Sesame Plant Disease Classification Using Deep Convolution Neural Networks
Abstract
1. Introduction
2. Related Work
3. Methodology
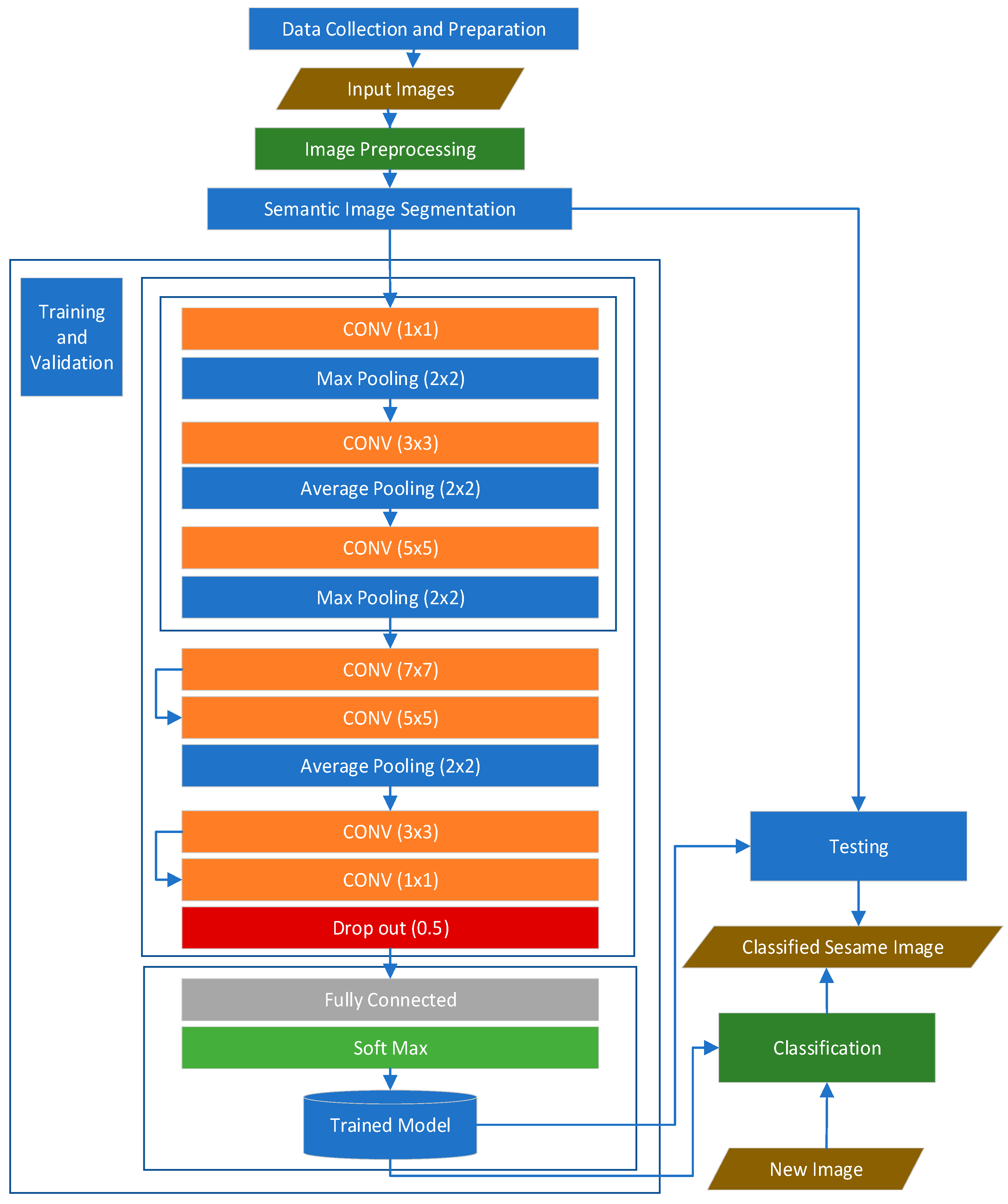
3.1. System Architecture
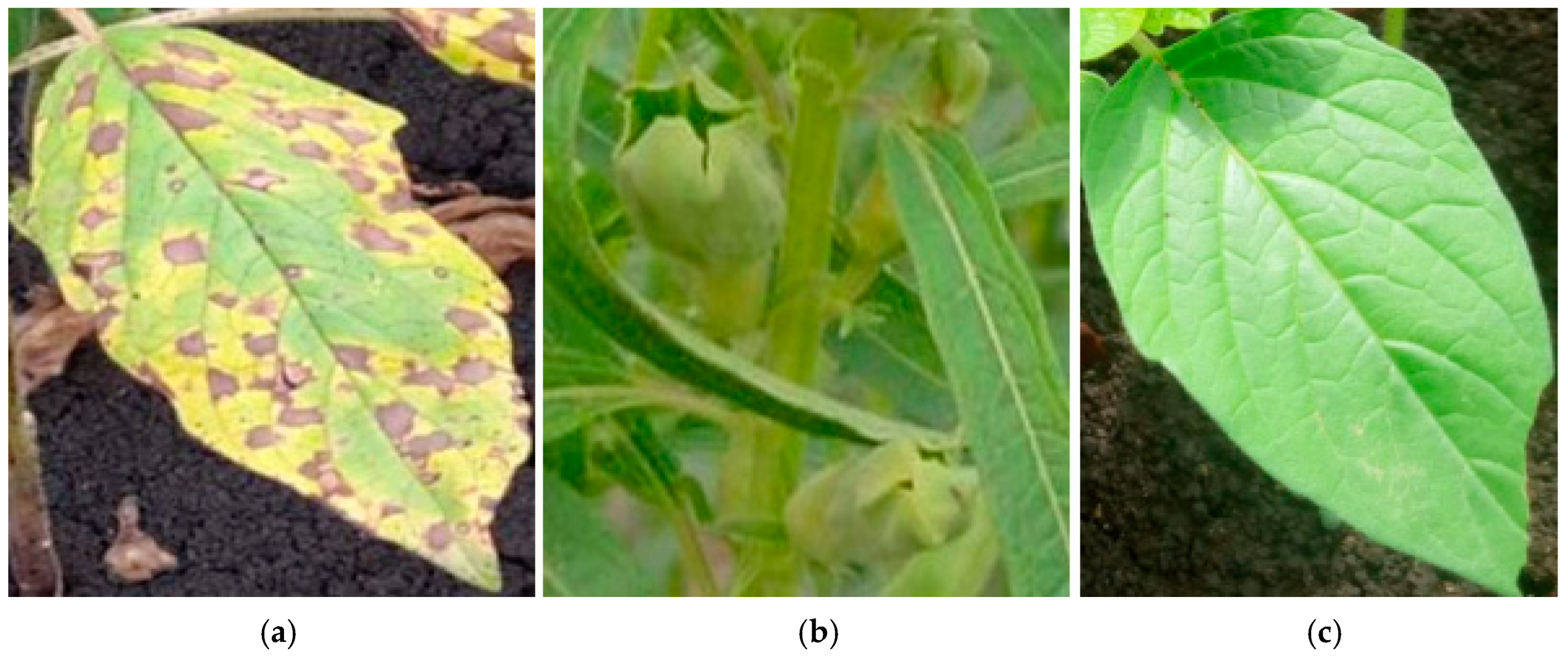
3.2. Data Collection and Preparation
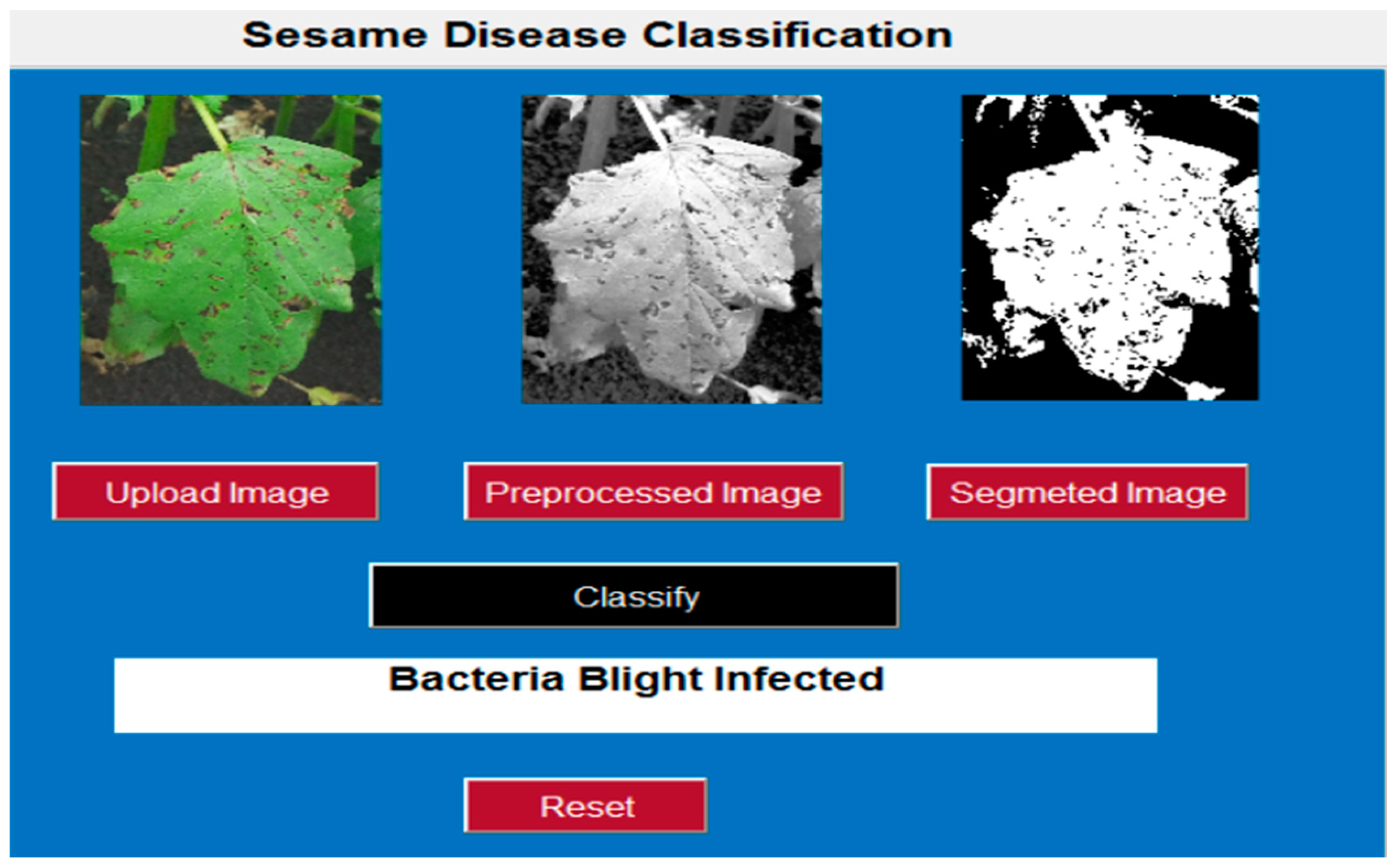
3.3. Image Preprocessing
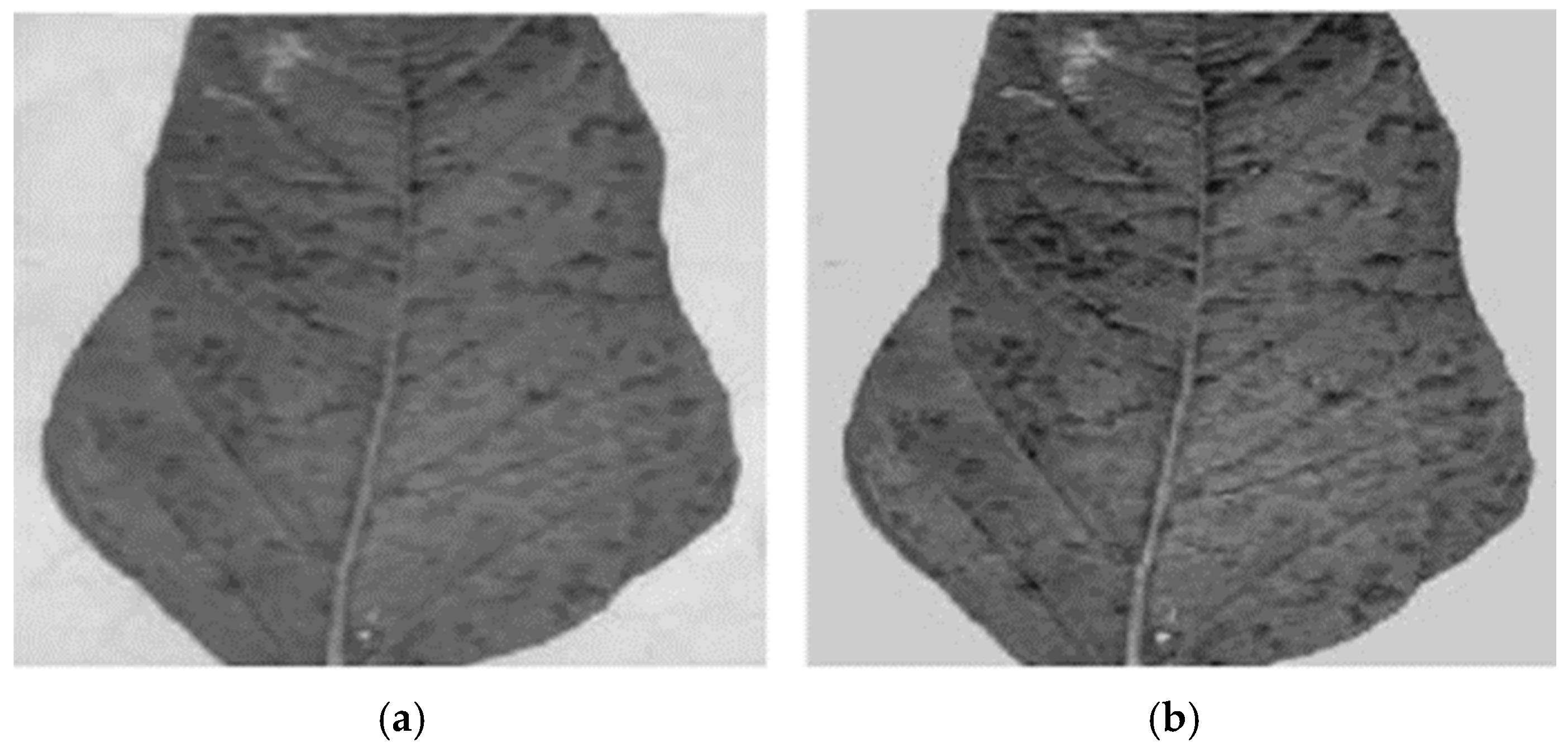
3.3.1. Contrast Stretching
3.3.2. Median Filtering
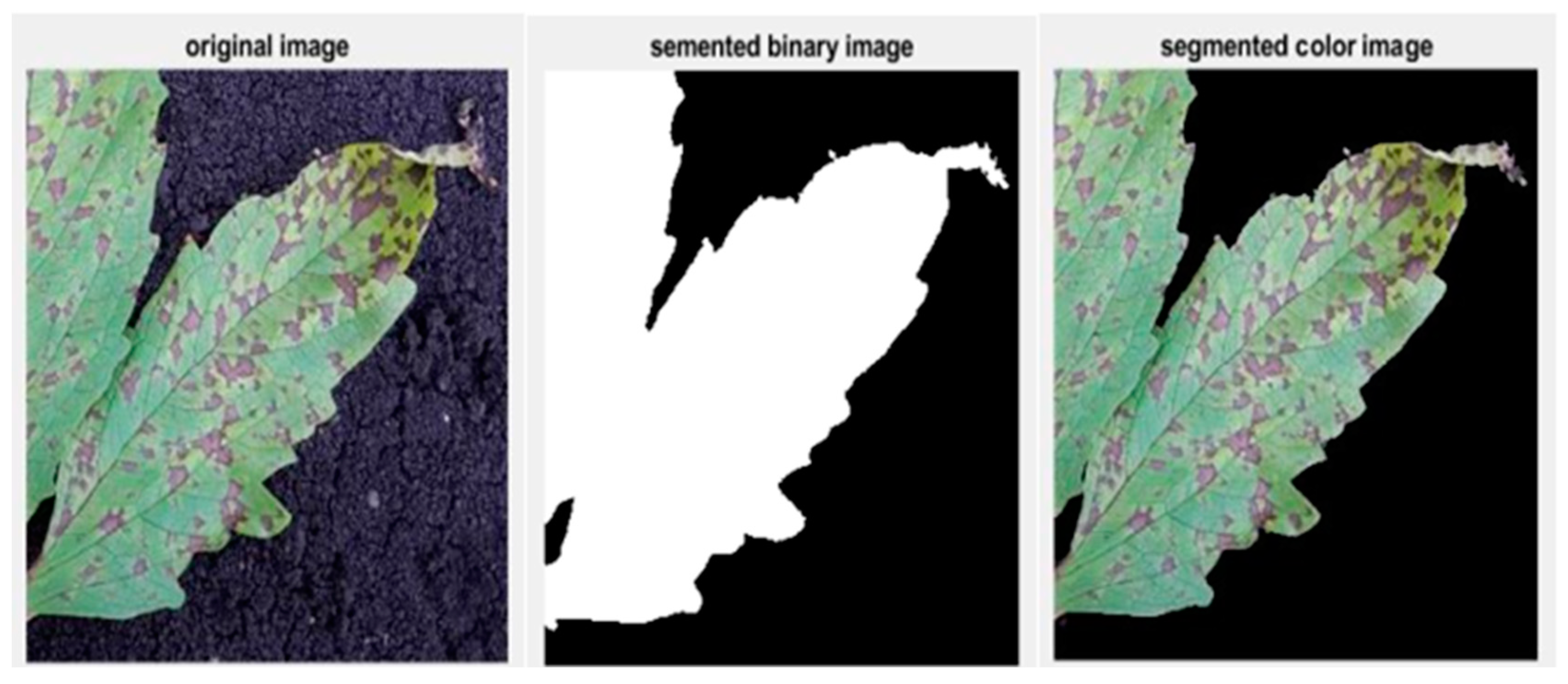
3.4. Image Segmentation
- Create pixel label images of the original sesame leaf images using the ground truth labeler;
- Load the training data and create a pixel label data store and an original image data store;
- Add the pre-trained VGG16 CNN model;
- Load the pre-trained SegNet semantic segmentation network;
- Train the SegNet network with our dataset images;
- Display and store the segmentation results.
3.5. Image Augmentation
3.6. Feature Extraction and Classification
3.7. Neural Network Training Algorithm
3.8. Performance Evaluation
4. Model Evaluation and Discussion
4.1. Performance of Proposed Model Trained with Original Images
4.2. Performance of Proposed Model Trained with Preprocessed and Segmented Images
4.3. Performance of Proposed Model Trained with Augmented Images
4.4. Discussion
4.4.1. Comparison with Inception-V3 Implementation Result
4.4.2. Comparison with Xception Implementation Result
4.4.3. Sample Classification of Sesame Plant Diseases
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reyad, S.; Albakry, B.S.S. Sesame oil, properties and advantages (Review Article). Futur. Biol. 2022, 1, 1–9. [Google Scholar] [CrossRef]
- Wei, P.; Zhao, F.; Wang, Z.; Wang, Q.; Chai, X.; Hou, G.; Meng, Q. Nutritional Value, Phytochemical Composition, Health Benefits, Development of Food, and Industrial Applications. Nutr. Rev. 2022, 14, 4079. [Google Scholar]
- Caruelle, D.; Shams, P.; Gustafsson, A.; Lervik-Olsen, L. Affective Computing in Marketing: Practical Implications and Research Opportunities Afforded by Emotionally Intelligent Machines. Mark. Lett. 2022, 33, 163–169. [Google Scholar] [CrossRef]
- Mequanenit, A.M.; Ayalew, A.M.; Salau, A.O.; Nibret, E.A.; Meshesha, M. Prediction of mung bean production using machine learning algorithms. Heliyon 2024, 10, e40971. [Google Scholar] [CrossRef]
- Vinnichek, L.; Pogorelova, E.; Dergunov, A. Oilseed market: Global trends. IOP Conf. Ser. Earth Environ. Sci. 2019, 274, 012030. [Google Scholar] [CrossRef]
- Meena, B.; Indiragandhi, P.; Ushakumari, R. Screening of sesame (Sesamum indicum L.) germplasm against major diseases. J. Pharmacogn. Phytochem. 2018, 7, 1466–1468. [Google Scholar]
- Kindeya, Y.B.; Golla, W.N.; Kebede, A.A.; Sibhatu, F.B. Survey and Identification of Major Sesame Diseases in Low Land Areas of Western Zone of Tigray. J. Biomater. 2018, 2, 58–64. [Google Scholar] [CrossRef]
- Ajay, A.; Reddy, K.V.S.R.; Kumar, A.A.; Someswar, C. Performance evaluation of twin-row planter for maize crop. J. Eco-Friendly Agric. 2024, 19, 214–218. [Google Scholar] [CrossRef]
- Development of Automatic Sesame Grain Grading System Using Image | 37212. Available online: https://www.longdom.org/proceedings/development-of-automatic-sesame-grain-grading-system-using-image-processing-techniques-37212.html (accessed on 15 December 2024).
- Alshawwa, I.A.; Elsharif, A.A.; Abu-Naser, S.S. An Expert System for Coconut Diseases Diagnosis. Int. J. Acad. Eng. Res. 2019, 3, 8–13. [Google Scholar]
- Marcos, A.P.; Rodovalho, N.L.S.; Backes, A.R. Coffee Leaf Rust Detection Using Convolutional Neural Network. In Proceedings of the 2019 XV Workshop de Visão Computacional (WVC), São Bernardo do Campo, Brazil, 9–11 September 2019; pp. 38–42. [Google Scholar] [CrossRef]
- Arivazhagan, S.; Ligi, S.V. Mango Leaf Diseases Identification Using Convolutional Neural Network. Int. J. Pure Appl. Math. 2018, 120, 11067–11079. [Google Scholar]
- Zhang, X.; Qiao, Y.; Meng, F.; Fan, C.; Zhang, M. Identification of maize leaf diseases using improved deep convolutional neural networks. IEEE Access 2018, 6, 30370–30377. [Google Scholar] [CrossRef]
- Yogananda, G.S.; Babu, A.J.; Julma, S.A.; Madhukar, G.; Manjula, B.M. Rice Plant Disease Classification using Transfer Learning of Deep ResNext Model. In Proceedings of the 2023 International Conference on Evolutionary Algorithms and Soft Computing Techniques (EASCT), Bengaluru, India, 20–21 October 2023; Volume XLII, pp. 18–20. [Google Scholar] [CrossRef]
- Afonso, M.; Blok, P.M.; Polder, G.; Van Der Wolf, J.M.; Kamp, J. Blackleg Detection in Potato Plants using Convolutional Neural Networks. IFAC-PapersOnLine 2019, 52, 6–11. [Google Scholar] [CrossRef]
- Idress, K.A.D.; Gadalla, O.A.A.; Öztekin, Y.B.; Baitu, G.P. Machine Learning-based for Automatic Detection of Corn-Plant Diseases Using Image Processing. Tarim Bilim. Derg. 2024, 30, 464–476. [Google Scholar] [CrossRef]
- Munteanu, C.; Lazarescu, V. Evolutionary contrast stretching and detail enhancement of satellite images. Proc. Mendel 1999, 99, 94–99. [Google Scholar]
- Sukassini, M.; Velmurugan, T. Noise removal using morphology and median filter methods in mammogram images. In Proceedings of the 3rd International Conference on Small and Medium Business, Hochiminh, Vietnam, 19–21 January 2016; pp. 413–419. [Google Scholar]
- Arce, G.R.; Paredes, J.L. Recursive weighted median filters admitting negative weights and their optimization. IEEE Trans. Signal Process. 2000, 48, 768–779. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, Y.; Georgiou, T.; Lew, M.S. A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr. 2018, 7, 87–93. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Toda, Y.; Okura, F. How convolutional neural networks diagnose plant disease. Plant Phenomics 2019, 2019, 9237136. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Biswas, K.; Kumar, S.; Banerjee, S.; Pandey, A.K. Smooth Maximum Unit: Smooth Activation Function for Deep Networks using Smoothing Maximum Technique. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 784–793. [Google Scholar] [CrossRef]
- Lai, L.; Liu, J. Support Vector Machine and Least Square Support Vector Machine Stock Forecasting Models. Comput. Sci. Inf. Technol. 2014, 2, 30–39. [Google Scholar] [CrossRef]
- Ahn, J.M.; Kim, S.; Ahn, K.-S.; Cho, S.-H.; Lee, K.B.; Kim, U.S. Correction: A deep learning model for the detection of both advanced and early glaucoma using fundus photography. PLoS ONE 2019, 14, e0211579. [Google Scholar] [CrossRef] [PubMed]

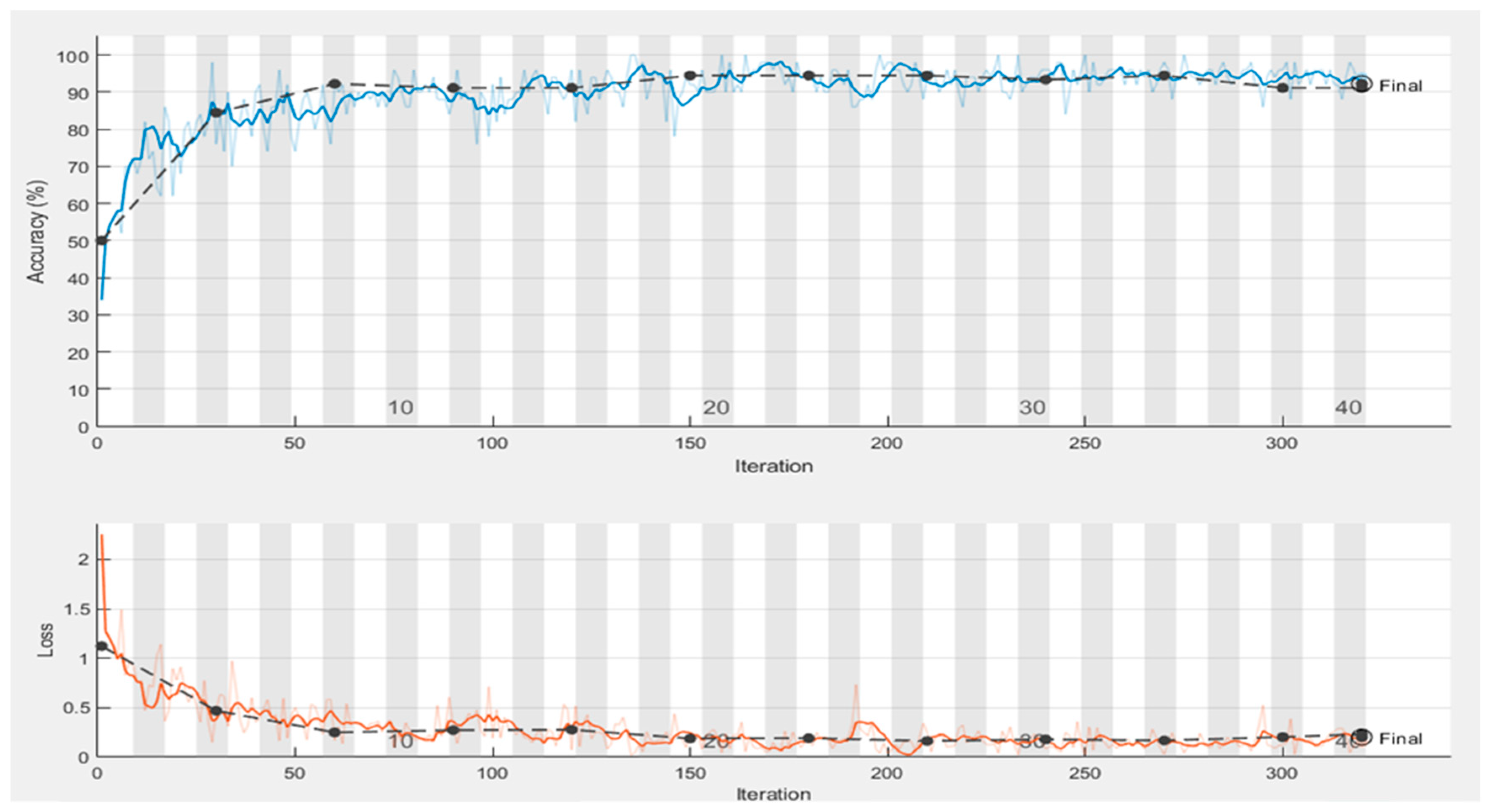
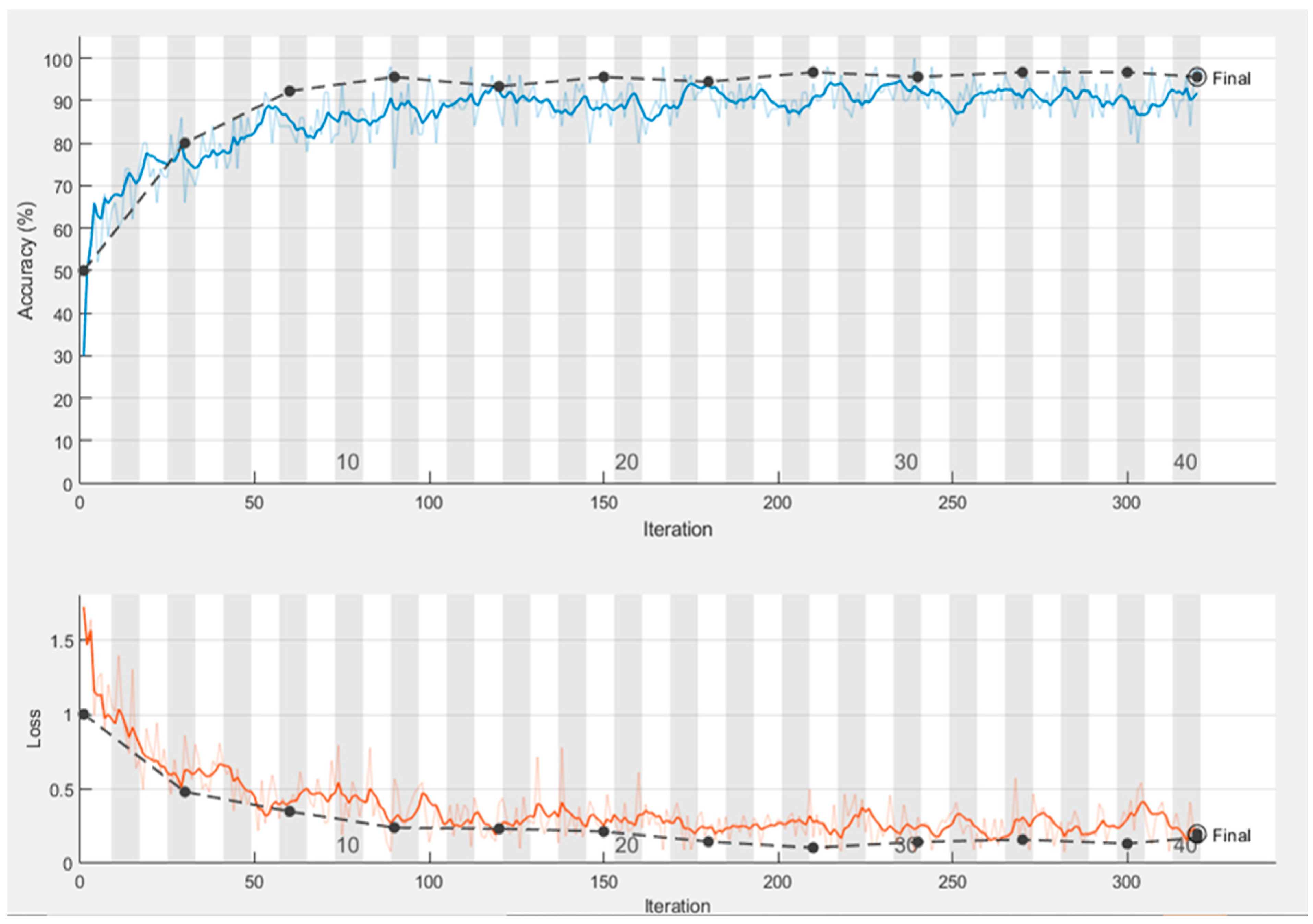
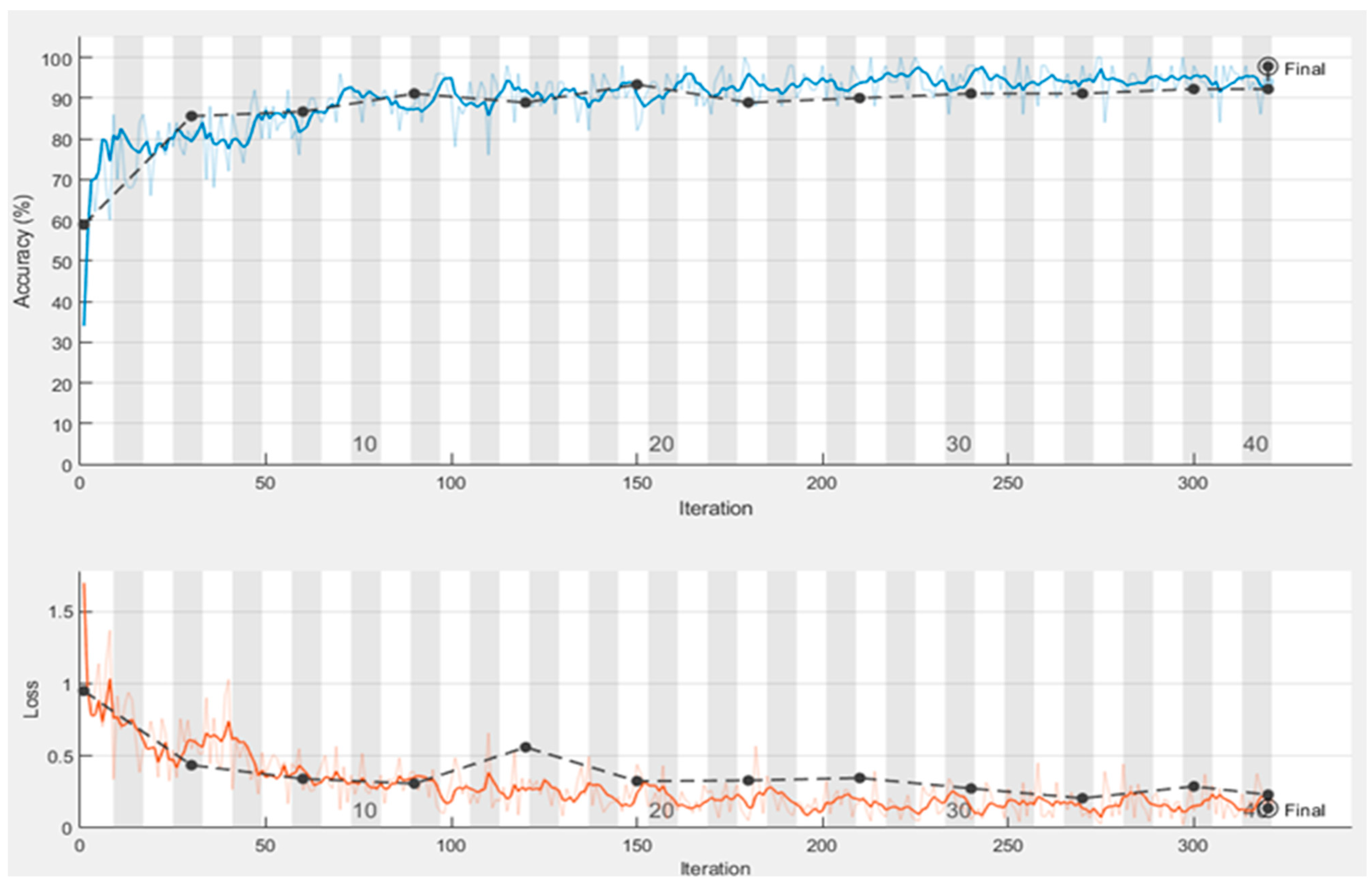

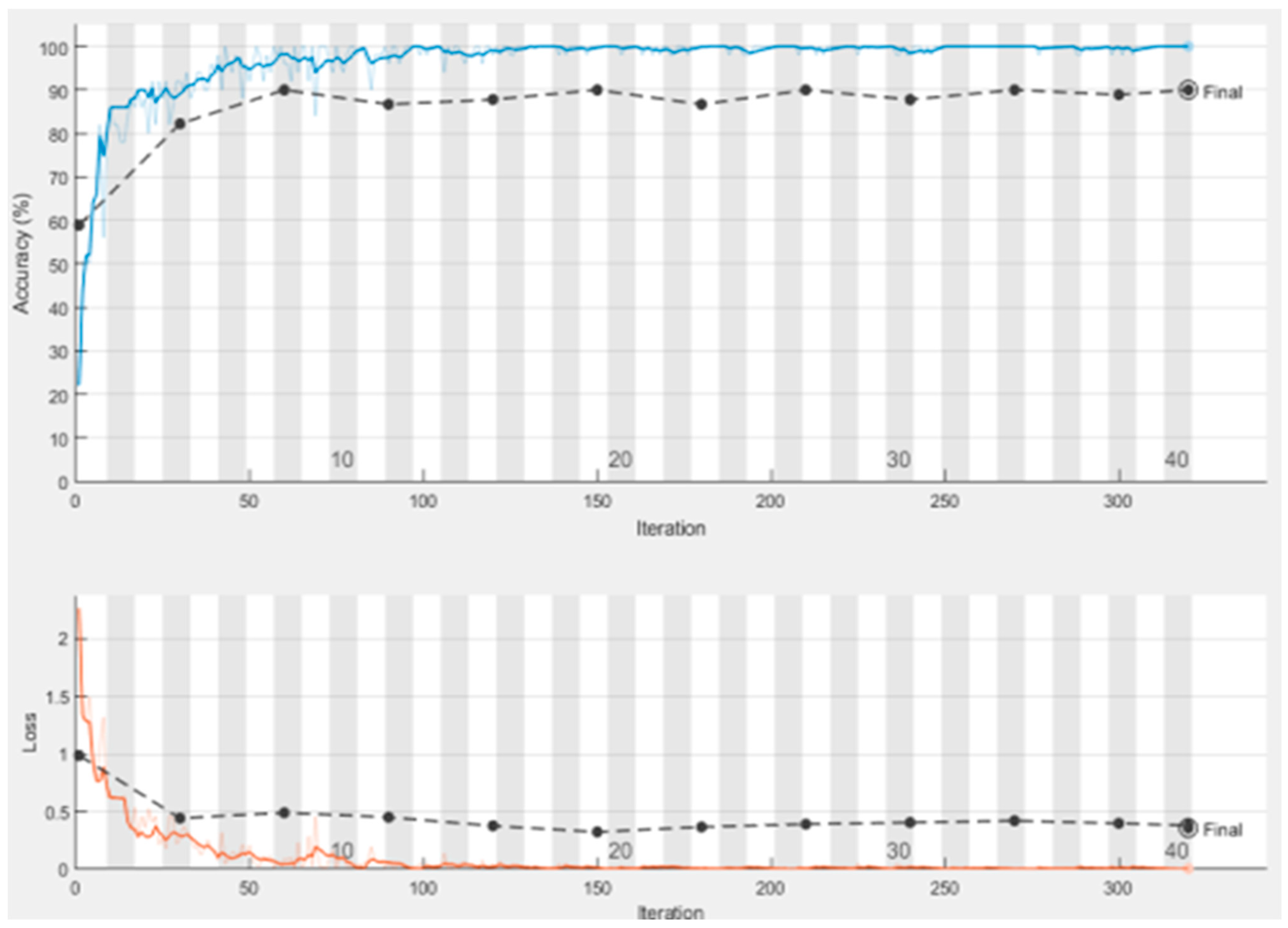
| Epoch | Iteration | Time Elapsed | Mini-Batch Accuracy | Validation Accuracy | Mini-Batch Loss | Validation Loss |
|---|---|---|---|---|---|---|
| 1 | 1 | 0:00:43 | 34.00% | 50.00% | 2.2525 | 1.1228 |
| 4 | 30 | 0:03:12 | 76.00% | 84.44% | 0.4858 | 0.4692 |
| 7 | 50 | 0:04:46 | 74.00% | 92.22% | 0.5023 | 0.2504 |
| 8 | 60 | 0:05:35 | 92.00% | 92.22% | 0.1682 | 0.2504 |
| 12 | 90 | 0:08:02 | 88.00% | 91.11% | 0.3048 | 0.2727 |
| 13 | 100 | 0:08:49 | 92.00% | 91.11% | 0.1922 | 0.2776 |
| 15 | 120 | 0:10:20 | 96.00% | 94.44% | 0.0737 | 0.1889 |
| 19 | 150 | 0:12:36 | 92.00% | 94.44% | 0.2408 | 0.1923 |
| 23 | 180 | 0:14:38 | 96.00% | 94.44% | 0.1399 | 0.1923 |
| 25 | 200 | 0:15:58 | 98.00% | 94.44% | 0.042 | 0.1641 |
| 27 | 210 | 0:16:40 | 96.00% | 93.33% | 0.0835 | 0.1793 |
| 30 | 240 | 0:18:40 | 94.00% | 94.44% | 0.0284 | 0.1692 |
| 32 | 250 | 0:19:19 | 100.00% | 94.44% | 0.0299 | 0.1692 |
| 34 | 270 | 0:20:38 | 98.00% | 91.11% | 0.1247 | 0.2036 |
| 38 | 300 | 0:22:38 | 94.00% | 91.11% | 0.1485 | 0.2319 |
| 40 | 320 | 0:24:01 | 94.00% | 91.11% | 0.1485 | 0.2319 |
| Test accuracy 0.9 | ||||||
| Epoch | Iteration | Time Elapsed | Mini-Batch Accuracy | Validation Accuracy | Mini-Batch Loss | Validation Loss |
|---|---|---|---|---|---|---|
| 1 | 1 | 0:00:48 | 28.00% | 60.00% | 1.7889 | 0.8766 |
| 4 | 30 | 0:03:12 | 78.00% | 81.11% | 0.5076 | 0.5075 |
| 7 | 50 | 0:04:36 | 88.00% | 87.78% | 0.2883 | 0.5753 |
| 8 | 60 | 0:05:20 | 86.00% | 87.78% | 0.394 | 0.5753 |
| 12 | 90 | 0:07:27 | 70.00% | 90.00% | 0.7554 | 0.4131 |
| 13 | 100 | 0:08:08 | 90.00% | 91.11% | 0.19 | 0.4551 |
| 15 | 120 | 0:09:30 | 98.00% | 91.11% | 0.0656 | 0.396 |
| 19 | 150 | 0:11:27 | 86.00% | 92.22% | 0.2381 | 0.4348 |
| 23 | 180 | 0:13:23 | 88.00% | 88.89% | 0.3693 | 0.5112 |
| 25 | 200 | 0:14:38 | 100.00% | 88.89% | 0.0447 | 0.5112 |
| 27 | 210 | 0:15:19 | 94.00% | 91.11% | 0.2933 | 0.4465 |
| 30 | 240 | 0:17:22 | 92.00% | 91.11% | 0.1948 | 0.4905 |
| 32 | 250 | 0:18:00 | 94.00% | 90.00% | 0.2031 | 0.4905 |
| 34 | 270 | 0:19:19 | 96.00% | 91.11% | 0.2284 | 0.4856 |
| 38 | 300 | 0:21:15 | 96.00% | 91.11% | 0.1672 | 0.5162 |
| 40 | 320 | 0:22:34 | 96.00% | 91.11% | 0.1672 | 0.5162 |
| Test accuracy 0.9444 | ||||||
| Epoch | Iteration | Time Elapsed | Mini-Batch Accuracy | Validation Accuracy | Mini-Batch Loss | Validation Loss |
|---|---|---|---|---|---|---|
| 1 | 1 | 0:00:28 | 38.00% | 56.67% | 1.9619 | 1.1099 |
| 4 | 30 | 0:02:53 | 86.00% | 86.67% | 0.3858 | 0.3753 |
| 7 | 50 | 0:04:10 | 74.00% | 84.44% | 0.7992 | 0.433 |
| 8 | 60 | 0:04:52 | 90.00% | 86.67% | 0.4536 | 0.3306 |
| 12 | 90 | 0:06:51 | 92.00% | 86.67% | 0.138 | 0.3306 |
| 13 | 100 | 0:07:29 | 74.00% | 84.44% | 1.0104 | 0.3789 |
| 15 | 120 | 0:08:47 | 90.00% | 91.11% | 0.2837 | 0.2539 |
| 19 | 150 | 0:10:47 | 74.00% | 86.67% | 0.1437 | 0.2942 |
| 23 | 180 | 0:12:39 | 92.00% | 88.89% | 0.2929 | 0.229 |
| 25 | 200 | 0:14:40 | 92.00% | 90.00% | 0.2897 | 0.2327 |
| 27 | 210 | 0:16:43 | 94.00% | 90.00% | 0.145 | 0.2327 |
| 30 | 240 | 0:16:43 | 96.00% | 94.44% | 0.1825 | 0.1961 |
| 32 | 250 | 0:18:13 | 92.00% | 94.44% | 0.1825 | 0.1961 |
| 34 | 270 | 0:18:49 | 92.00% | 93.33% | 0.1449 | 0.2185 |
| 38 | 300 | 0:21:13 | 98.00% | 92.22% | 0.0466 | 0.209 |
| 40 | 320 | 0:23:13 | 98.00% | 92.22% | 0.0466 | 0.209 |
| Test accuracy 0.9667 | ||||||
| Epoch | Iteration | Time Elapsed | Mini-Batch Accuracy | Validation Accuracy | Mini-Batch Loss | Validation Loss |
|---|---|---|---|---|---|---|
| 1 | 1 | 00:00:38 | 38.00% | 43.33% | 1.4999 | 1.1412 |
| 4 | 30 | 00:02:46 | 70.00% | 76.67% | 0.6124 | 0.4623 |
| 7 | 50 | 00:04:09 | 88.00% | 87.78% | 0.387 | 0.3339 |
| 8 | 60 | 00:04:53 | 92.00% | 87.78% | 0.2985 | 0.3339 |
| 12 | 90 | 00:06:58 | 92.00% | 90.00% | 0.2654 | 0.2376 |
| 13 | 100 | 00:07:38 | 90.00% | 87.78% | 0.3719 | 0.3521 |
| 15 | 120 | 00:09:05 | 96.00% | 90.00% | 0.1376 | 0.2287 |
| 19 | 150 | 00:11:10 | 94.00% | 91.11% | 0.2833 | 0.2157 |
| 23 | 180 | 00:13:17 | 92.00% | 92.22% | 0.1218 | 0.1865 |
| 25 | 200 | 00:14:38 | 92.00% | 88.89% | 0.2385 | 0.229 |
| 27 | 210 | 00:15:28 | 96.00% | 91.11% | 0.2519 | 0.2152 |
| 30 | 240 | 00:17:28 | 96.00% | 91.11% | 0.1583 | 0.2295 |
| 32 | 250 | 00:18:06 | 92.00% | 91.11% | 0.0896 | 0.2295 |
| 34 | 270 | 00:19:25 | 92.00% | 93.33% | 0.1573 | 0.205 |
| 38 | 300 | 00:21:22 | 90.00% | 87.78% | 0.2623 | 0.2446 |
| 40 | 320 | 00:22:40 | 92.00% | 87.78% | 0.1778 | 0.2446 |
| Test accuracy 0.8889 | ||||||
| Epoch | Iteration | Time Elapsed (hh:mm:ss) | Mini-Batch Accuracy | Validation Accuracy | Mini-Batch Loss | Validation Loss |
|---|---|---|---|---|---|---|
| 1 | 1 | 00:00:22 | 34.00% | 58.89% | 1.7006 | 0.9504 |
| 4 | 30 | 00:02:19 | 80.00% | 85.56% | 0.5491 | 0.4368 |
| 7 | 50 | 00:04:00 | 88.00% | 88.89% | 0.367 | 0.3421 |
| 8 | 60 | 00:04:18 | 88.00% | 86.67% | 0.2642 | 0.3088 |
| 12 | 90 | 00:06:18 | 88.00% | 91.11% | 0.388 | 0.5609 |
| 13 | 100 | 00:07:06 | 90.00% | 88.89% | 0.2456 | 0.3295 |
| 15 | 120 | 00:08:15 | 92.00% | 93.33% | 0.4419 | 0.3259 |
| 19 | 150 | 00:10:14 | 94.00% | 88.89% | 0.0522 | 0.3477 |
| 23 | 180 | 00:12:13 | 92.00% | 90.00% | 0.1239 | 0.3477 |
| 25 | 200 | 00:13:41 | 94.00% | 90.00% | 0.0715 | 0.2749 |
| 27 | 210 | 00:14:12 | 94.00% | 91.11% | 0.1271 | 0.2749 |
| 30 | 240 | 00:16:48 | 90.00% | 91.11% | 0.2741 | 0.2749 |
| 32 | 250 | 00:17:18 | 90.00% | 91.11% | 0.2714 | 0.2749 |
| 35 | 270 | 00:19:10 | 98.00% | 92.22% | 0.118 | 0.2396 |
| 38 | 300 | 00:20:11 | 98.00% | 92.22% | 0.185 | 0.2316 |
| 40 | 320 | 00:21:34 | 94.00% | 92.22% | 0.185 | 0.2316 |
| Test accuracy 0.9333 | ||||||
| Trained with the Original Images | ||||||
|---|---|---|---|---|---|---|
| Model | Precision | Recall | F1-Score | Testing Accuracy | Training Accuracy | Validation Accuracy |
| Proposed model | 0.90 | 0.90 | 0.90 | 90.0% | 94.0% | 92.2% |
| InceptionV3 | 0.73 | 0.73 | 0.73 | 73.3% | 82.0% | 75.6% |
| Xception | 0.89 | 0.89 | 0.89 | 88.9% | 92.0% | 86.7% |
| Trained with Preprocessed and Segmented Images | ||||||
| Model | Precision | Recall | F1-Score | Testing Accuracy | Training Accuracy | Validation Accuracy |
| Proposed model | 0.94 | 0.94 | 0.94 | 94.4% | 96.0% | 95.6% |
| InceptionV3 | 0.82 | 0.82 | 0.82 | 82.2% | 92.2% | 86.7% |
| Xception | 0.90 | 0.90 | 0.90 | 90.0% | 92.0% | 90.0% |
| Trained with Augmented Images | ||||||
| Model | Precision | Recall | F1-Score | Testing Accuracy | Training Accuracy | Validation Accuracy |
| Proposed model | 0.97 | 0.97 | 0.97 | 96.7% | 98.0% | 97.7% |
| InceptionV3 | 0.89 | 0.89 | 0.89 | 88.9% | 92.0% | 88.9% |
| Xception | 0.93 | 0.93 | 0.93 | 93.3% | 94.0% | 90.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nibret, E.A.; Mequanenit, A.M.; Ayalew, A.M.; Kusrini, K.; Martínez-Béjar, R. Sesame Plant Disease Classification Using Deep Convolution Neural Networks. Appl. Sci. 2025, 15, 2124. https://doi.org/10.3390/app15042124
Nibret EA, Mequanenit AM, Ayalew AM, Kusrini K, Martínez-Béjar R. Sesame Plant Disease Classification Using Deep Convolution Neural Networks. Applied Sciences. 2025; 15(4):2124. https://doi.org/10.3390/app15042124
Chicago/Turabian StyleNibret, Eyerusalem Alebachew, Azanu Mirolgn Mequanenit, Aleka Melese Ayalew, Kusrini Kusrini, and Rodrigo Martínez-Béjar. 2025. "Sesame Plant Disease Classification Using Deep Convolution Neural Networks" Applied Sciences 15, no. 4: 2124. https://doi.org/10.3390/app15042124
APA StyleNibret, E. A., Mequanenit, A. M., Ayalew, A. M., Kusrini, K., & Martínez-Béjar, R. (2025). Sesame Plant Disease Classification Using Deep Convolution Neural Networks. Applied Sciences, 15(4), 2124. https://doi.org/10.3390/app15042124







