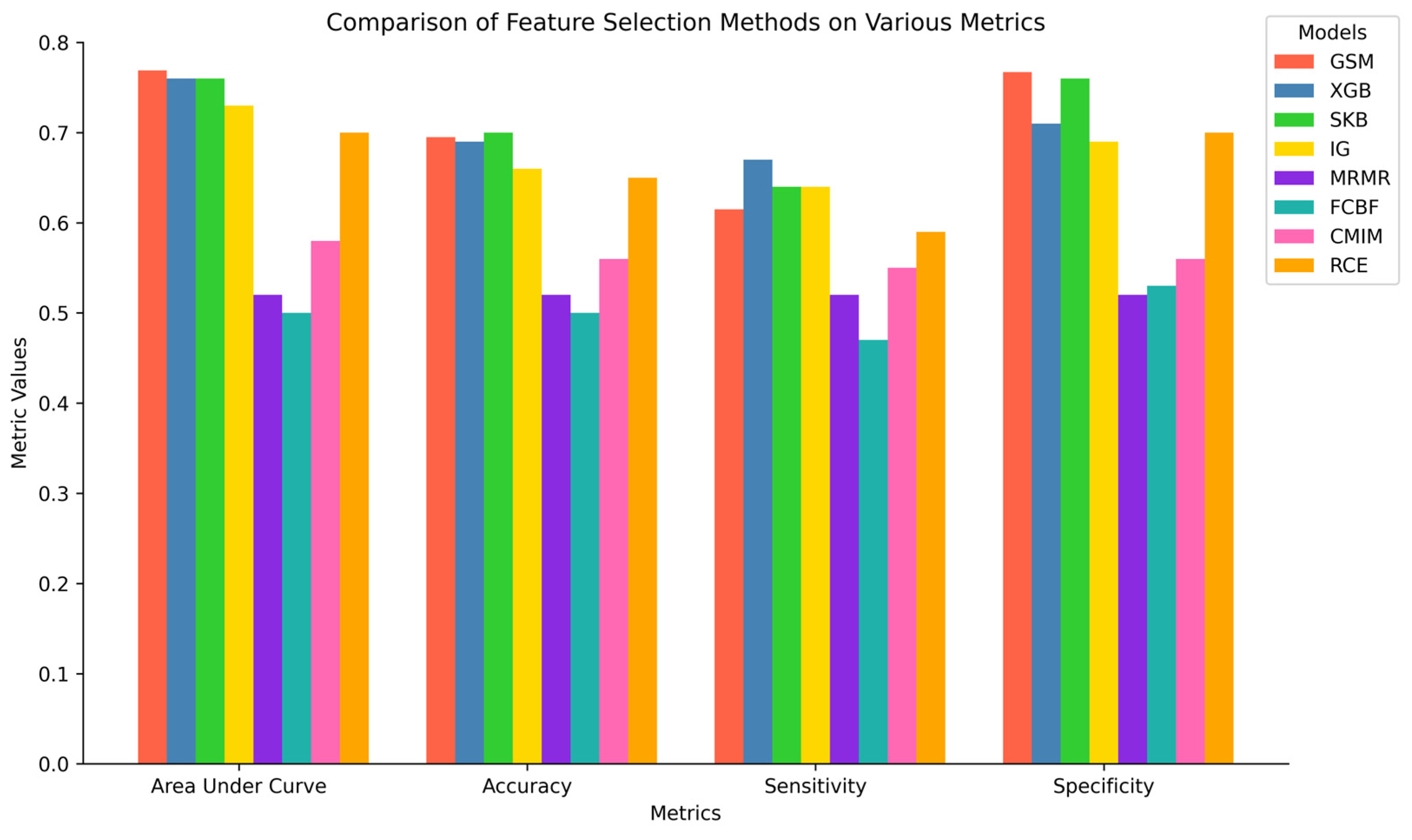
3.1. Performance Evaluation of the Proposed EC-Nomenclature-Based G-S-M Approach
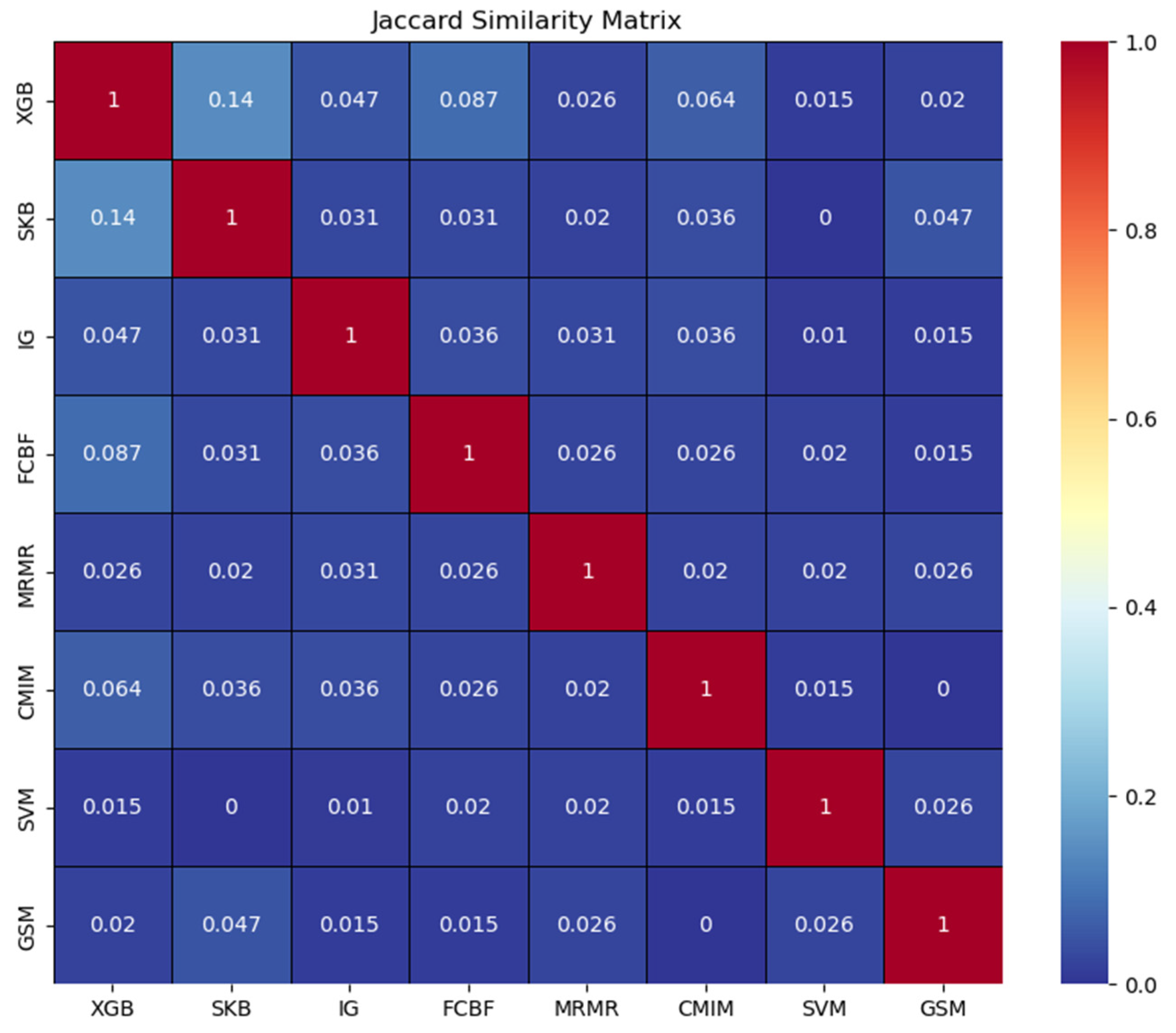
3.2. Comparative Performance Evaluation of G-S-M with Traditional Feature Selection Methods
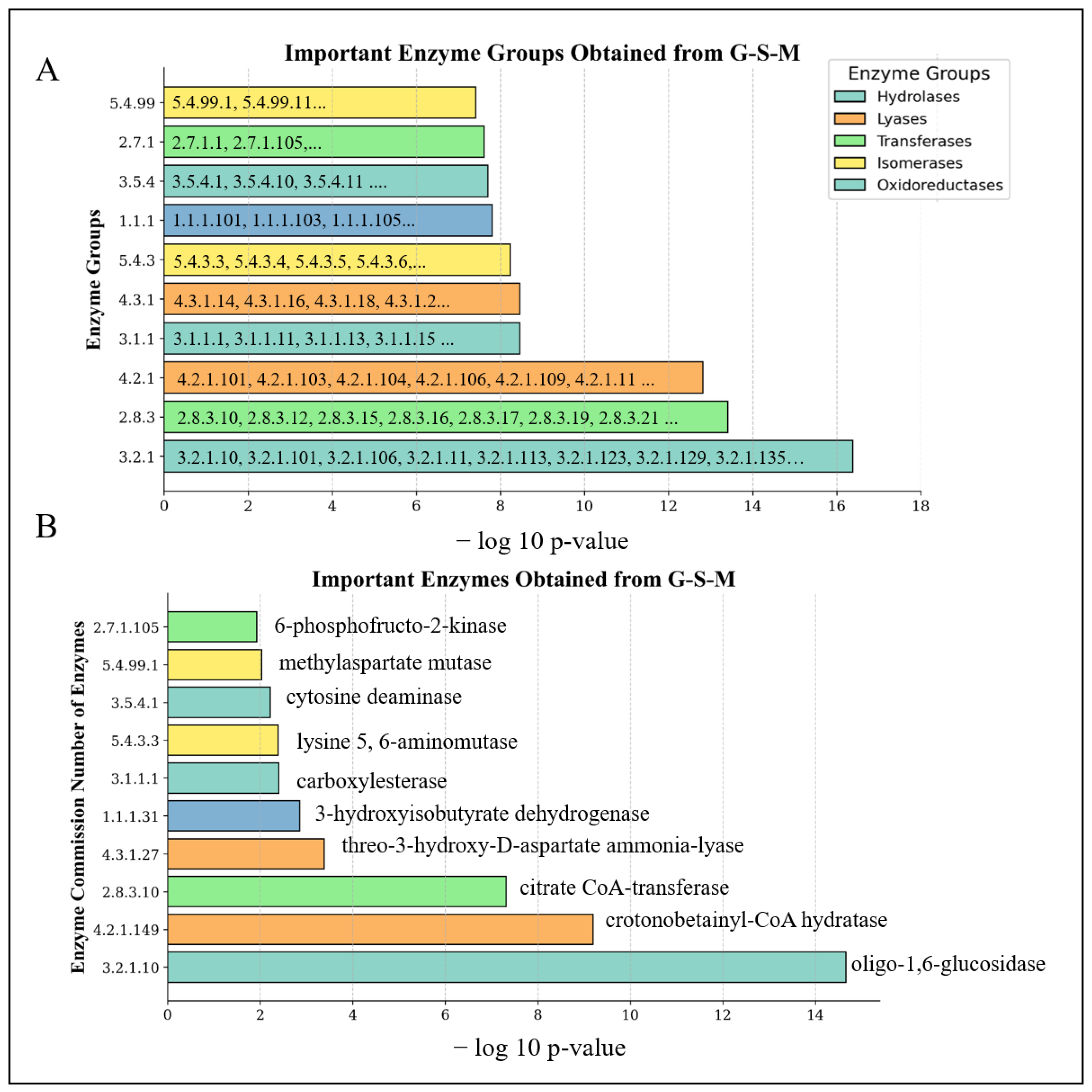
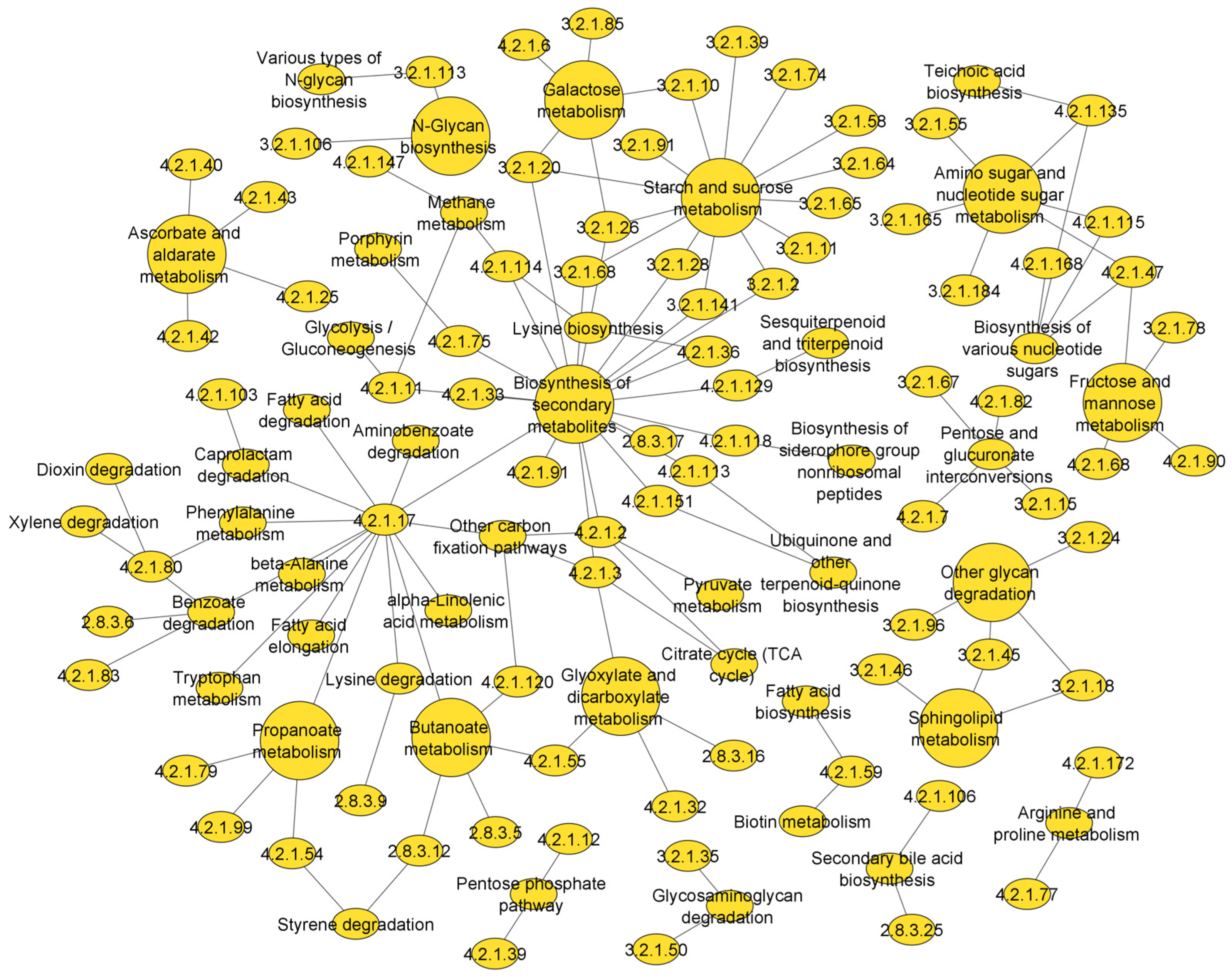
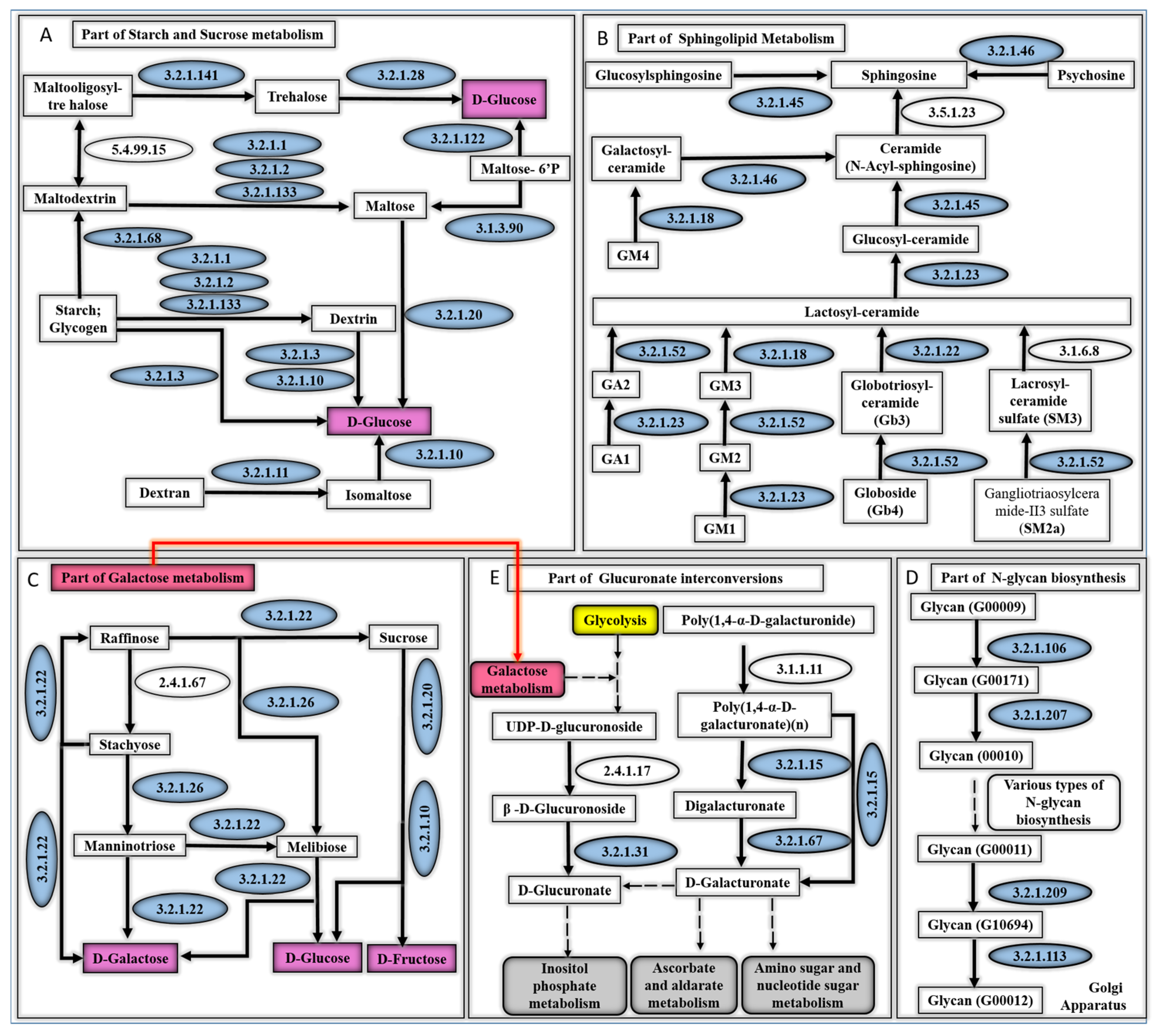
3.3. Metabolic Pathways That Are Associated with the Top Scoring Enzyme Groups
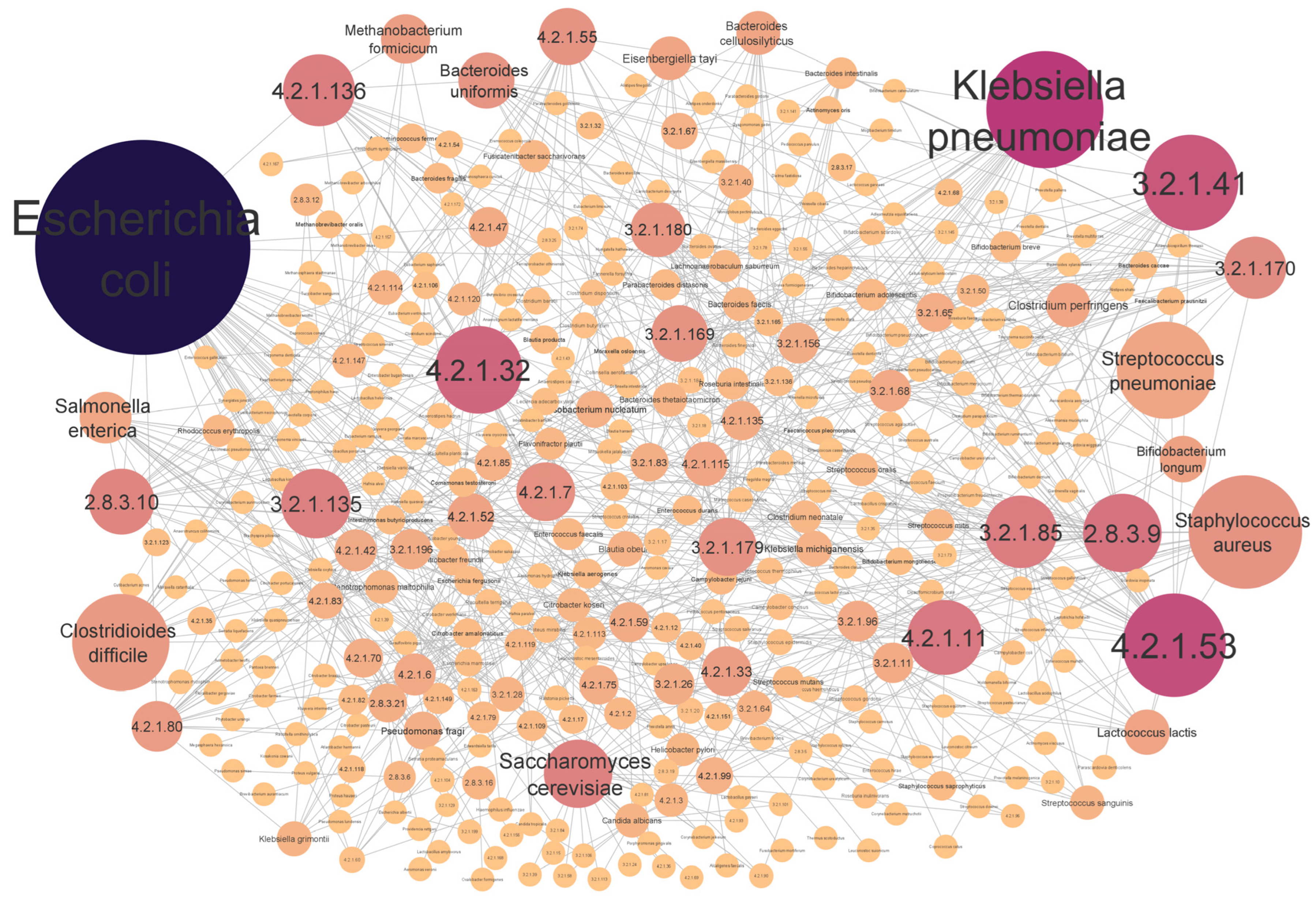
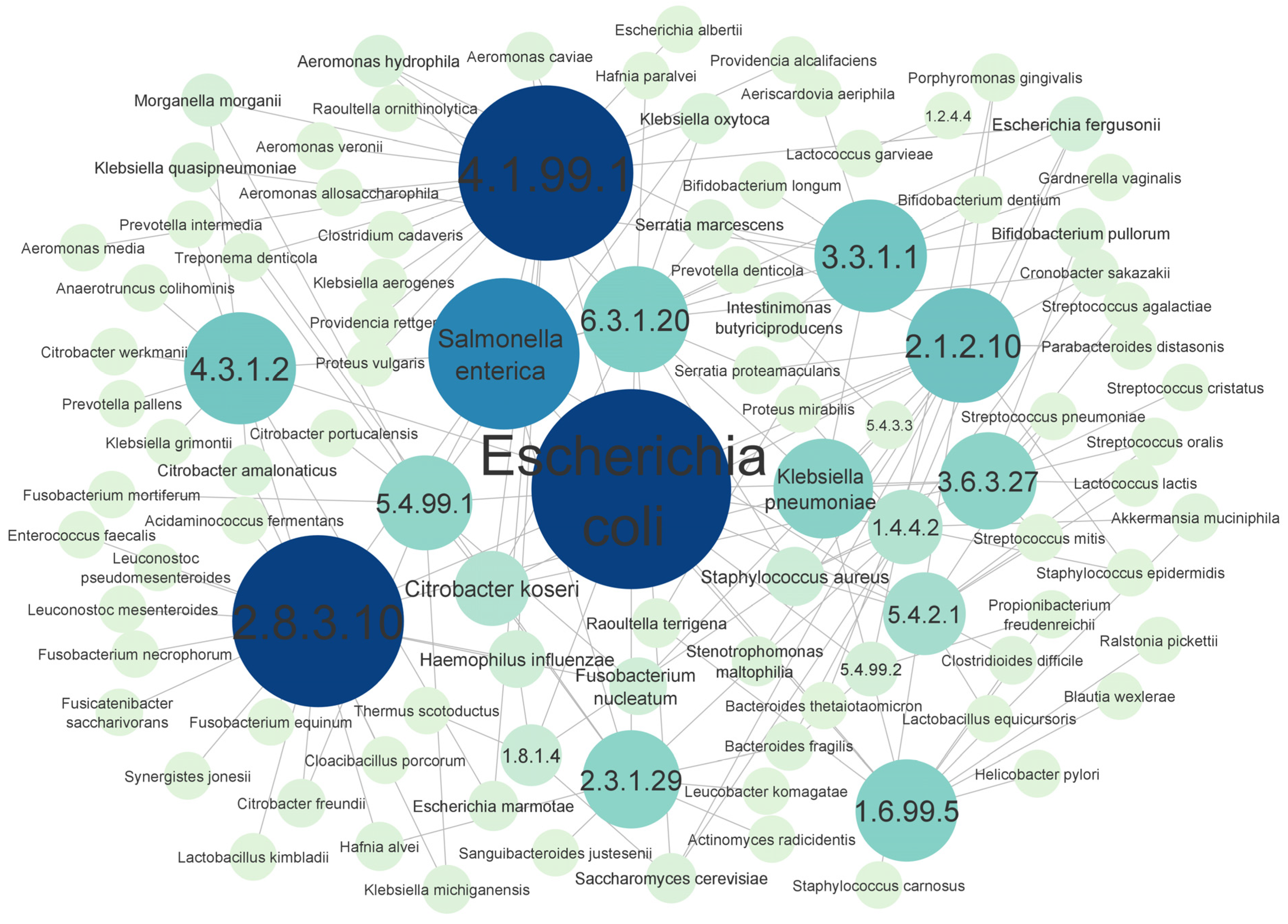
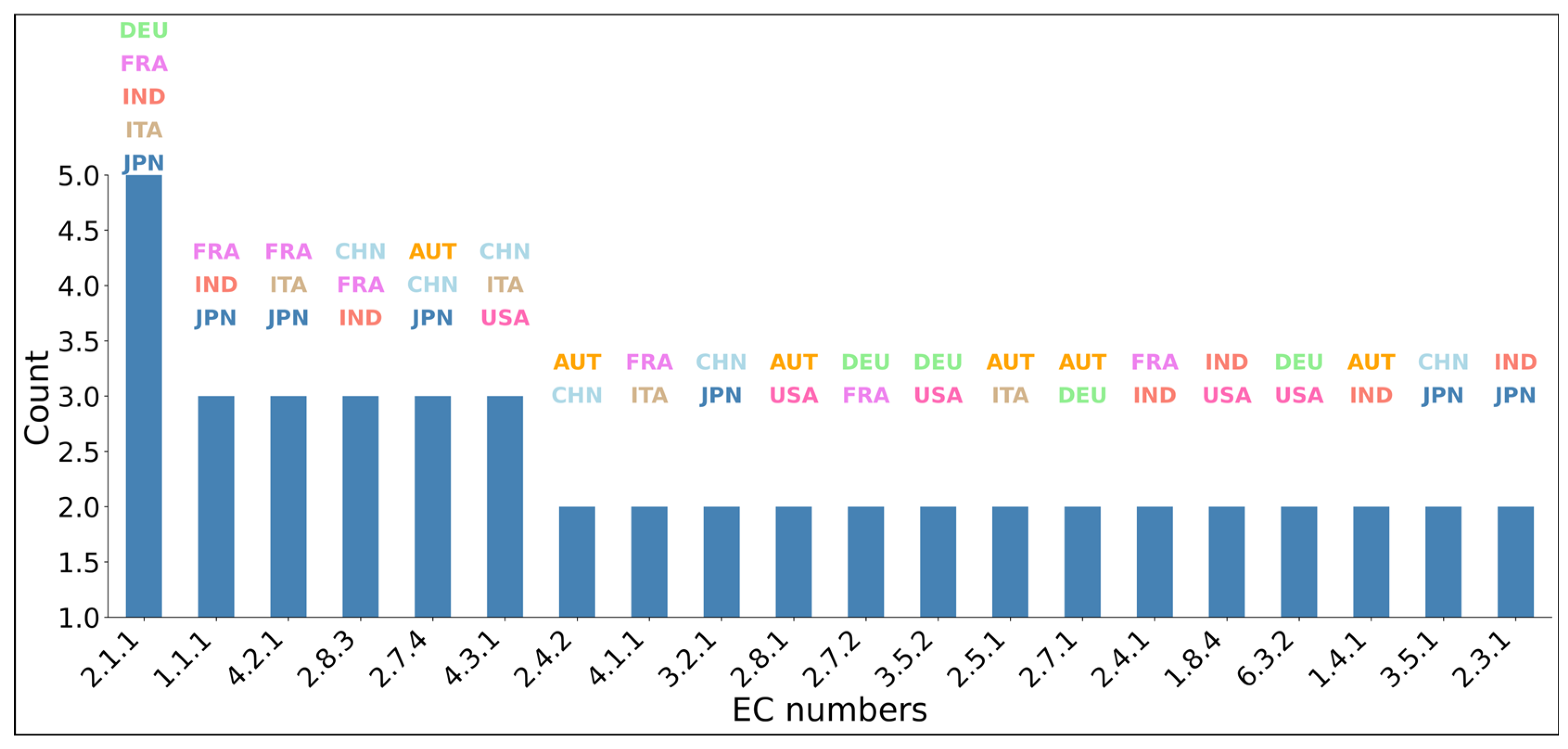
3.4. Top Scored Enzyme-Associated Species Obtained from CRC Dataset
4. Discussion
4.1. Computational Performance Evaluation of the EC-Nomenclature-Based G-S-M Model
4.2. Comparative Performance Evaluation of the EC-Nomenclature-Based G-S-M Model
4.3. Metabolic Pathways of Top Scoring Enzymes
4.4. The Microorganisms That Synthesize the Enzymes Identified by the EC-Nomenclature-Based G-S-M and Their Association with CRC Development
4.5. Study Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Ma, X.; Chakravarti, D.; Shalapour, S.; DePinho, R.A. Genetic and biological hallmarks of colorectal cancer. Genes Dev. 2021, 35, 787–820. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-De-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Ryan, B.M.; Wolff, R.K.; Valeri, N.; Khan, M.; Robinson, D.; Paone, A.; Bowman, E.D.; Lundgreen, A.; Caan, B.; Potter, J.; et al. An analysis of genetic factors related to risk of inflammatory bowel disease and colon cancer. Cancer Epidemiol. 2014, 38, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.; Yu, J. Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 2023, 20, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome In Colorectal Cancer Progression. Front. Immunol. 2022, 12, 807648. [Google Scholar] [CrossRef]
- McNally, L.; Brown, S.P. Building the microbiome in health and disease: Niche construction and social conflict in bacteria. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140298. [Google Scholar] [CrossRef]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70 (Suppl. 1), S38–S44. [Google Scholar] [CrossRef]
- Scarpellini, E.; Ianiro, G.; Attili, F.; Bassanelli, C.; De Santis, A.; Gasbarrini, A. The human gut microbiota and virome: Potential therapeutic implications. Dig. Liver Dis. 2015, 47, 1007–1012. [Google Scholar] [CrossRef]
- Stearns, J.C.; Lynch, M.D.J.; Senadheera, D.B.; Tenenbaum, H.C.; Goldberg, M.B.; Cvitkovitch, D.G.; Croitoru, K.; Moreno-Hagelsieb, G.; Neufeld, J.D. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011, 1, 170. [Google Scholar] [CrossRef]
- Matamoros, S.; Gras-Leguen, C.; Le Vacon, F.; Potel, G.; de La Cochetiere, M.-F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013, 21, 167–173. [Google Scholar] [CrossRef]
- Yadav, D.; Ghosh, T.S.; Mande, S.S. Global investigation of composition and interaction networks in gut microbiomes of individuals belonging to diverse geographies and age-groups. Gut Pathog. 2016, 8, 17. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Knight, R. Dietary effects on human gut microbiome diversity. Br. J. Nutr. 2015, 113, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Chi, L.; Zhu, Y.; Shi, X.; Tu, P.; Li, B.; Yin, J.; Gao, N.; Shen, W.; Schnabl, B. An Introduction to Next Generation Sequencing Bioinformatic Analysis in Gut Microbiome Studies. Biomolecules 2021, 11, 530. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Nam, N.N.; Do, H.D.K.; Trinh, K.T.L.; Lee, N.Y. Metagenomics: An Effective Approach for Exploring Microbial Diversity and Functions. Foods 2023, 12, 2140. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Niwa, H.; Uchida-Fujii, E.; Nukada, T. Establishment and assessment of an amplicon sequencing method targeting the 16S-ITS-23S rRNA operon for analysis of the equine gut microbiome. Sci. Rep. 2021, 11, 11884. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zeng, Z.; Xu, M.; Sun, F.; Yang, L.; Bi, X.; Lin, Y.; Gao, Y.; Hao, H.; et al. Advances in Metagenomics and Its Application in Environmental Microorganisms. Front. Microbiol. 2021, 12, 766364. [Google Scholar] [CrossRef]
- Blanco-Míguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M.; et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 2023, 41, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 2021, 10, e65088. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Kim, C. The Importance of Biomarker Testing in the Treatment of Advanced Non-Small Cell Lung Cancer: A Podcast. Oncol. Ther. 2024, 12, 223–231. [Google Scholar] [CrossRef]
- Perscheid, C. Integrative biomarker detection on high-dimensional gene expression data sets: A survey on prior knowledge approaches. Briefings Bioinform. 2021, 22, bbaa151. [Google Scholar] [CrossRef]
- Yousef, M.; Inal, Y.; Gungor, B.B.; Allmer, J. G-S-M: A Comprehensive Framework for Integrative Feature Selection in Omics Data Analysis and Beyond. bioRxiv 2024. [Google Scholar] [CrossRef]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Piñero, J.; Queralt-Rosinach, N.; Bravo, À.; Deu-Pons, J.; Bauer-Mehren, A.; Baron, M.; Sanz, F.; Furlong, L.I. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database 2015, 2015, bav028. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Hubbard, T.J.P.; Ailey, B.; Brenner, S.E.; Murzin, A.G.; Chothia, C. SCOP, Structural classification of proteins database: Applications to evaluation of the effectiveness of sequence alignment methods and statistics of protein structural data. Acta Crystallogr. Sect. D Struct. Biol. 1998, 54, 1147–1154. [Google Scholar] [CrossRef]
- Orengo, C.; Michie, A.; Jones, S.; Jones, D.; Swindells, M.; Thornton, J. CATH—A hierarchic classification of protein domain structures. Structure 1997, 5, 1093–1109. [Google Scholar] [CrossRef]
- Matsuta, Y.; Ito, M.; Tohsato, Y. ECOH: An Enzyme Commission number predictor using mutual information and a support vector machine. Bioinformatics 2013, 29, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Yousef, M.; Abdallah, L.; Allmer, J. maTE: Discovering expressed interactions between microRNAs and their targets. Bioinformatics 2019, 35, 4020–4028. [Google Scholar] [CrossRef]
- Yousef, M.; Goy, G.; Bakir-Gungor, B. miRModuleNet: Detecting miRNA-mRNA Regulatory Modules. Front. Genet. 2022, 13, 767455. [Google Scholar] [CrossRef] [PubMed]
- Yousef, M.; Ülgen, E.; Sezerman, O.U. CogNet: Classification of gene expression data based on ranked active-subnetwork-oriented KEGG pathway enrichment analysis. PeerJ Comput. Sci. 2021, 7, e336. [Google Scholar] [CrossRef]
- Yousef, M.; Ozdemir, F.; Jaaber, A.; Allmer, J.; Bakir-Gungor, B. PriPath: Identifying Dysregulated Pathways from Differential Gene Expression via Grouping, Scoring and Modeling with an Embedded Machine Learning Approach. Preprint, 2022. [Google Scholar] [CrossRef]
- Jabeer, A.; Temiz, M.; Bakir-Gungor, B.; Yousef, M. miRdisNET: Discovering microRNA biomarkers that are associated with diseases utilizing biological knowledge-based machine learning. Front. Genet. 2023, 13, 1076554. [Google Scholar] [CrossRef]
- Ersoz, N.S.; Bakir-Gungor, B.; Yousef, M. GeNetOntology: Identifying affected gene ontology terms via grouping, scoring, and modeling of gene expression data utilizing biological knowledge-based machine learning. Front. Genet. 2023, 14, 1139082. [Google Scholar] [CrossRef]
- Söylemez, Ü.G.; Yousef, M.; Bakir-Gungor, B. AMP-GSM: Prediction of Antimicrobial Peptides via a Grouping–Scoring–Modeling Approach. Appl. Sci. 2023, 13, 5106. [Google Scholar] [CrossRef]
- Yousef, M.; Kumar, A.; Bakir-Gungor, B. Application of Biological Domain Knowledge Based Feature Selection on Gene Expression Data. Entropy 2021, 23, 2. [Google Scholar] [CrossRef]
- Pudjihartono, N.; Fadason, T.; Kempa-Liehr, A.W.; O’Sullivan, J.M. A Review of Feature Selection Methods for Machine Learning-Based Disease Risk Prediction. Front. Bioinform. 2022, 2, 927312. [Google Scholar] [CrossRef]
- Kuzudisli, C.; Bakir-Gungor, B.; Bulut, N.; Qaqish, B.; Yousef, M. Review of feature selection approaches based on grouping of features. PeerJ 2023, 11, e15666. [Google Scholar] [CrossRef]
- Prasetiyowati, M.I.; Maulidevi, N.U.; Surendro, K. Determining threshold value on information gain feature selection to increase speed and prediction accuracy of random forest. J. Big Data 2021, 8, 84. [Google Scholar] [CrossRef]
- Radovic, M.; Ghalwash, M.; Filipovic, N.; Obradovic, Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinform. 2017, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Gopika, N.; A. Meena Kowshalaya, M.E. Correlation Based Feature Selection Algorithm for Machine Learning. In Proceedings of the 2018 3rd International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 15–16 October 2018; pp. 692–695. [Google Scholar] [CrossRef]
- Bakir-Gungor, B.; Hacılar, H.; Jabeer, A.; Nalbantoglu, O.U.; Aran, O.; Yousef, M. Inflammatory bowel disease biomarkers of human gut microbiota selected via different feature selection methods. PeerJ 2022, 10, e13205. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Guha, R.; Sarkar, R.; Abraham, A. A wrapper-filter feature selection technique based on ant colony optimization. Neural Comput. Appl. 2020, 32, 7839–7857. [Google Scholar] [CrossRef]
- Yousef, M.; Jung, S.; Showe, L.C.; Showe, M.K. Recursive Cluster Elimination (RCE) for classification and feature selection from gene expression data. BMC Bioinform. 2007, 8, 144. [Google Scholar] [CrossRef]
- Kuzudisli, C.; Bakir-Gungor, B.; Qaqish, B.; Yousef, M. RCE-IFE: Recursive cluster elimination with intra-cluster feature elimination. bioRxiv 2024. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Chang, Q. Feature selection methods for big data bioinformatics: A survey from the search perspective. Methods 2016, 111, 21–31. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Mathieu, A.; Leclercq, M.; Sanabria, M.; Perin, O.; Droit, A. Machine Learning and Deep Learning Applications in Metagenomic Taxonomy and Functional Annotation. Front. Microbiol. 2022, 13, 811495. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Ahern, A.; Carbone, C.; Temko, A.; Claesson, M.J.; Gasbarrini, A.; Tortora, G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 635–648. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Turukalo, T.L.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Pödör, Z.; Hekfusz, M. Comparing Feature Selection Methods on Metagenomic Data using Random Forest Classifier. Trans. Mach. Learn. Artif. Intell. 2024, 12, 175–187. [Google Scholar] [CrossRef]
- Bakir-Gungor, B.; Bulut, O.; Jabeer, A.; Nalbantoglu, O.U.; Yousef, M. Discovering Potential Taxonomic Biomarkers of Type 2 Diabetes From Human Gut Microbiota via Different Feature Selection Methods. Front. Microbiol. 2021, 12, 628426. [Google Scholar] [CrossRef]
- Bakir-Gungor, B.; Temiz, M.; Inal, Y.; Cicekyurt, E.; Yousef, M. CCPred: Global and population-specific colorectal cancer prediction and metagenomic biomarker identification at different molecular levels using machine learning techniques. Comput. Biol. Med. 2024, 182, 109098. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Coker, O.O.; Nakatsu, G.; Wu, W.K.K.; Zhao, L.; Chen, Z.; Chan, F.K.L.; Kristiansen, K.; Sung, J.J.Y.; Wong, S.H.; et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 2018, 6, 70. [Google Scholar] [CrossRef]
- Xu, X.; Ocansey, D.K.W.; Hang, S.; Wang, B.; Amoah, S.; Yi, C.; Zhang, X.; Liu, L.; Mao, F. The gut metagenomics and metabolomics signature in patients with inflammatory bowel disease. Gut Pathog. 2022, 14, 26. [Google Scholar] [CrossRef]
- Jacobs, J.P.; Lagishetty, V.; Hauer, M.C.; Labus, J.S.; Dong, T.S.; Toma, R.; Vuyisich, M.; Naliboff, B.D.; Lackner, J.M.; Gupta, A.; et al. Multi-omics profiles of the intestinal microbiome in irritable bowel syndrome and its bowel habit subtypes. Microbiome 2023, 11, 5. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Dougherty, M.W.; Jobin, C. Intestinal bacteria and colorectal cancer: Etiology and treatment. Gut Microbes 2023, 15, 2185028. [Google Scholar] [CrossRef]
- Hera, M.R.; Liu, S.; Wei, W.; Rodriguez, J.S.; Ma, C.; Koslicki, D. Metagenomic functional profiling: To sketch or not to sketch? Bioinformatics 2024, 40, ii165–ii173. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Chai, E.Z.P.; Siveen, K.S.; Shanmugam, M.K.; Arfuso, F.; Sethi, G. Analysis of the intricate relationship between chronic inflammation and cancer. Biochem. J. 2015, 468, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hung, R.J.; Ulrich, C.M.; Goode, E.L.; Brhane, Y.; Muir, K.; Chan, A.T.; Le Marchand, L.; Schildkraut, J.; Witte, J.S.; Eeles, R.; et al. Cross Cancer Genomic Investigation of Inflammation Pathway for Five Common Cancers: Lung, Ovary, Prostate, Breast, and Colorectal Cancer. JNCI J. Natl. Cancer Inst. 2015, 107, djv246. [Google Scholar] [CrossRef]
- Pandey, H.; Tang, D.W.T.; Wong, S.H.; Lal, D. Gut Microbiota in Colorectal Cancer: Biological Role and Therapeutic Opportunities. Cancers 2023, 15, 866. [Google Scholar] [CrossRef]
- Fedirko, V.; Tramacere, I.; Bagnardi, V.; Rota, M.; Scotti, L.; Islami, F.; Negri, E.; Straif, K.; Romieu, I.; La Vecchia, C.; et al. Alcohol drinking and colorectal cancer risk: An overall and dose–response meta-analysis of published studies. Ann. Oncol. 2011, 22, 1958–1972. [Google Scholar] [CrossRef] [PubMed]
- Little, C.H.; Combet, E.; McMillan, D.C.; Horgan, P.G.; Roxburgh, C.S.D. The role of dietary polyphenols in the moderation of the inflammatory response in early stage colorectal cancer. Crit. Rev. Food Sci. Nutr. 2017, 57, 2310–2320. [Google Scholar] [CrossRef]
- Shivappa, N.; Zucchetto, A.; Montella, M.; Serraino, D.; Steck, S.E.; La Vecchia, C.; Hébert, J.R. Inflammatory potential of diet and risk of colorectal cancer: A case–control study from Italy. Br. J. Nutr. 2015, 114, 152–158. [Google Scholar] [CrossRef]
- Tojjari, A.; Choucair, K.; Sadeghipour, A.; Saeed, A.; Saeed, A. Anti-Inflammatory and Immune Properties of Polyunsaturated Fatty Acids (PUFAs) and Their Impact on Colorectal Cancer (CRC) Prevention and Treatment. Cancers 2023, 15, 4294. [Google Scholar] [CrossRef]
- Thanikachalam, K.; Khan, G. Colorectal Cancer and Nutrition. Nutrients 2019, 11, 164. [Google Scholar] [CrossRef]
- Rohrhofer, J.; Zwirzitz, B.; Selberherr, E.; Untersmayr, E. The Impact of Dietary Sphingolipids on Intestinal Microbiota and Gastrointestinal Immune Homeostasis. Front. Immunol. 2021, 12, 635704. [Google Scholar] [CrossRef] [PubMed]
- Ersöz, N.Ş.; Adan, A. Cytotoxic Effects of Resveratrol and Its Combinations with Ceramide Metabolism Inhibitors on FLT3 Positive Acute Myeloid Leukemia. Erzincan Üniversitesi Fen Bilim. Enstitüsü Derg. 2020, 13, 1205–1216. [Google Scholar] [CrossRef]
- Ersöz, N.Ş.; Adan, A. Resveratrol triggers anti-proliferative and apoptotic effects in FLT3-ITD-positive acute myeloid leukemia cells via inhibiting ceramide catabolism enzymes. Med Oncol. 2022, 39, 35. [Google Scholar] [CrossRef]
- Ersöz, N.Ş.; Adan, A. Resveratrol Targets Sphingolipid Metabolism to Induce Growth Inhibition in FLT3 ITD Acute Myeloid Leukemia. Proceedings 2019, 40, 4. [Google Scholar] [CrossRef]
- Ersöz, N.Ş.; Adan, A. Differential in vitro anti-leukemic activity of resveratrol combined with serine palmitoyltransferase inhibitor myriocin in FMS-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) carrying AML cells. Cytotechnology 2022, 74, 271–281. [Google Scholar] [CrossRef]
- Johnson, E.L.; Heaver, S.L.; Waters, J.L.; Kim, B.I.; Bretin, A.; Goodman, A.L.; Gewirtz, A.T.; Worgall, T.S.; Ley, R.E. Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun. 2020, 11, 2471. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Bryan, P.-F.; Karla, C.; Edgar Alejandro, M.-T.; Sara Elva, E.-P.; Gemma, F.; Luz, C. Sphingolipids as Mediators in the Crosstalk between Microbiota and Intestinal Cells: Implications for Inflammatory Bowel Disease. Mediat. Inflamm. 2016, 2016, 9890141. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhi, F. Lower Level of Bacteroides in the Gut Microbiota Is Associated with Inflammatory Bowel Disease: A Meta-Analysis. BioMed Res. Int. 2016, 2016, 5828959. [Google Scholar] [CrossRef]
- Brown, E.M.; Ke, X.; Hitchcock, D.; Jeanfavre, S.; Avila-Pacheco, J.; Nakata, T.; Arthur, T.D.; Fornelos, N.; Heim, C.; Franzosa, E.A.; et al. Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host Microbe 2019, 25, 668–680.e7. [Google Scholar] [CrossRef]
- Lee-Sarwar, K.; Kelly, R.S.; Lasky-Su, J.; Moody, D.B.; Mola, A.R.; Cheng, T.-Y.; Comstock, L.E.; Zeiger, R.S.; O’Connor, G.T.; Sandel, M.T.; et al. Intestinal microbial-derived sphingolipids are inversely associated with childhood food allergy. J. Allergy Clin. Immunol. 2018, 142, 335–338.e9. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, M.; Kostic, A.D.; Xavier, R.J. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe 2015, 17, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Sano, R.; Trindade, V.M.; Tessitore, A.; D’Azzo, A.; Vieira, M.B.; Giugliani, R.; Coelho, J.C. GM1-ganglioside degradation and biosynthesis in human and murine GM1-gangliosidosis. Clin. Chim. Acta 2005, 354, 131–139. [Google Scholar] [CrossRef]
- Kytzia, H.; Hinrichs, U.; Maire, I.; Suzuki, K.; Sandhoff, K. Variant of GM2-gangliosidosis with hexosaminidase A having a severely changed substrate specificity. EMBO J. 1983, 2, 1201–1205. [Google Scholar] [CrossRef] [PubMed]
- Kolter, T.; Sandhoff, K. Sphingolipid metabolism diseases. Biochim. Biophys. Acta (BBA)-Biomembr. 2006, 1758, 2057–2079. [Google Scholar] [CrossRef]
- Jmoudiak, M.; Futerman, A.H. Gaucher disease: Pathological mechanisms and modern management. Br. J. Haematol. 2005, 129, 178–188. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Jiang, Q.; Yin, Y. Butyrate in Energy Metabolism: There Is Still More to Learn. Trends Endocrinol. Metab. 2021, 32, 159–169. [Google Scholar] [CrossRef]
- Geuking, M.B.; Köller, Y.; Rupp, S.; McCoy, K.D. The interplay between the gut microbiota and the immune system. Gut Microbes 2014, 5, 411–418. [Google Scholar] [CrossRef]
- Chung, H.; Kasper, D.L. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr. Opin. Immunol. 2010, 22, 455–460. [Google Scholar] [CrossRef]
- Krishnan, S.; Alden, N.; Lee, K. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr. Opin. Biotechnol. 2015, 36, 137–145. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef] [PubMed]
- Serino, M.; Blasco-Baque, V.; Nicolas, S.; Burcelin, R. Far from the eyes, close to the heart: Dysbiosis of gut microbiota and cardiovascular consequences. Curr. Cardiol. Rep. 2014, 16, 540. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-G.; Udayanga, K.G.S.; Totsuka, N.; Weinberg, J.B.; Núñez, G.; Shibuya, A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe 2014, 15, 95–102. [Google Scholar] [CrossRef]
- Yang, W.; Cong, Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell. Mol. Immunol. 2021, 18, 866–877. [Google Scholar] [CrossRef]
- Wang, X.; Fang, Y.; Liang, W.; Cai, Y.; Wong, C.C.; Wang, J.; Wang, N.; Lau, H.C.-H.; Jiao, Y.; Zhou, X.; et al. Gut–liver translocation of pathogen Klebsiella pneumoniae promotes hepatocellular carcinoma in mice. Nat. Microbiol. 2025, 10, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Fantini, M.C.; Guadagni, I. From inflammation to colitis-associated colorectal cancer in inflammatory bowel disease: Pathogenesis and impact of current therapies. Dig. Liver Dis. 2021, 53, 558–565. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Kitamoto, S.; Kuffa, P.; Kamada, N. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest. Res. 2016, 14, 127–138. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Peloquin, J.M.; Nguyen, D.D. The microbiota and inflammatory bowel disease: Insights from animal models. Anaerobe 2013, 24, 102–106. [Google Scholar] [CrossRef]
- Tomasello, G.; Tralongo, P.; Damiani, P.; Sinagra, E.; Di Trapani, B.; Zeenny, M.N.; Hussein, I.H.; Jurjus, A.; Leone, A. Dismicrobism in inflammatory bowel disease and colorectal cancer: Changes in response of colocytes. World J. Gastroenterol. 2014, 20, 18121–18130. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Dhar, R.; Pethusamy, K.; Seethy, A.; Srivastava, T.; Sah, R.; Sharma, J.; Karmakar, S. Exploring the Role of Gut Microbiome in Colon Cancer. Appl. Biochem. Biotechnol. 2021, 193, 1780–1799. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.; Wu, R.; Tokumaru, Y.; Terracina, K.P.; Takabe, K. The Role of the Microbiome on the Pathogenesis and Treatment of Colorectal Cancer. Cancers 2022, 14, 5685. [Google Scholar] [CrossRef] [PubMed]
- Rezaee, M.A.; Nouri, R.; Hasani, A.; Shirazi, K.M.; Alivand, M.R.; Sepehri, B.; Sotoodeh, S.; Hemmati, F. Escherichia coli and Colorectal Cancer: Unfolding the Enigmatic Relationship. Curr. Pharm. Biotechnol. 2022, 23, 1257–1268. [Google Scholar] [CrossRef]
- Bonnet, M.; Buc, E.; Sauvanet, P.; Darcha, C.; Dubois, D.; Pereira, B.; Déchelotte, P.; Bonnet, R.; Pezet, D.; Darfeuille-Michaud, A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin. Cancer Res. 2014, 20, 859–867. [Google Scholar] [CrossRef]
- Wassenaar, T.M. E. coli and colorectal cancer: A complex relationship that deserves a critical mindset. Crit. Rev. Microbiol. 2018, 44, 619–632. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Schaapveld, M.; Kramers, J.; Mooij, S.; Neefjes-Borst, E.A.; van Pelt, W.; Neefjes, J. Increased colon cancer risk after severe Salmonella infection. PLoS ONE 2018, 13, e0189721. [Google Scholar] [CrossRef]
- Martin, O.C.; Bergonzini, A.; D’Amico, F.; Chen, P.; Shay, J.W.; Dupuy, J.; Svensson, M.; Masucci, M.G.; Frisan, T. Infection with genotoxin-producing Salmonella enterica synergises with loss of the tumour suppressor APC in promoting genomic instability via the PI3K pathway in colonic epithelial cells. Cell. Microbiol. 2019, 21, e13099. [Google Scholar] [CrossRef]
- Patel, R.K.; Cardeiro, M.; Frankel, L.; Kim, E.; Takabe, K.; Rashid, O.M. Incidence of Colorectal Cancer After Intestinal Infection Due to Clostridioides difficile. World J. Oncol. 2024, 15, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Coleman, O.I.; Nunes, T. Role of the Microbiota in Colorectal Cancer: Updates on Microbial Associations and Therapeutic Implications. BioResearch Open Access 2016, 5, 279–288. [Google Scholar] [CrossRef]
- Narayanan, V.; Peppelenbosch, M.P.; Konstantinov, S.R. Human Fecal Microbiome–Based Biomarkers for Colorectal Cancer. Cancer Prev. Res. 2014, 7, 1108–1111. [Google Scholar] [CrossRef]
- Karampatakis, T.; Tsergouli, K.; Behzadi, P. Carbapenem-Resistant Klebsiella pneumoniae: Virulence Factors, Molecular Epidemiology and Latest Updates in Treatment Options. Antibiotics 2023, 12, 234. [Google Scholar] [CrossRef] [PubMed]
- Dubois, R.N. Role of inflammation and inflammatory mediators in colorectal cancer. Trans. Am. Clin. Climatol. Assoc. 2014, 125, 358–372, discussion 372–373. [Google Scholar] [PubMed]
- Zhang, Q.; Su, X.; Zhang, C.; Chen, W.; Wang, Y.; Yang, X.; Liu, D.; Zhang, Y.; Yang, R. Klebsiella pneumoniae Induces Inflammatory Bowel Disease Through Caspase-11–Mediated IL18 in the Gut Epithelial Cells. Cell. Mol. Gastroenterol. Hepatol. 2022, 15, 613–632. [Google Scholar] [CrossRef] [PubMed]
- Strakova, N.; Korena, K.; Karpiskova, R. Klebsiella pneumoniae producing bacterial toxin colibactin as a risk of colorectal cancer development—A systematic review. Toxicon Off. J. Int. Soc. Toxinol. 2021, 197, 126–135. [Google Scholar] [CrossRef]
- Chiang, M.-K.; Hsiao, P.-Y.; Liu, Y.-Y.; Tang, H.-L.; Chiou, C.-S.; Lu, M.-C.; Lai, Y.-C. Two ST11 Klebsiella pneumoniae strains exacerbate colorectal tumorigenesis in a colitis-associated mouse model. Gut Microbes 2021, 13, 1980348. [Google Scholar] [CrossRef]



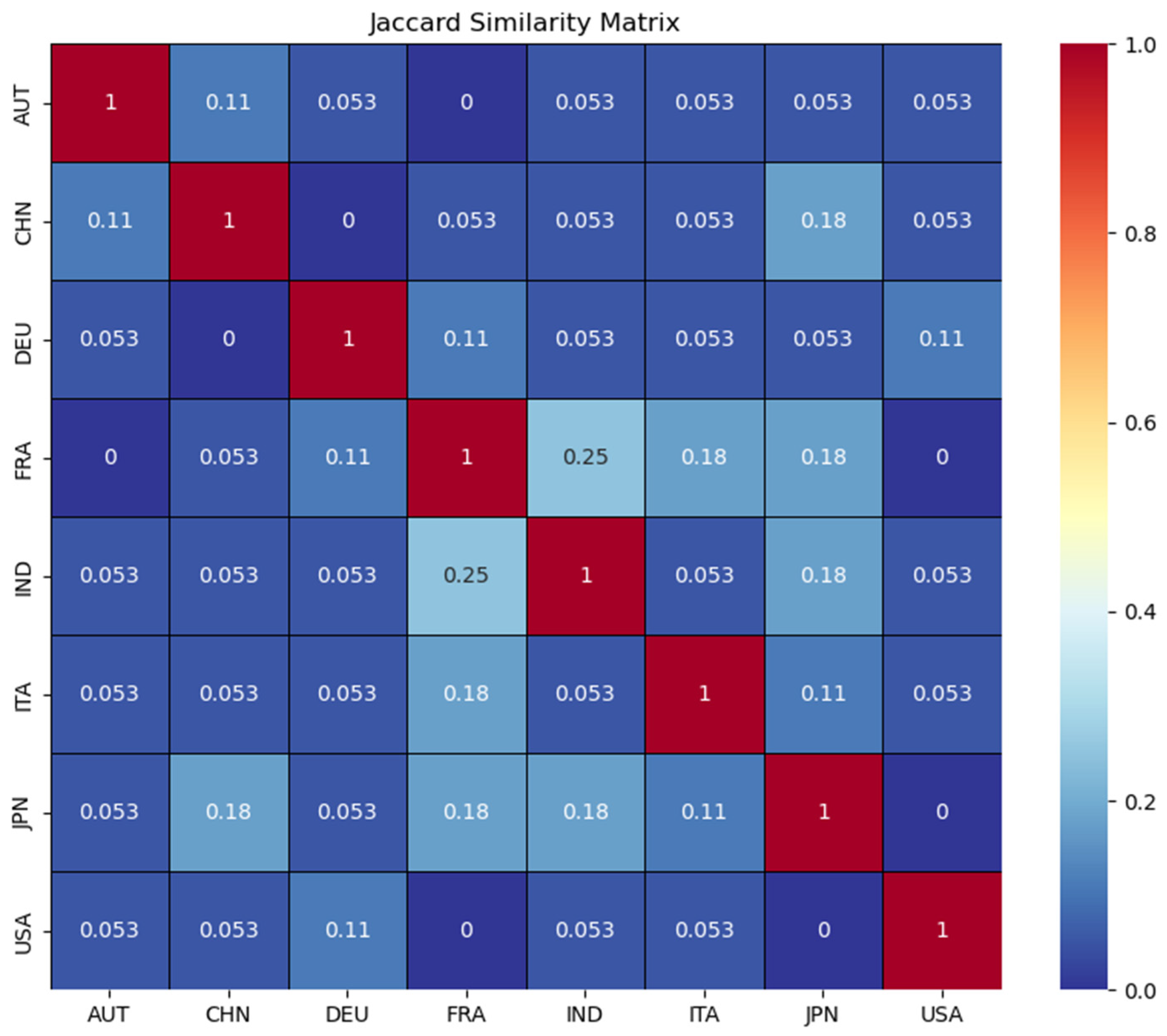

| Country | # of Controls | # of CRC Patient | Total |
|---|---|---|---|
| Austria (AUT) | 61 | 46 | 107 |
| China (CHN) | 53 | 75 | 128 |
| Germany (DEU) | 65 | 60 | 125 |
| France (FRA) | 61 | 53 | 114 |
| Indian (IND) | 30 | 30 | 60 |
| Italy (ITA) | 49 | 57 | 106 |
| Japan (JP)/(JPN) | 291 | 227 | 518 |
| United State of America (USA) | 52 | 52 | 104 |
| Total | 662 | 600 | 1262 |
| # of Groups | # of Enzymes | AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|
| 1 | 59.5 ± 26.517 | 0.728 ± 0.031 | 0.673 ± 0.028 | 0.749 ± 0.08 | 0.588 ± 0.058 |
| 2 | 90.7 ± 38.257 | 0.769 ± 0.041 | 0.695 ± 0.037 | 0.767 ± 0.050 | 0.615 ± 0.049 |
| 3 | 147.9 ± 45.261 | 0.763 ± 0.031 | 0.704 ± 0.032 | 0.769 ± 0.054 | 0.632 ± 0.033 |
| 4 | 200.9 ± 52.821 | 0.765 ± 0.032 | 0.704 ± 0.035 | 0.773 ± 0.039 | 0.627 ± 0.052 |
| 5 | 244.9 ± 64.824 | 0.770 ± 0.029 | 0.706 ± 0.029 | 0.773 ± 0.034 | 0.630 ± 0.057 |
| 6 | 281.9 ± 68.709 | 0.763 ± 0.033 | 0.694 ± 0.026 | 0.766 ± 0.024 | 0.613 ± 0.046 |
| 7 | 330.4 ± 79.269 | 0.761 ± 0.029 | 0.683 ± 0.036 | 0.760 ± 0.034 | 0.598 ± 0.060 |
| 8 | 372.1 ± 82.131 | 0.766 ± 0.034 | 0.692 ± 0.037 | 0.755 ± 0.038 | 0.622 ± 0.050 |
| 9 | 391.1 ± 84.069 | 0.767 ± 0.026 | 0.706 ± 0.032 | 0.772 ± 0.030 | 0.633 ± 0.048 |
| 10 | 457.0 ± 79.538 | 0.763 ± 0.035 | 0.688 ± 0.045 | 0.739 ± 0.047 | 0.632 ± 0.062 |
| Enzyme Group | Enzyme Group Name | p-Value | # of Enzymes | Enzymes (EC) |
|---|---|---|---|---|
| 3.2.1 | Glycosidases | 4.13 × 10−17 | 74 | 3.2.1.1, 3.2.1.10, 3.2.1.101… |
| 2.8.3 | CoA-transferase | 3.86 × 10−14 | 12 | 2.8.3.10, 2.8.3.12, 2.8.3.15… |
| 4.2.1 | Hydro-lyases | 1.52 × 10−13 | 73 | 4.2.1.101, 4.2.1.103, 4.2.1.104… |
| 3.1.1 | Carboxylic-ester Hydrolases | 3.43 × 10−9 | 28 | 3.1.1.1, 3.1.1.11, 3.1.1.13… |
| 4.3.1 | Ammonia-lyases | 3.43 × 10−9 | 16 | 4.3.1.14, 4.3.1.16, 4.3.1.18… |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakir-Gungor, B.; Ersoz, N.S.; Yousef, M. Integrating Biological Domain Knowledge with Machine Learning for Identifying Colorectal-Cancer-Associated Microbial Enzymes in Metagenomic Data. Appl. Sci. 2025, 15, 2940. https://doi.org/10.3390/app15062940
Bakir-Gungor B, Ersoz NS, Yousef M. Integrating Biological Domain Knowledge with Machine Learning for Identifying Colorectal-Cancer-Associated Microbial Enzymes in Metagenomic Data. Applied Sciences. 2025; 15(6):2940. https://doi.org/10.3390/app15062940
Chicago/Turabian StyleBakir-Gungor, Burcu, Nur Sebnem Ersoz, and Malik Yousef. 2025. "Integrating Biological Domain Knowledge with Machine Learning for Identifying Colorectal-Cancer-Associated Microbial Enzymes in Metagenomic Data" Applied Sciences 15, no. 6: 2940. https://doi.org/10.3390/app15062940
APA StyleBakir-Gungor, B., Ersoz, N. S., & Yousef, M. (2025). Integrating Biological Domain Knowledge with Machine Learning for Identifying Colorectal-Cancer-Associated Microbial Enzymes in Metagenomic Data. Applied Sciences, 15(6), 2940. https://doi.org/10.3390/app15062940







