1. Introduction
As social productivity rapidly develops and quality of life continues to improve [
1], there is a growing demand for medical imaging diagnostics among patients. Medical imaging (including X-rays, CT scans, pathological images, and MRI) serves as crucial reference material for imaging physicians’ diagnostic work, as shown in
Figure 1. Traditionally, the task of interpreting medical images has primarily fallen to radiologists and clinicians. However, current deep learning methods still face numerous challenges in medical image classification, such as insufficient model generalization, sensitivity to noisy data, and high computational resource consumption. Addressing these issues is crucial for improving the accuracy and efficiency of medical image classification.
Machine learning algorithms can rapidly detect anomalies through image feature analysis, effectively complementing manual diagnostic limitations and alleviating physicians’ workload. Nevertheless, medical imaging data often suffer from restricted dimensions and poor signal-to-noise ratios, which poses challenges for deep learning models in precisely identifying abnormalities using only visual information [
2]. Thus, the task of accurately and efficiently categorizing medical images to support physicians in diagnosing diseases continues to be a significant challenge within medical image analysis. Consequently, this research seeks to improve medical image classification performance by refining the architecture of the model and attention modules, targeting higher classification accuracy and greater robustness to noisy data.
Although deep learning technology has shown great potential in medical image analysis, its application in clinical practice has raised a series of ethical issues, particularly regarding data privacy. Medical image analysis faces the challenge of small sample sizes, and the traditional solution is to aggregate data from multiple sites. However, due to strict privacy protection policies, such as HIPAA (Health Insurance Portability and Accountability Act) and GDPR (General Data Protection Regulation), directly sharing medical image data is not feasible. Federated learning, as a solution for collaborative training without sharing data, can effectively protect data privacy and demonstrates its effectiveness and potential in addressing data privacy issues in medical image analysis [
3,
4].
The rapid advancement of deep learning in recent years has led to remarkable progress in multiple areas, including image classification, object detection, and semantic segmentation. Computer-assisted diagnostic technology employs deep learning methods to analyze medical images and patient data [
5], enabling both condition assessment and clinical decision support. The AlexNet model’s excellent performance on ImageNet dataset classification tasks marked a breakthrough in deep learning for image classification [
6]. Subsequently, VGG-Net improved classification accuracy by increasing network depth and reducing parameter count [
7]. GoogLeNet reduced parameter count through the Inception module and optimized computational resource usage [
8]. Nonetheless, these methods face limitations in medical image classification, including the oversight of multi-scale feature extraction and a lack of model interpretability. When adapted to medical image classification tasks, their performance typically lags significantly behind their initial target tasks.
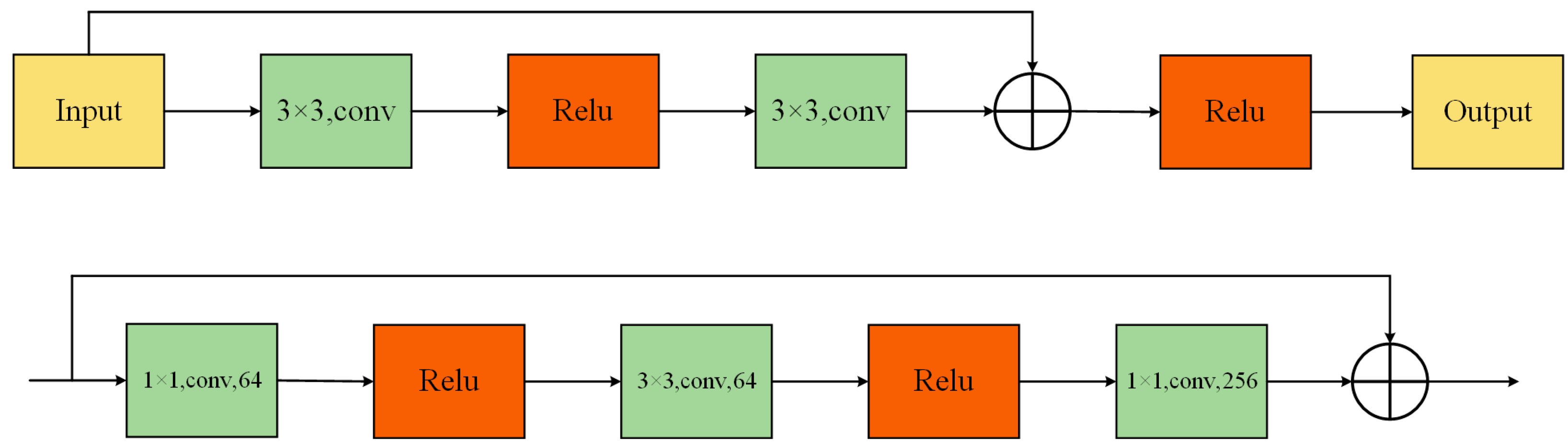
Currently, mainstream methods in medical image analysis are still based on ResNet and its variants. Given its modular design and robust feature extraction capabilities, ResNet demonstrates remarkable adaptability to diverse tasks in medical image processing [
9]. Nonetheless, ResNet was primarily conceived for image classification, characterized by restricted receptive fields and a lack of cross-channel and cross-spatial interaction functionalities, which could hinder its effectiveness in direct medical imaging scenarios. Therefore, manual adjustments to ResNet are usually required for specific tasks. Although such manual adjustments can improve performance to some extent, they also limit generalizability and efficiency in broader medical applications. For instance, Res2Net [
10] enhanced ResNet through improved multi-scale feature representation, ResNeXt [
11] achieved higher accuracy using grouped convolutions, and CBAM [
12] strengthened feature representation through the introduction of spatial attention modules. Beyond ResNet and its variants, the emergence of ViT (Vison in Transformers) has opened new avenues for medical image processing, and more research has begun to utilize ViT and its derivatives in this domain. MC-ViT [
13] uses two sub-networks to predict slice-level pathological information and whole-slide-level thymoma subtypes, significantly improving the accuracy of thymoma subtype classification. The MaxCerVixT [
14] model combines the multi-axis vision transformer (MaxViT) architecture, ConvNeXtv2 blocks, and GRN-based MLP layers, effectively improving the accuracy and inference speed of cervical cancer detection.
ResGANet [
15], introduced in 2022, enhances feature representation through grouped attention mechanisms, yet still exhibits limitations in addressing rotation invariance and local feature capture. Therefore, although existing methods have improved in accuracy, they are still insufficient in addressing key challenges in medical image analysis, such as rotation invariance, local feature capture, and cross-channel interaction. To overcome these limitations, we propose the Residual Group Dual-Channel Attention Network (ResGDANet), which substantially enhances model performance in medical classification tasks by incorporating Dual-Channel Attention Fusion (DCAF) and RMT modules [
16], showing stronger adaptability and robustness, especially in terms of rotation invariance, local feature capture, and cross-channel interaction.
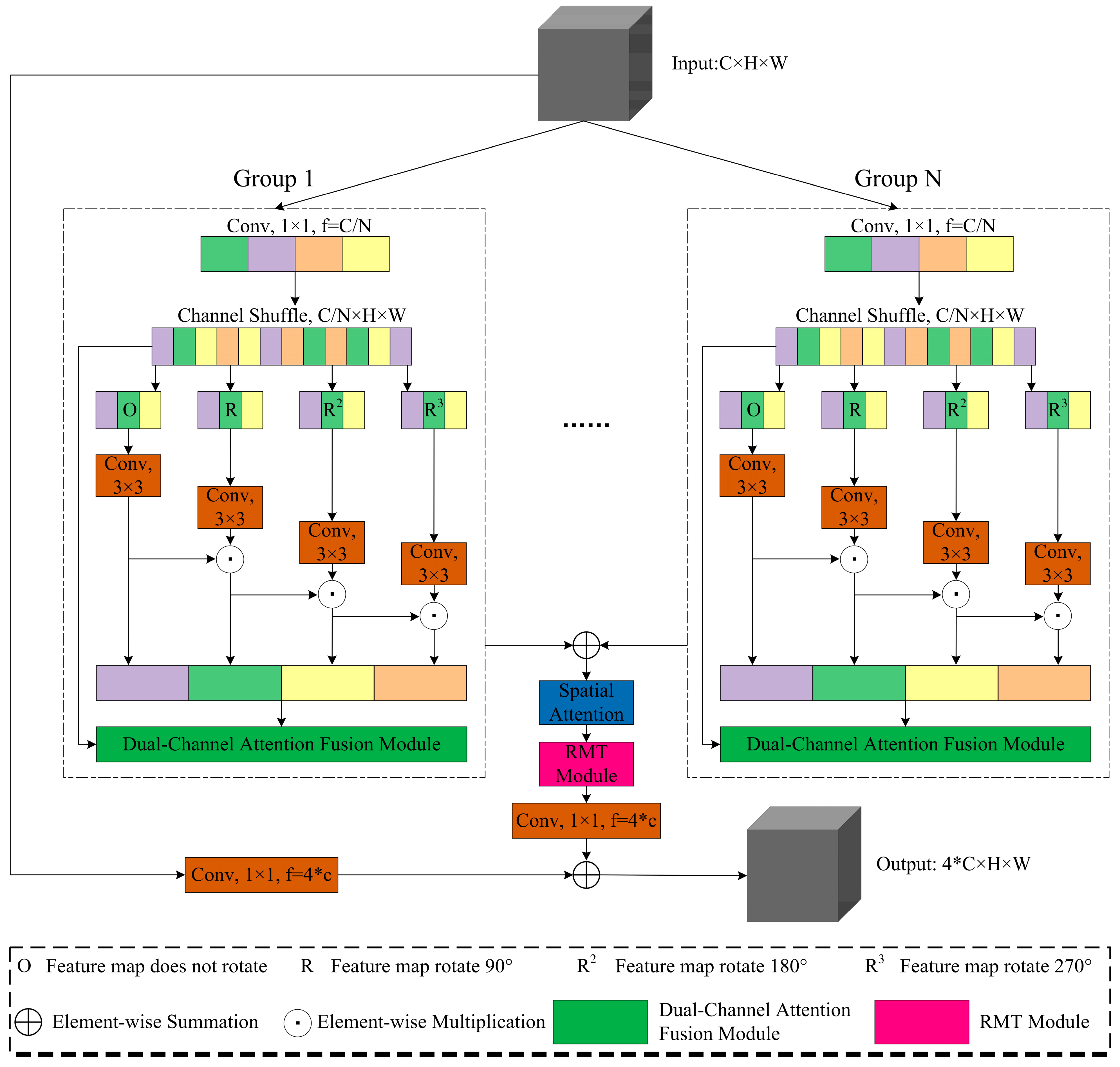
This paper introduces ResGDANet, an innovative medical image classification method built upon ResGANet, and our proposed attention module designed for enhanced feature representation. The primary contributions of this work are:
- (1)
Dual-Channel Attention Fusion (DCAF) module: Employs a dual-branch architecture combining global average pooling and max pooling to enhance the network’s capacity for capturing locally significant features, while integrating channel attention and coordinate attention to improve the robustness and accuracy of feature representations.
- (2)
RMT module: By leveraging the retention mechanism of Retentive Networks, the RMT module allows the model to efficiently capture and preserve critical information in the data when facing rotating objects. Simultaneously, it incorporates the global context modeling ability of Vision Transformers to comprehend the connections between different components, leading to more effective feature extraction. Through the Manhattan Self-Attention (MaSA) mechanism, the model performs better in handling rotation invariance issues, significantly improving the performance of medical image classification tasks.
The first chapter emphasizes the research background and significance of image classification while providing an overview of recent developments in convolutional neural networks within the field of medical image analysis. The second chapter highlights current work pertaining to medical image classification tasks, encompassing residual networks, their various derivatives, and attention mechanisms. The third chapter presents ResGDANet, designed for image classification based on ResGANet, which begins by introducing the network model’s components, then elaborates on the model’s design and implementation methodology, including ResGANet, the Dual-Channel Attention Fusion (DCAF) module, and the RMT module. The fourth chapter begins with a comprehensive description of the experimental dataset and evaluation metrics. Subsequent experimental validation shows that the ResGDA model outperforms the original model and other leading image classification models in terms of feature extraction ability and medical image classification accuracy. In conclusion, a comprehensive overview of this research and its findings is presented, highlighting existing limitations and suggesting potential directions and opportunities for future research and enhancements.
3. Methodology
3.1. Dual-Channel Attention Fusion Module
Through learning from data, attention mechanisms can weight multi-dimensional features according to their importance, enhancing critical features while suppressing interference. Hou et al. [
33] proposed embedding positional information into channel attention, using one-dimensional global average pooling in the horizontal and vertical directions to compress information, reducing positional information loss from 2D pooling. Nevertheless, this technique employs average pooling exclusively to derive global region data, potentially overlooking locally salient features in lesion areas, such as small lesions, edge details, or texture features [
34]. In medical image analysis, since many pathological regions share visual characteristics with normal tissues, it is essential to preserve fine local features to support subsequent tasks such as lesion classification, localization, and diagnosis.
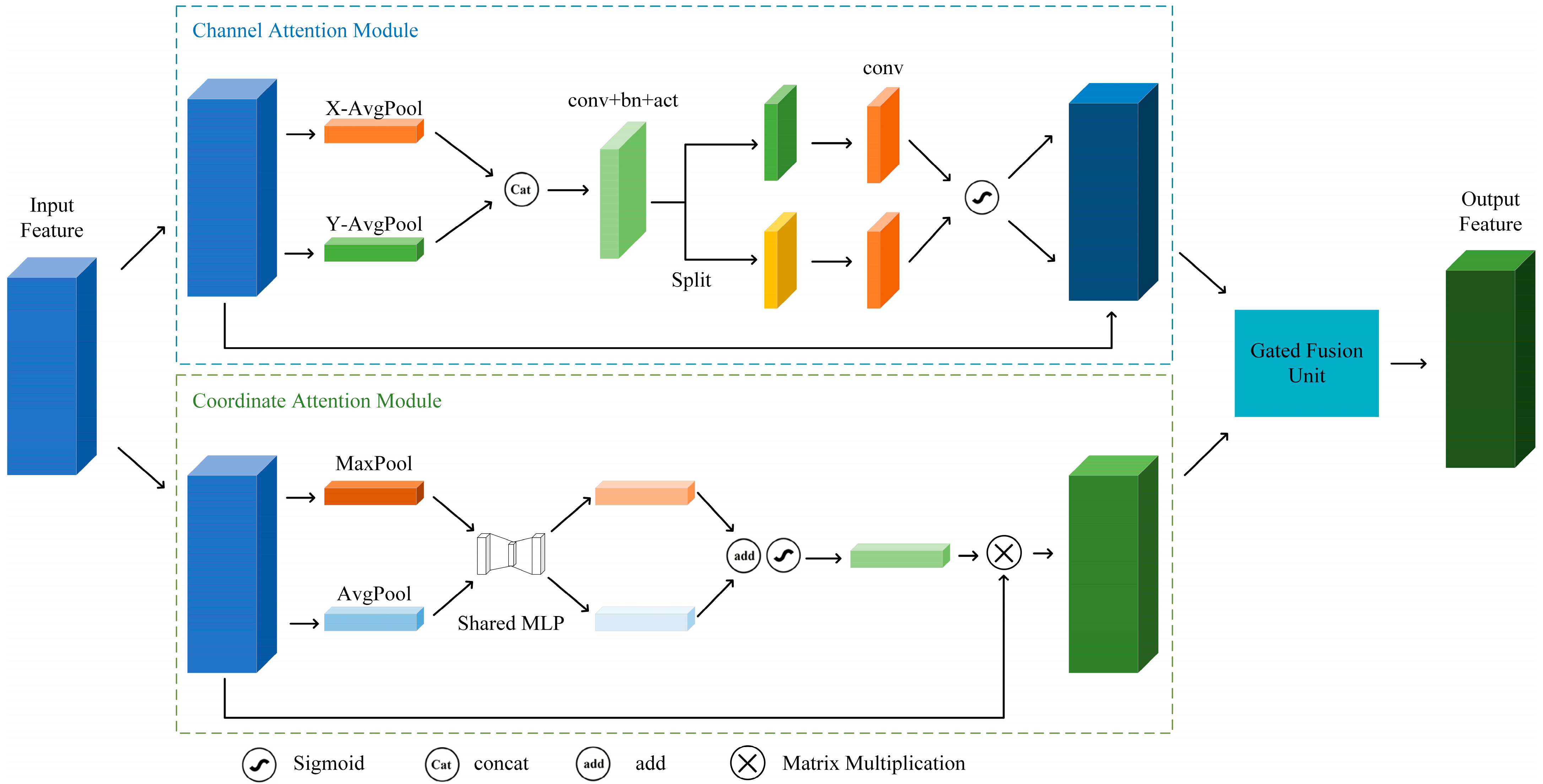
To address this challenge, we designed the Dual-Channel Attention Fusion (DCAF) module, introducing a dual-branch structure combining global average pooling and max pooling to enhance the capture of significant local features in medical images, as shown in
Figure 3. Besides maintaining the original global average pooling branch, we incorporated a max pooling branch to capture prominent local features. By fusing feature information from both branches, the model can simultaneously utilize global contextual information and local salient features, thereby further improving the accuracy and robustness of classification tasks.
For input feature map
, where
is the batch size,
is the number of channels, and
and
are the height and width of the feature map, the spatial information is first aggregated through global average pooling and global max pooling:
and
represent the global average pooling and global max pooling results, respectively, with dimensions
. The outputs of these two branches generate channel attention maps through a shared-weight multilayer perceptron (MLP):
where
is the channel attention map,
and
are the weight matrices of MLP,
is the ReLU activation function, and
is the Sigmoid activation function. The reduction ratio, denoted as
, signifies that the number of channels in the input feature map is first reduced to
via a
convolution and subsequently restored to the original channel count.
The other branch is coordinate attention, which first calculates global information for each position in spatial directions through global average pooling operations on the feature map’s spatial dimensions. Specifically, for input feature map
with dimensions
, global average pooling is performed in the horizontal and vertical directions to obtain feature maps
and
in Equation (4):
This operation maintains global information on the feature map along horizontal and vertical directions, enabling the network to comprehend object distribution in both horizontal and vertical orientations.
To merge information from height and width directions, we transpose the dimensions of
to
, then concatenate it with
along spatial dimensions to obtain feature map
:
After batch normalization and activation operations on the convolved feature map,
is split along spatial dimensions into two tensor representations,
and
, as shown in Equations (6) and (7).
Then, we pass
and
through
convolution layers to restore the channel number to
, and generate attention weights
and
through the Sigmoid activation function in Equation (8):
These weights reflect the spatial importance of each position in the feature map. Finally, we multiply the generated attention weights
and
with the input
yield output feature map:
The symbol in the formula denotes element-wise multiplication.
To effectively integrate channel attention with coordinate attention, the DCAF module combines their outputs using adaptive weights. Initially, the coordinate attention and channel attention branches generate feature maps
and
, respectively:
In Equation (10), and refer to the outputs of the coordinate attention module and the channel attention module, respectively. Their relationship with Equations (3) and (9) is as follows: calculates obtained through the coordinate attention module, while calculates obtained through the channel attention module.
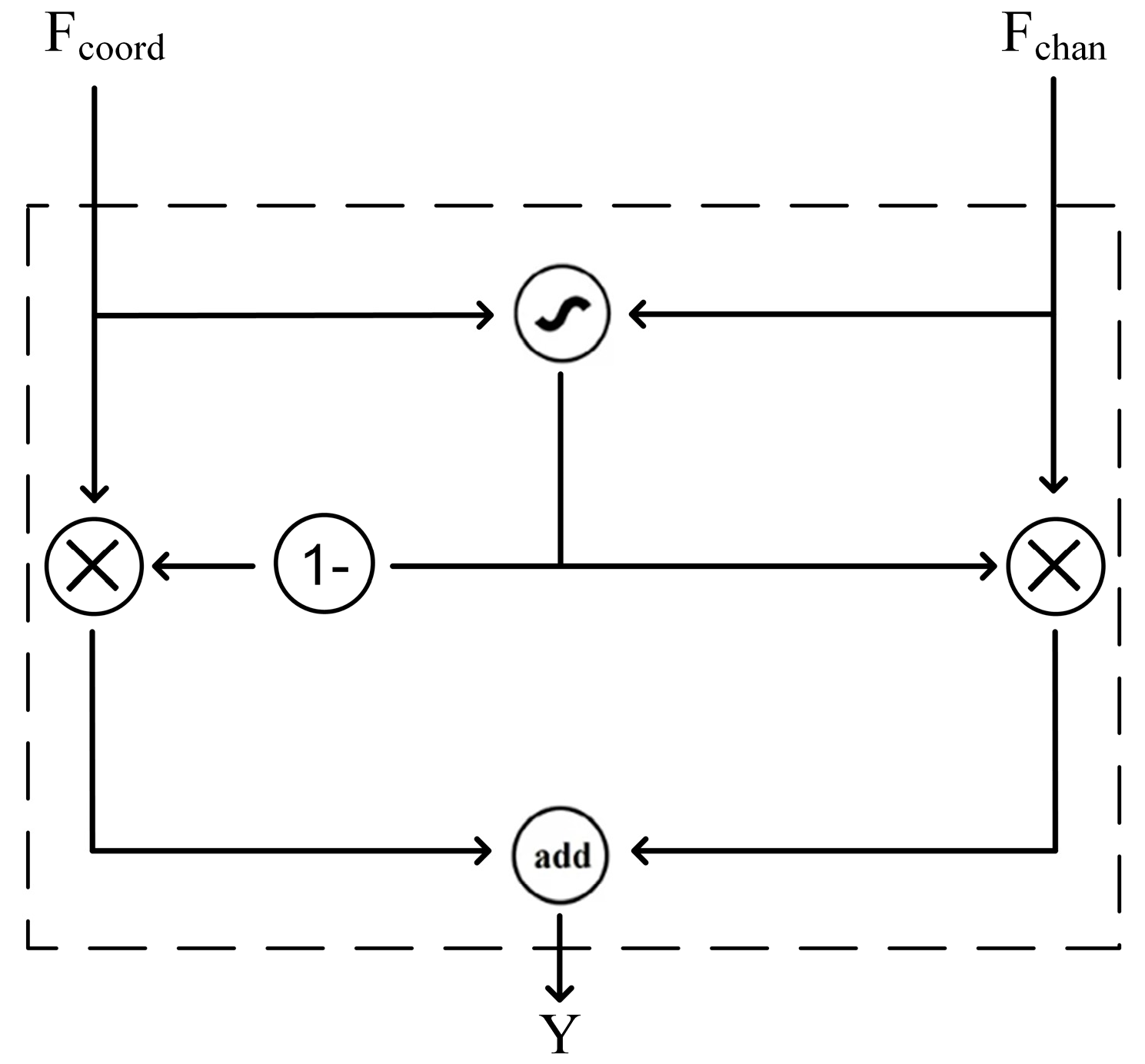
Finally,
and
pass through the Gated Fusion Unit in
Figure 4. This module combines the feature maps of coordinate attention and channel attention through an adaptive weighting mechanism, effectively enhancing the model’s performance in medical image classification. Through this operation, the network can better focus on locally important information and exhibit stronger adaptability in complex medical images. The module fuses the outputs of the two branches using an adaptive weight
, resulting in
in Equation (12):
where
is the adaptive fusion weight and
is the final output feature map.
The detailed pseudocode description of the DCAF module can be found in Algorithm A1 in
Appendix A.
3.2. RMT Module
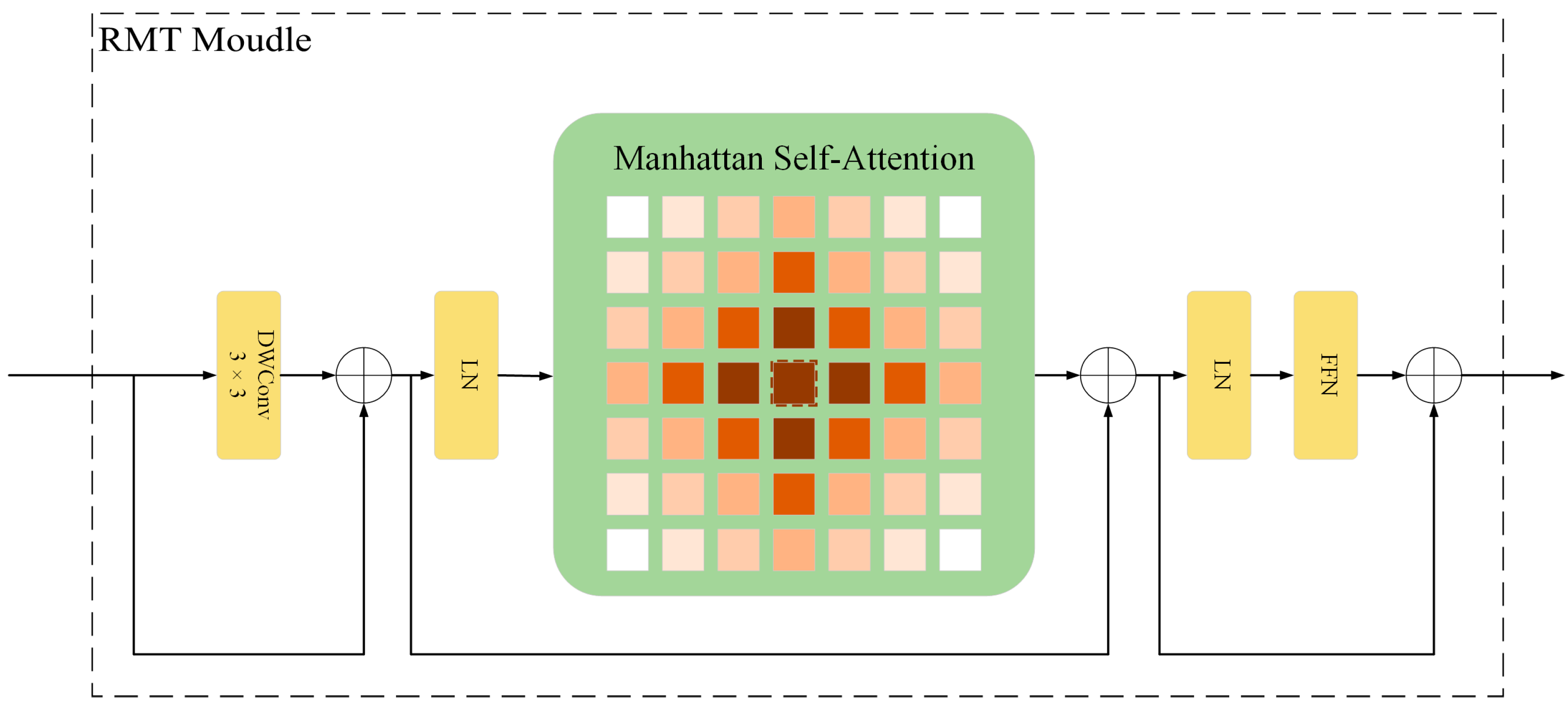
Medical image lesions exhibit significant variations in morphology and orientation, while traditional deep learning models lack rotational invariance learning capabilities, making it difficult to accurately identify lesion features at different angles. In response to this issue, we introduce the RMT module [
16], shown in
Figure 5, which integrates the global context modeling strengths of Vision Transformers and the retention mechanism from Retentive Networks, enabling the effective incorporation of spatial prior knowledge and the extraction of rotation-invariant features.
Through self-attention mechanisms alongside explicit spatial prior information, the RMT module learns stable features under rotational transformations, enhancing its ability to extract features from lesions of various morphologies. The module employs a two-dimensional distance-based decay matrix and utilizes the Manhattan Self-Attention (MaSA) mechanism for computation along both image axes, reducing the computational burden associated with extensive annotations. This design is particularly suitable for capturing morphologically diverse lesion features in medical images.
The RMT module’s output can be expressed as:
Here,
is the output feature of the RMT module,
is the output of the Manhattan self-attention mechanism,
is the output of the local context enhancement module based on depthwise convolution (DWConv), and
is the value matrix of the input feature map. LCE denotes the local context enhancement module employing Depthwise Separable Convolution (DWConv), while MaSA decomposes into two image axes, as shown in Equation (14). To be precise, the attention scores are derived individually for the horizontal and vertical dimensions. Subsequently, the attention weights are modified using one-dimensional bidirectional decay matrices.
In Equation (14), and represent the attention weights computed along the horizontal and vertical axes, respectively. and are the query matrices along the two axes, and are the key matrices along the two axes, and are the decay matrices along the two axes, and denotes element-wise multiplication (Hadamard product).
The RMT module integrates spatial prior information explicitly into the self-attention mechanism, strengthening its ability to interpret spatial relationships. Through the application of spatial priors and attentional allocation mechanisms, the module achieves reduced computational overhead while preserving linear complexity. The rotational invariance property improves the robustness of lesions in any direction, significantly enhancing the detection accuracy of abnormal tissues in medical images. This characteristic offers significant advantages in processing lesions of various angles and morphologies, helping to improve the accuracy of medical image classification.
3.3. Residual Group Dual-Attention Network
This study proposes ResGDANet (Residual Group Dual-Attention Network) based on ResGANet, improving the original model through integrated attention modules.
In
Figure 6, the network first groups the input image matrix
according to channel number
. For broader feature extraction, basic feature transformation is performed following channel reorganization. For each coordinate
in the matrix, the transformation is represented as:
where
represents the matrix coordinate points, and
[
35].
The transformed feature map passes through ResNet bottleneck blocks, operating with
convolution kernels to enlarge the model’s receptive field, providing a foundation for subsequent attention weight allocation. Using
to denote the
convolution operation, with output represented as
in Equation (16):
In Equation (16), represents the s-th group of the input feature map, and denotes the initial convolution output.
Following basic feature extraction, the Dual-Channel Attention Fusion (DCAF) module is incorporated to enhance feature representation. The module utilizes a dual-branch architecture combining max pooling and global average pooling, fusing channel attention and coordinate attention via adaptive weighting as in Equation (12).
The spatial attention module generates spatial feature descriptors through global average pooling (GAP) and global max pooling (GMP), enhancing spatial dimension feature relationships.
In Equation (17), represents the spatial attention weight, and denotes the Sigmoid activation function.
Following this, the attention scores for the RMT module are computed as per Equation (14). The final output feature is:
This design enables the RMT module to extract features at stable resolution while maintaining low computational complexity, enhancing feature rotational invariance and thereby improving medical image classification performance.
4. Experiments
4.1. Experimental Settings
The experiments were carried out on a system running Ubuntu 22.04, equipped with an RTX 4090 GPU with 24 GB of video memory, sourced from NVIDIA. The implementation of the experiments was carried out using the PyTorch(1.9.0+cu111) framework, with cross-entropy loss function, the Adam [
28] optimizer, a batch size of 16, a learning rate of 3.5 × 10
−5, and 120 training epochs. The detailed hyperparameter settings are presented in
Table 1.
4.2. Dataset and Image Preprocessing
COVID19-CT [
36]: This dataset comprises medical images and COVID-19-related publications collected by He et al. [
37] from medRxiv2 and bioRxiv3. It includes 349 CT scan images confirmed as COVID-19 positive and 397 CT scan images classified as normal or negative for other diseases. The dimensions of the images vary between 143 × 76 and 1637 × 1225. Following the original data division strategy, we divided the dataset into three parts—training set, validation set, and test set—in a ratio of 0.6:0.15:0.25.
ISIC2018 [
38]: In the evaluation of the ResGDANet model, we used the ISIC2018 skin lesion diagnosis dataset. This dataset consists of seven categories, totaling 10,015 images. It includes 6705 melanocytic nevus images, 115 dermatofibroma images, 1113 melanoma images, 327 actinic keratosis images, 1099 benign keratosis images, 514 basal cell carcinoma images, and 142 vascular lesion images. All images in the dataset have a size of 650 × 450 pixels. We divided the dataset into training, validation, and testing sets in a ratio of 0.6:0.15:0.25 using stratified sampling.
Examples of ISIC2018 and COVID19-CT images are shown in
Figure 7.
For image preprocessing, we applied normalization and data augmentation techniques to enhance the model’s generalization ability and avoid overfitting. All preprocessing operations were performed after the dataset was divided to ensure that no information would leak between the training, validation, and test sets.
To improve the model’s generalization ability, various data augmentation methods were applied to the training set during the data preprocessing phase. These methods included random horizontal flipping and random vertical flipping, each with a probability of 50%; random rotation with an angle range of −15° to 15°, to simulate rotation variations of images in natural environments; and random adjustments of the image’s brightness, contrast, and saturation, with adjustment ranges from 0.8 to 1.2, to improve the model’s adaptability to images under different lighting conditions. It is important to emphasize that data augmentation was only applied to the training set, while the validation and test sets retained the original data to ensure the fairness and consistency of performance evaluation.
All images were normalized. The normalization used the mean and standard deviation of the ImageNet dataset: mean of [0.485, 0.456, 0.406] and standard deviation of [0.229, 0.224, 0.225]. The specific normalization formula is
where
is the original pixel value and
is the normalized pixel value. In addition, for images smaller than 224 × 224, proportional scaling was first applied to adjust the shorter side of the image to 256 pixels, followed by a crop of a 224 × 224 region from the center of the scaled image to ensure the consistency and adaptability of the input image.
4.3. Evaluation Metrics
This study employs accuracy, precision, recall, and F1 score as evaluation metrics.
Table 2 presents the confusion matrix. The matrix comprises True Positive (TP), False Negative (FN), False Positive (FP), and True Negative (TN) classes, which can be seen in
Table 2.
We use accuracy as one of the performance metrics for the experiment. Accuracy refers to the percentage of correct malware identification and overall classification performance. Due to the imbalanced nature of the medical image dataset, we also use precision, recall, and F1-score as evaluation metrics. These metrics are suitable for evaluating imbalanced deep-learning classification problems. Precision represents the positive predictive rate, while recall represents the true positive rate. The F1 score combines precision and recall into a single metric. The evaluation metrics are calculated using the following Equations (18)–(21):
TP, TN, FP, and FN represent the number of true positive, true negative, false positive, and false negative samples, respectively.
4.4. Ablation Research and Visualization
4.4.1. Ablation Studies in Different Groups
We conducted ablation experiments on multiple group settings of ResGDANet, employing the COVID19-CT dataset for evaluation. Results are presented for the ResGDANet model with grouping (G) values of 1, 2, and 4.
Table 3 displays the experimental results for accuracy, precision, recall, and F1-score (F1). Experimental results demonstrate superior classification accuracy when using group sizes of 2 or 4, which are shown in
Table 3.
Empirical studies further indicate that a higher number of groups leads to a reduction in the network’s inference speed. Using network configurations with two and four groups allows for achieving the optimal balance among various metrics in subsequent experiments. These findings confirm that expanding the number of groups within the ResGDANet enhances the classification performance for medical images.
4.4.2. Ablation Experiment of Attention Module
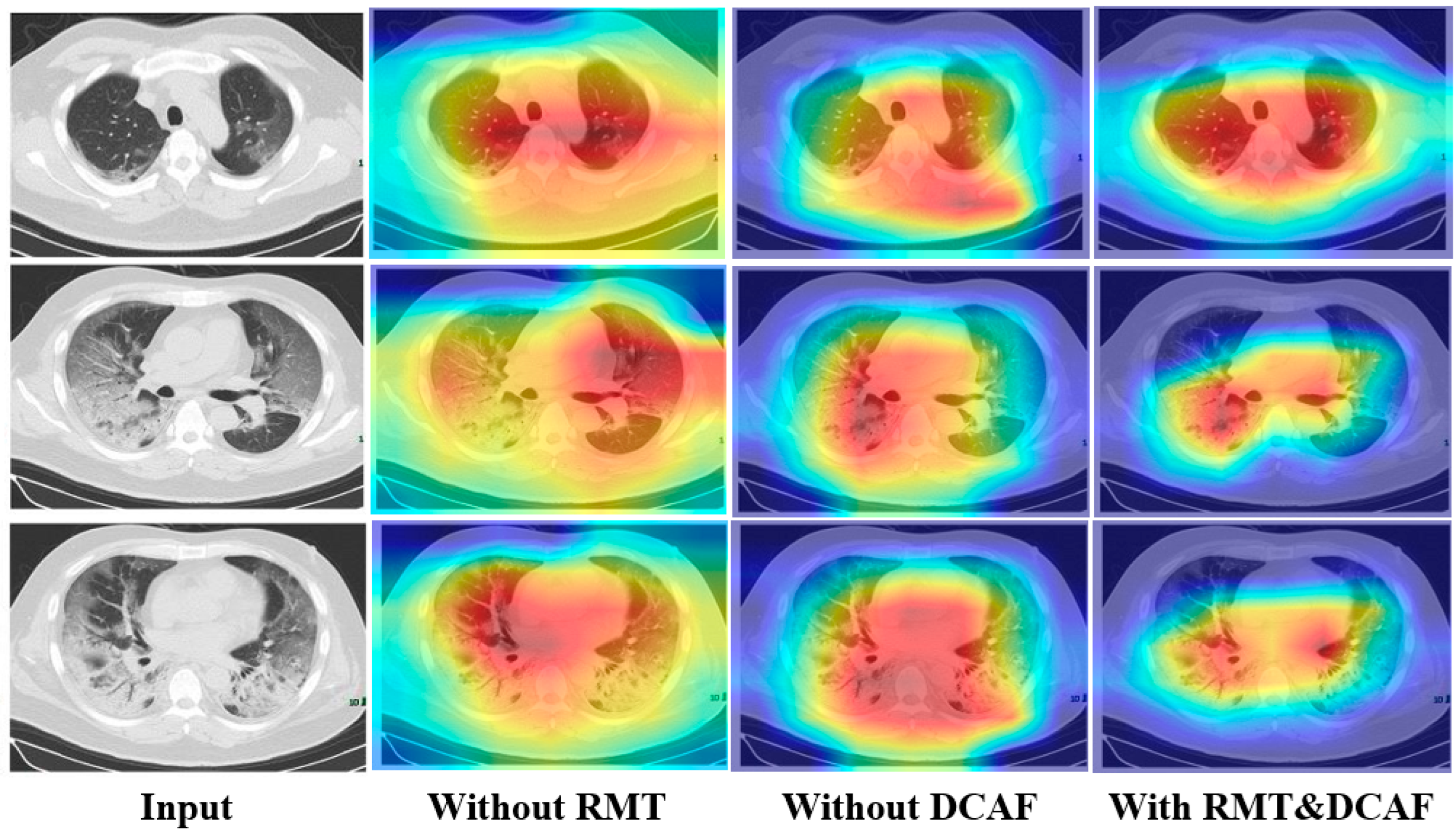
To more intuitively and visually validate the classification performance of the proposed ResGDANet model in this paper, we used Grad-CAM [
39] to visualize the attention generated by ResGDANet without attention modules, without DCAF, without RMT, and with both attention modules.
Figure 8 demonstrates that without attention modules, the network’s focus is limited to specific portions of the target region. With the addition of the DCAF module, the region of interest expands, though it shows misalignment with the target area. The combination of both attention modules outperforms the individual modules, enabling more precise localization and coverage of target objects. The visualization performance achieved by incorporating only the spatial attention module and RMT module is marginally inferior to that of using solely the DCAF module. The RMT module exhibits a weaker ability to localize boundary regions compared to the DCAF module. However, when compared to visualizations without attention modules, the RMT module shows enhanced attention to the target region.
To further validate the effectiveness of each strategy and module, we also conducted ablation experiments on the ISIC2018 dataset. The experimental results, presented in
Table 4, primarily validate the effectiveness of various attention modules.
Table 4 demonstrates that the incorporation of both RMT and Dual-Channel Attention Fusion modules yields superior performance compared to the baseline without attention modules.
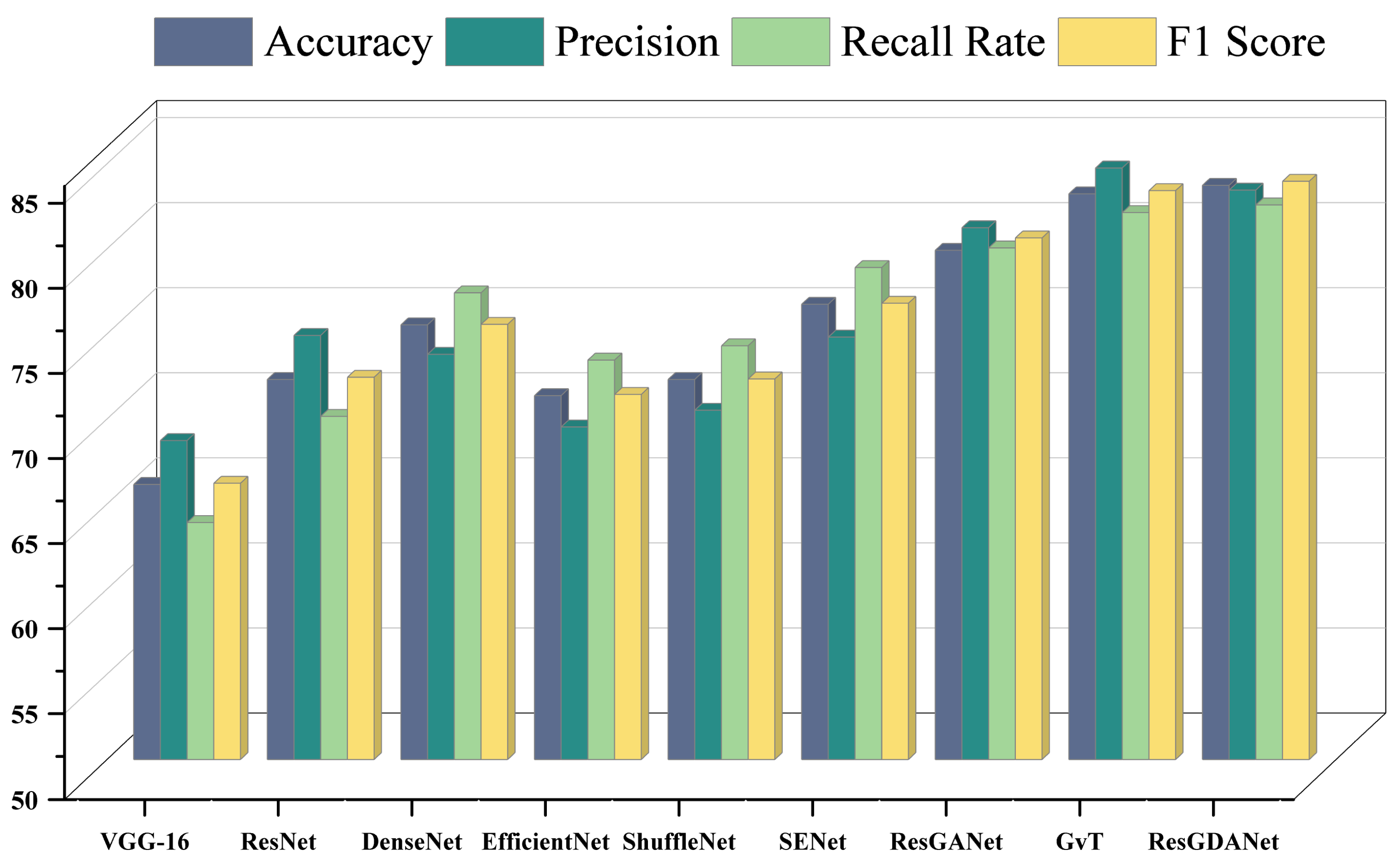
4.5. Model Comparison Experiment
We examined the performance metrics of various advanced classification techniques on the COVID19-CT dataset. The comparison included VGG-16 [
7], ResNet [
9], ResNeXt [
11], Res2Net [
10], DenseNet [
18], EfficientNet [
40], ShuffleNet 1.0 × (G = 4) [
41], SENet [
20], GvT [
21], and ResGANet [
15]. All models were implemented in their original form, sharing the same operational environment and essential hyperparameters. The experimental results are shown in
Table 5 and
Figure 9.
As shown in
Table 5, the approach introduced in this study surpasses other models in classification tasks, further demonstrating that our method can effectively diagnose COVID19-CT images. The highest values for each metric are indicated in bold.
Similar to ResNet and DenseNet, we use residual connections and dense connections to improve the efficiency of feature reuse and gradient flow. ResNet tackles deep network degradation through residual structures, whereas DenseNet achieves efficient feature reuse through dense connectivity. Additionally, like ResNeXt and Res2Net, we utilize group convolution and multi-scale feature representations to enhance model expressiveness. ResNeXt enhances network width by introducing the concept of cardinality, while Res2Net captures multi-scale information by building hierarchical residual connections. By incorporating these advantages, we achieve superior classification performance.
Making reference to ResGANet, we utilize attention modules to improve the feature representation abilities of convolutional neural networks. ResGANet adaptively recalibrates feature responses using channel and spatial attention mechanisms, whereas EfficientNet balances network depth, width, and resolution through compound scaling. Relative to ResGANet, our network demonstrated a 3.82% enhancement in classification accuracy (79.92% vs. 83.74%). This is mainly attributed to the introduction of the DCAF module and RMT module, which enhance the model’s ability to capture multi-scale features and model global contextual information.
GvT employs the Talking-Heads mechanism, enhancing model expressiveness through inter-head communication in multi-head attention. However, due to the complex structure of the GvT model, the training process requires more computational resources and time, which may result in its limited performance on small datasets. Our method performs better on small datasets, particularly improving by 0.41% compared to GvT in medical image classification tasks (83.33% vs. 83.74%). Although GvT scores slightly higher than ResGDANet in precision (84.76% vs. 83.46%), ResGDANet performs better in terms of F1 score (83.98% vs. 83.43%), indicating that ResGDANet has an advantage in balancing precision and recall.
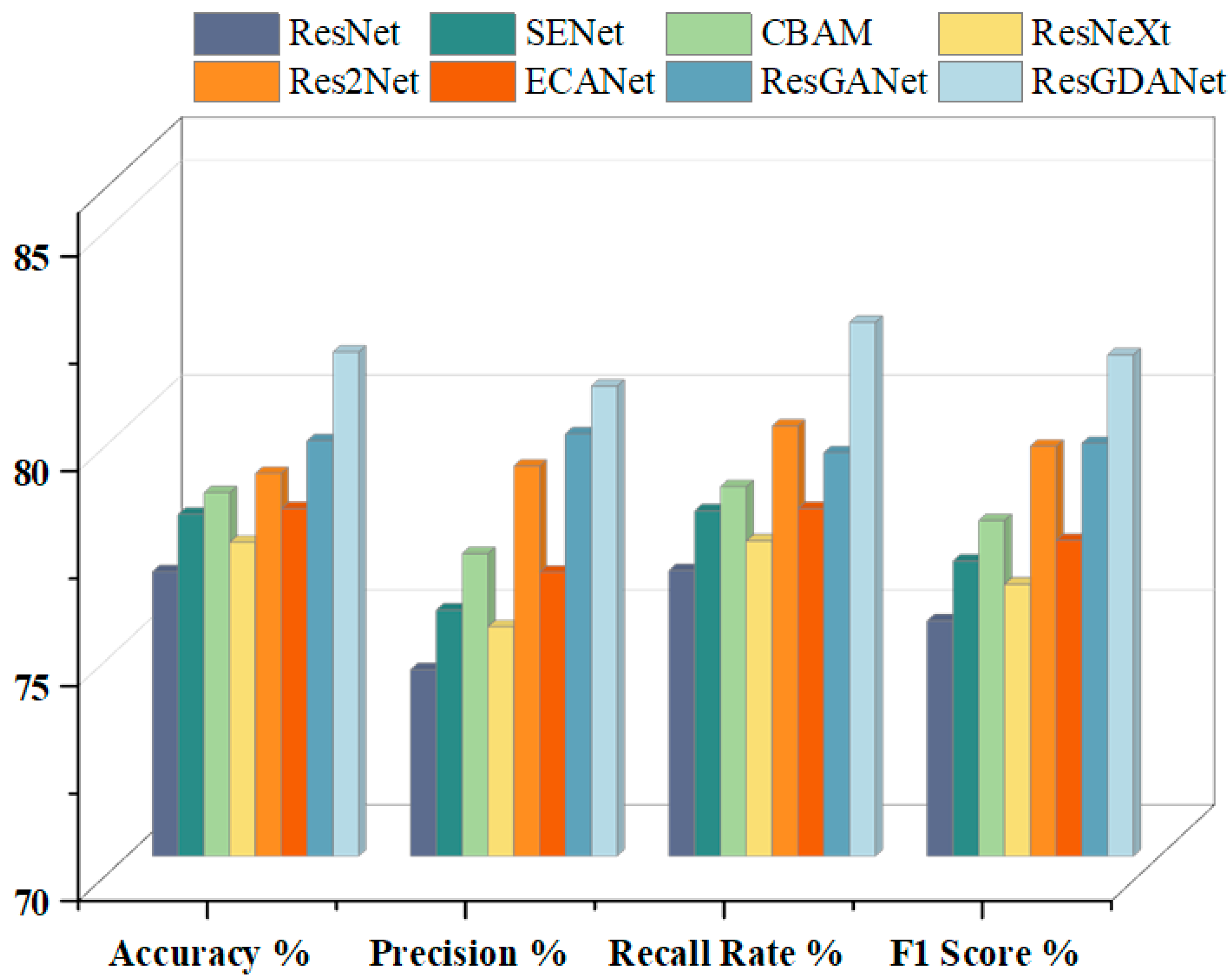
To further validate the proposed model, we also compared the classification performance of different models on the ISIC2018 dataset. The experimental results are shown in
Table 6 and
Figure 10.
The results show that ResGDANet has advantages in medical image classification. In comparison to CBAM and SENet, we built attention modules to enhance the feature representation capability of convolutional neural networks. Our network achieved accuracy improvements of 3.77% over SENet-50 (77.96% vs. 81.73%) and 3.27% over CBAM-50 (78.46% vs. 81.73%). ResGDANet demonstrated a 2.06% accuracy improvement over ResGANet (79.67% vs. 81.73%). These experimental results confirm that incorporating the DCAF module and RMT module or refining features into multiple groups, enhances classification model performance to varying degrees. ResGANet incorporates both these features and achieved the highest classification accuracy (81.73%) on the ISIC2018 dataset.
Figure 11 displays the confusion matrices of ResGDANet on the ISIC2018 and COVID19-CT datasets. Overall, the classifier demonstrates significant performance across all labels. In the ISIC2018 dataset, the most frequent misclassification is predicting MEL as NV. However, there is a reasonable explanation for this: the sample size of minority categories is small, leading to poorer performance in these categories. A solution to this issue is to increase the number of images in the database, including more data sources.
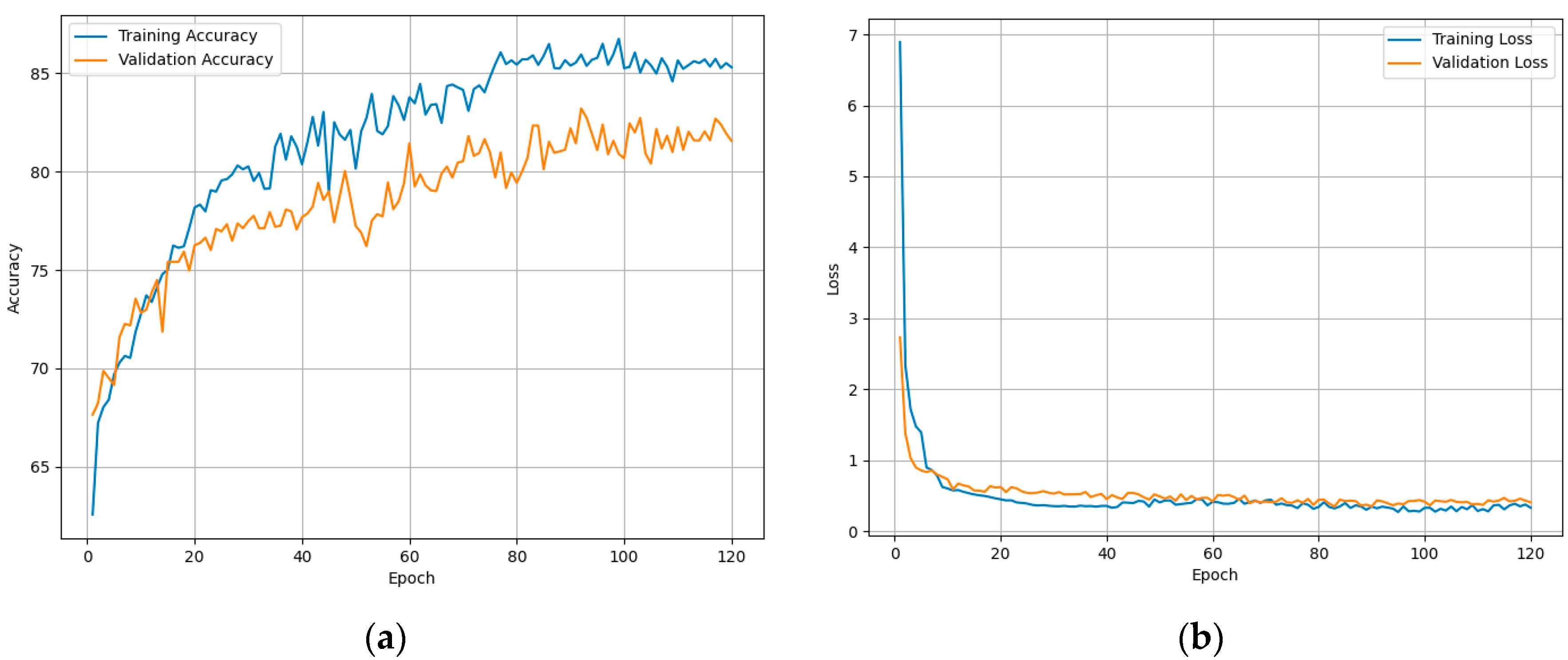
Figure 12 demonstrates the model fitting while training to assess the classification performance of ResGDANet. The figure shows that the model rapidly reaches an optimal state, with no signs of overfitting on the training or validation sets.