Featured Application
This research exposes a practical application for using coloured phenolics as chemical fingerprints for authentication and characterization of red wines. A relatively fast and cost-effective method using high-performance liquid chromatography coupled with a diode array detector (HPLC-DAD) combined with multivariate statistical techniques is presented for obtaining origin discrimination of red wines. This approach enhances quality control and verification within the wine industry and can be extended to authenticate other plant-based derived products.
Abstract
Anthocyanins are important bioactive compounds crucial for the sensory characteristics of red wines. Anthocyanin profiles of 205 monovarietal red wines from the Canary Islands were investigated. Eleven anthocyanins were identified and determined using HPLC-DAD. Anthocyanin concentrations of red wines produced in Canary Islands fell within the usual range observed in red wines from other regions. Red wines elaborated with international grape cultivars presented, in general, higher mean concentrations than those elaborated using autochthonous cultivars. The influence of grape cultivar, production island, denomination of origin, and wine aging on the anthocyanin concentration was studied, leading to the deduction that aging was the parameter with the highest influence. A high number of significant correlations between the anthocyanins determined were found out supporting a common organic synthetic way for these coloured phenolics. Application of multivariate analysis techniques, such as principal component analysis and discriminant analysis, tended to classify the red wine samples according to grape cultivar, geographical production areas, and aging. This study could contribute to the quality control and verification within the wine industry, which is an interesting tool in the prevention of fraud and for increasing consumer confidence.
1. Introduction
Phenolic compounds are secondary metabolites produced by plants, many of which have been identified as bioactive substances with recognized antioxidant properties, which implies a high potential for applications in the health field [1]. Their natural occurrence has been used for statistical differentiation in food products, according to cultivar in plants [2] or bee pollen [3], as well as according to geographical origin in olive oil [4] or tomato derivatives [5].
Anthocyanins are a class of phenolic compounds classified as pigments and responsible for the red, purple, and blue colours in many fruits and vegetables [6]. In grapevines (Vitis vinifera), anthocyanins are predominantly located in the berry skins and play a crucial role for determining the colour and quality of red wines. These compounds contribute to the sensory attributes of red wines including hue, intensity, and colour stability [7].
The biosynthesis of anthocyanins in grape berries involves the flavonoid pathway, originating from the amino acid phenylalanine through the action of phenylalanine ammonia-lyase [6]. Key enzymes in this pathway include chalcone synthase, chalcone isomerase, flavanone 3-hydroxylase, dihydroflavonol 4-reductase, anthocyanidin synthase, and uridine diphosphate-glucose flavonoid 3-o-glucosyltransferase (UFGT) [8]. The final glycosylation step catalysed by UFGT is essential for anthocyanin stability and accumulation in grape skins [9].
Anthocyanin composition in grapes is influenced by genetic factors, environmental conditions, and viticultural practices [10]. Different grape varieties exhibit distinct anthocyanin profiles, characterized by variations in the types and proportions of anthocyanin derivatives such as malvidin, delphinidin, cyanidin, peonidin, and petunidin glucosides [11]. Environmental factors like temperature, light exposure, soil composition, and water availability can significantly affect synthesis and accumulation of anthocyanins [12]. Excessively high temperatures generally lead to pigment degradation [13]. An increased solar radiation (light exposure) enhances anthocyanin biosynthesis, whereas excessive shading can reduce their concentration in grape skins [14]. Similarly, moderate water deficit conditions have been associated with increased anthocyanin content due to the activation of stress-related metabolic pathways [15].
In winemaking, the anthocyanin extraction from grape skins into the must is a critical process that determines the colour intensity and hue of the resulting wine. Winemaking techniques such as maceration time, fermentation temperature, and the use of enzymes can have an influence on anthocyanin extraction and stability [7]. Additionally, anthocyanins can undergo various reactions during wine aging, leading to the formation of more stable pigments like polymeric anthocyanins, which contribute to the long-term colour stability of red wines [16].
There is an increasing need for wine authentication in the market, driving research to propose more effective chemical markers. The Canary Islands wine sector represents an essential economic activity, contributing to local development through employment and tourism. However, like many wine-producing regions, it faces challenges related to authenticity and fraud, which can compromise consumer trust and the value of its Denominations of Origin (DOs). Ensuring wine authenticity through reliable analytical methods is crucial for safeguarding their identity. Authentication strategies, such as anthocyanin profiling, provide a robust tool for verifying wine origin and strengthening quality control systems, ultimately benefiting both producers and consumers. Phenolic profiles [17] have been particularly used for differentiating wines based on grape cultivars and regions of origin. Advanced analytical techniques, such as mass spectrometry [18] or nuclear magnetic resonance [19], have been employed for phenolic analysis. However, these analytical techniques have high costs, with the result that high-performance liquid chromatography (HPLC) coupled with a diode array detector (DAD) was a preferred method due to its low cost-effectiveness ratio in addition to its precision, accessibility, and robustness [20]. The analysis of anthocyanin fingerprints using HPLC-DAD is probably a reliable approach for linking wine characteristics to grape cultivars and winemaking practices [21].
In this study, HPLC-DAD was applied as a useful tool for contributing to wine authenticity and enhancing quality control within the wine industry. To the best of our knowledge, this is the first study strictly focused on the anthocyanin composition of monovarietal red wines from the Canary Islands (Spain), including a substantial number of samples from all the producer islands. The influence of grape cultivar, production island, Denomination of Origin (DO), and wine aging on anthocyanin profiles was considered. Correlation studies were applied to discover relationships between the analysed anthocyanins. Multivariate analysis techniques were applied to attempt to classify the monovarietal red wines according to grape cultivar, island of origin, DO, and wine aging. This research represents a comprehensive assessment of monovarietal red wines from all major wine-producing islands of the archipelago. Unlike blended wines, monovarietal wines provide a clearer fingerprint of anthocyanin composition associated with specific grape cultivars, offering deeper insights into varietal differentiation and regional authenticity.
2. Materials and Methods
2.1. Wine Samples
A total of 205 bottled monovarietal red wines were selected from the Canary Islands (Spain) for this study. Red wines were produced from ten grape cultivars such as Listán Negro (LN, n = 93); Baboso (B, n = 30); Vijariego (V, n = 17); Negramoll (N, n = 13); Listán Prieto (LP n = 14); Syrah (S, n = 12); Tintilla (T, n = 9); Castellana (C, n = 7); Rubí Cabernet (R, n = 5); and Merlot (M, n = 5). The distribution of the red wine samples according to the six islands of precedence was designed to proportionally represent the red wine production in each area, considering that certain islands mainly produce white wines. As a result, Tenerife Island was the most represented in the set of red wines (n = 130). This island comprises five Denominations of Origin (DO): Abona (A, n = 46); Tacoronte-Acentejo (TA, n = 45); Valle de la Orotava (O, n = 27); Ycoden-Daute-Isora (Y, n = 18); and Valle de Güímar (G, n = 10). The remaining red wines came from the other islands, from more to less production presented as El Hierro (H, n = 18); La Palma (LP, n = 15); Gran Canaria (GC, n = 13); Lanzarote (LZ, n = 7); and La Gomera (GO, n = 6). The red wine samples were collected in eight vintages classified in three groups according to the aging: young wines (≤1 year, n = 125); short aged (1–3 years, n = 73); and aged wines (≥3 years, n = 7).
2.2. Analytical Methods
HPLC was utilized to separate phenolic compounds using a Waters 2690 Separation Module equipped with a Waters 996 DAD and interfaced with a Waters data-processing station running Millennium32 software (version 3.0.1). Chromatograms were recorded at 520 nm wavelength.
Red wine samples were previously filtered through a 0.45 μm membrane filter, and then 15 μL aliquots were injected onto a Nova-Pak C18 reversed-phase column (3.9 × 150 mm, 4 μm particle size; Waters) maintained at 30 °C. The chromatographic conditions were adapted from Ibern-Gómez et al. [22]. Solvent A was Milli-Q grade water, and solvent B was gradient-grade acetonitrile (Sigma-Aldrich, St. Louis, MO, USA); both solvents were acidified with 0.2% trifluoroacetic acid (spectrophotometric grade, ≥99%, Sigma-Aldrich). The flow rate was set at 1.5 mL/min. The linear gradient for solvent B was programmed as follows: 0 min, 0%; 2 min, 2%; 8 min, 8%; 15 min, 15%; 20 min, 23%; and returning to 0% at 25 min.
Detection was performed using the DAD, with peak identification based on their retention times and ultraviolet and visible spectra. Anthocyanin compounds were identified by direct comparison with commercial standards when they were available. Additional confirmation was achieved by spiking red wine samples with the available phenolic standards. Retention times obtained with the chromatographic conditions used by us are detailed in Table S1 from Supplementary Materials. Other compounds were tentatively identified by comparing their relative retention times and spectral data with those published under similar chromatographic conditions [23,24,25].
Anthocyanins were detected at 520 nm and quantified using oenin as the standard (Sigma-Aldrich), expressed as mg of oenin equivalents/L. The calibration curve was constructed over the concentration range observed in the samples and was generated by plotting peak area (absorbance) vs. concentration (mg/L). Standards had a linear response within the studied concentration ranges, with correlation coefficient (r) always ≥0.999. The anthocyanin compounds identified (abbreviated name was included between brackets) were the following: Delphinidin-3-glucoside (D-3-glc); Cyanidin-3-glucoside (C-3-glc); Petunidin-3-glucoside (Pt-3-glc); Peonidin-3-glucoside (Pe-3-glc); Malvidin-3-glucoside (M-3-glc, oenin); Cyanidin-(6-acetyl)-3-glucoside (C-6-ac-3-glc); Petunidin-(6-acetyl)-3-glucoside (Pt-6-ac-3-glc); Peonidin-(6-acetyl)-3-glucoside (Pe-6-ac-3-glc); Malvidin-(6-acetyl)-3-glucoside (M-6-ac-3-glc); Peonidin-(6-coumaroyl)-3-glucoside (Pe-6-co-3-glc); Malvidin-(6-coumaroyl)-3-glucoside (M-6-co-3-glc).
Using these eleven individual anthocyanin compounds, several groups with different anthocyanin derivatives were calculated as follows:
Glucosides without oenin (Glc/oenin) = D-3-glc + C-3-glc + Pt-3-glc + Pe-3-glc
Glucosides with oenin (Glc) = Glc/oenin + M-3-glc (oenin)
Acetylated Anthocyanins (Acet) = C-6-ac-3-glc + Pt-6-ac-3-glc + Pe-6-ac-3-glc + M-6-ac-3-glc
Coumaroyled Anthocyanins (Cou) = Pe-6-co-3-glc + M-6-co-3-glc
Total Anthocyanins (Tot) = Glc + Acet + Cou
Anthocyanin glucosides were grouped in two distinct ways, as described by two different equations: (1) Excluding malvidin-3-O-glucoside (oenin), the most abundant anthocyanin in wine; this approach minimizes the effect of its predominance overshadowing observations related to less abundant pigments; and (2) including all individual glucosides in order to know the behaviour of the overall glucosides together.
2.3. Statistical Analysis
All statistical analyses were conducted using SPSS version 18. One-way ANOVA tests were performed considering a p-value threshold ≤ 0.05 to establish statistical significance. Duncan’s multiple range test was utilized for post hoc comparisons to pinpoint specific group differences. Bivariate correlations were examined using Pearson’s correlation coefficient to highlight positive or inverse associations between the anthocyanins analysed.
Principal Component Analysis (PCA) was carried out to reduce data dimensionality and identify underlying patterns by transforming the original variables into a new set of uncorrelated variables called principal components. This method facilitated the visualization of data structure and helped in detecting clusters and trends among the red wines analysed.
Linear Discriminant Analysis (LDA), a stepwise method, was employed to identify the most significant anthocyanins contributing to group separation. Additionally, analyses incorporating all variables were conducted to explore potential interactions and relationships among factors. Probabilities were calculated based on the sample size of each population and using an intra-group variance matrix. This approach enabled the evaluation of group differences and the identification of variables influencing classification. Moreover, cross-validation analyses were performed for both methods, stepwise and using all variables.
3. Results
For a better understanding, Section 3 was divided into the following three subsections: univariate analysis, correlation analysis and multivariate analysis. The cultivars, geographical origin, and anthocyanins were abbreviated according to previous indications from Section 2.
3.1. Univariate Analysis
Anthocyanin concentrations (mean and standard deviation), variance analysis (One-way ANOVA), and Duncan’s test for comparing mean concentration values were calculated for red wines grouped according to qualitative variables such as grape cultivar, geographical origin, and aging.
3.1.1. Cultivar Analysis
The anthocyanin profile according to cultivar is detailed in Table 1. As expected, M-3-glc (oenin) was the anthocyanin with the highest mean concentration, generally followed by Pe-3-glc or Pe-3-glc, depending on the grape cultivar.

Table 1.
Mean and standard deviation of anthocyanin concentration (mg/L) according to the grape cultivar.
The highest mean Glc concentrations were observed in C, LN, and R cultivars, with significant differences (p < 0.05) when their mean concentrations were compared with the mean concentration of V cultivar. Moreover, if the Glc was considered without M-3-gluc or oenin (the major anthocyanin glucoside), one can deduce that the R, C, and S cultivar had higher mean concentrations than T, V, and N cultivars. However, the behaviour was different for each individual glucoside. Thus, C-3-glc and Pe-3-glc did not show significant differences in the mean concentrations obtained among the cultivars considered, while the R, followed by C and S cultivars, presented the highest mean D-3-glc, Pt-3-glc, and M-3-glc concentrations, with significant differences compared with the mean concentrations observed in many cultivars. Analogously, R and S cultivars had the highest mean Acet concentrations, with significant differences with respect to those mean concentrations obtained in the rest of the cultivars, except for M and T cultivars. C-6-ac-3-glc permitted the differentiation of two groups of grape cultivars. A group formed by V, LN, and N had lower concentrations (≤2 mg/L) than the rest of the cultivars. For the other acetylated derivatives (Pe-6-ac-3-glc; Pt-6-ac-3-glc; M-6-ac-3-glc), the foreign cultivars (S, R, and M) and T cultivars presented, in general, higher mean concentrations than those mean concentrations found in the red wines from the rest of the grape cultivars. The same behaviour was observed in the individual coumaryl anthocyanin derivatives. Further, when the individual coumaryl derivatives were considered together (Cou), the V cultivar had higher concentrations than the mean concentrations found for red wines from LN, R, and C cultivars.
3.1.2. Geographical Origin
In relation to the geographical origin, two factors were considered: island of precedence and DO into the Tenerife Island.
Island of Precedence
Some interesting differences were observed when the distribution of anthocyanins concentration was studied as a function of the precedence island (Table 2). The highest mean concentration of Glc/oenin (without M-3-glc) was found in the red wines from Tenerife, followed by those from Lanzarote island. The red wines produced in both islands had clearly higher Glc/oenin than the mean concentrations found in the rest of the islands considered, with significant differences for many of them. Similar results were observed for all the individual glucosides, D-3-glc, Pe-3-glc, Pt-3-glc, and M-3-glc, except for C-3-glc, which did not show significant differences among the mean concentrations obtained by precedence island. When the concentrations of total glucosides included M-3-gluc, the red wines from Lanzarote, Tenerife, and La Gomera islands had higher (p < 0.05) mean concentrations than the mean concentrations obtained in the red wines produced in the rest of islands (La Palma, Gran Canaria, and El Hierro). The total acetylated anthocyanins, C-6-ac-3-glc, and M-6-ac-3-glc did not present significant differences among their mean concentrations according to the island of production. Red wines from Lanzarote Island showed the highest mean Pe-6-ac-3-glc and Pt-6-ac-3-glc concentrations, with significant differences in relation to the mean concentrations obtained for red wines of La Gomera and La Palma, as well as La Gomera islands, respectively.

Table 2.
Mean and standard deviation (between brackets) of anthocyanin concentration (mg/L) according to the precedence island.
Considering the individual coumaroyl derivatives, the mean concentration of M-6-cou-3-glc found in red wines from Tenerife Island was higher than the mean concentrations obtained in the rest of islands, with significant differences only with the red wines from La Palma, while Pe-6-cou-3-glc did not show significant differences according to the precedence island. In contrast, the concentration of Cou in red wines of El Hierro was interestingly higher than the mean concentrations obtained in the red wines from the rest of the islands.
Mean Tot concentrations according to the island of production agrees with the results obtained for the groups of anthocyanin compounds studied, except Cou.
Denomination of Origin into Tenerife Island
Tenerife island included 5 DOs which have different edaphoclimatic characteristics which could affect the anthocyanin concentrations of the red wines produced. Results (mean and standard deviation and variance analysis) according to the DO into the Tenerife Island are included in Table S2. Considering the individual anthocyanin glucosides, the red wines from DO G, followed by the DO O, had higher mean D-3-glc and C-3-glc concentrations than those mean concentrations obtained in the other DOs. Significant differences were found when the mean C-3-glc and D-3-glc concentrations in red wines produced from DO G were compared with those mean concentrations obtained for the other DOs, as well as for DO TA and DO Y, respectively. No significant differences were found among the mean Pe-3-glc, Pt-3-glc, and M-3-glc concentrations found in the red wines according to the DO. Similar to individual glucosides, the highest mean Glc/oenin concentration was observed in red wines from DO G, with significant differences in relation to those concentrations found for DO TA and DO Y. While a different behaviour was observed for Glc, red wines from DO O had the highest mean Glc concentrations, with significant differences with respect to red wines from DO A and DO O.
All the acetylated derivatives behaved in a similar way according to the DOs; their mean concentrations in red wines were ordinated in accordance with the following sequence: DO G > DO A > DO O > DO TA > DO Y, observing two groups statistically well-differentiated in most cases. Thus, red wines produced in DO G and DO A had higher mean acetylated derivative concentrations that those red wines from DO O, DO TA, and DO Y. The coumaroyl derivatives (M-6-cou-3-glc, Pe-6-cou-3-glc, and Cou) had a performance resemblance to most of the previous derivatives. One can see how red wines produced in DO G and DO A showed the largest mean concentrations, with many significant differences when compared with the mean concentrations in the other DOs. Mean Tot concentrations according to the DOs of Tenerife agreed in general with the results obtained for the groups of compounds previously studied.
Figure 1 shows details about the main anthocyanin groups, categorized by their chemical structure and DO. The mean Glc concentrations were higher in red wines from DO O, DO TA, and DO Y compared to those from the south of the island of Tenerife, DO G, and DO A. Significant differences were only observed between DO O and southern Tenerife DOs (G and A). Interestingly, the mean Acet concentrations show the opposite trend, with significantly higher values in DO A and DO G compared to all the northern Tenerife DOs (O, TA, and Y). Regarding Cou concentrations, the values are similar across most DOs, except for DO G, which shows significant differences compared to DO O.
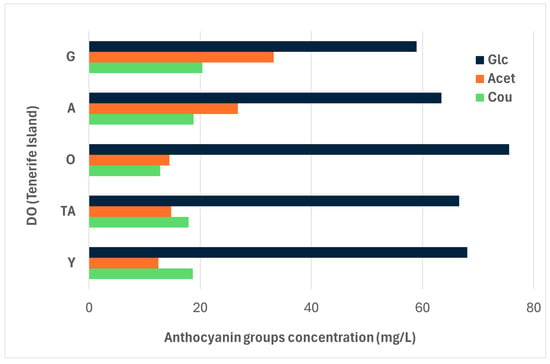
Figure 1.
Groups of anthocyanin compounds (mg/L) in red wines according to DO in Tenerife. * Denomination of Origin (Tenerife Island) abbreviations—A: Abona; TA: Tacoronte-Acentejo); O: Valle de la Orotava; Y: Ycoden-Daute-Isora; G: Valle de Güímar. ** Anthocyanin compounds abbreviations—Glc: Glucosides with oenin; Acet: Acetylated Anthocyanins; Cou: Coumaroyled Anthocyanins.
3.1.3. Aging Analysis
As can be observed from Table S3 (Supplementary Materials), mean concentration of Tot, Glc, Glc/oenin, and individual glucosides (D-3-glc, Pe-3-glc, Pe-3-gl, M-3-glc), clearly decreased with aging; in particular, the mean concentration of red wines > 3 years of aging was markedly lower than the rest of the red wines. Coumaroil anthocyanin derivatives had an opposite behaviour; the mean concentrations significantly increased with aging. It is worth highlighting that the mean concentrations for individual coumaroyl derivatives had no significant differences. Neither the four acetyl derivatives (6-ac-3-glc) presented clear tendencies of their mean values according to aging; in fact, Pe-6-ac-3-glc and M-6-ac-3-glc did not show significant differences among mean concentrations according to aging. Figure 2 shows the mean Tot, Glc, and Cou concentrations as a function of aging, where one can clearly see that the mean Tot and Glc concentrations significantly decrease with aging, while mean Cou concentration increases. Further, the mean Acet concentration has no clear tendency according to aging.
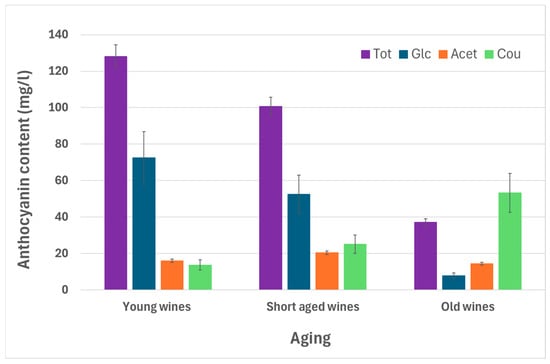
Figure 2.
Total and grouped anthocyanin derivatives mean concentrations according to aging. * Anthocyanin compounds abbreviations—Glc: Glucosides with oenin; Acet: Acetylated Anthocyanins; Cou: Coumaroyled Anthocyanins; Tot: Total Anthocyanins (sum of Glc + Acet + Cou based on individual data).
3.2. Correlation Study
The correlation analysis of the anthocyanins (11) analysed as well as the obtained parameters for calculation in the analysed red wines showed many significant relationships, as shown in Table 3. Most of relationships established among the anthocyanin compounds were highly significant (p < 0.001). No significant correlations were only found between C-3-glc with Pt-6-ace-3-glc, Peo-6-ace-3-glc, M-3-glc, Pe-6-cou-3-glc, M-6-cou-3-glc, and Acet; Pe-3-glc with Pe-6-cou-3-glc; and C-6-ace-3-glc and M-6-ace-3glc with Glc and Cou.

Table 3.
Correlations among the anthocyanin concentrations obtained in the red wine analyzed.
All the correlations were positive, except those including the total coumaroyl derivatives. In fact, most of anthocyanin glucosides demonstrated strong positive correlations between them, indicating that these compounds tend to increase their concentrations together. D-3-glc, Pe-3-glc, and M-3-glc showed highly significant correlations, with r > 0.88 for the three correlations. Moderate correlations (r = 0.64–0.67) with a lower significance were found between these three glucosides and Pe-3-glc, while weak correlations were found with C-3-glc. Figure 3 shows the highly significant correlation between D-3-glc and Pe-3-glc, which are non-major anthocyanin compounds. In this Figure, it can be observed how the cloud points representing the red wines are aligned, defining the equation from the corresponding regression line indicated in the Figure. In contrast, significant inverse correlations were observed between Glc and Cou compounds with individual and calculated anthocyanins. Between them, it could be emphasized the inverse correlation between Cou and Glc concentrations due to its high significance (r = −0.924) (Figure 4). In addition, D-3-glc (r = −0.393, p < 0.001), Pt-3-glc (r = −0.454, p < 0.001), Pe-3-glc (r = −0.386, p < 0.001), and M-3-glc (r = −0.485, p < 0.001) showed inverse relationships with Cou, indicating that when the concentration of these non-acylated anthocyanins increases, the concentration of Cou decreases.
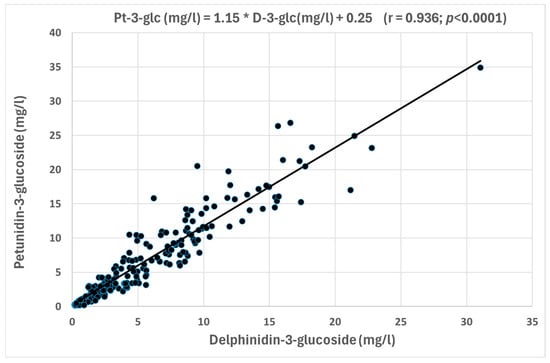
Figure 3.
Correlation between D-3-glc and Pe-3-glc.
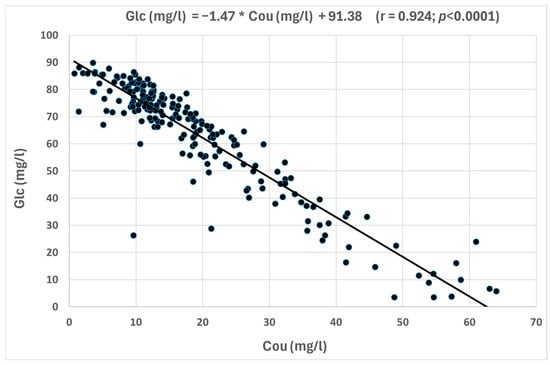
Figure 4.
Correlations between calculated Glc and Cou concentrations anthocyanin groups.
Therefore, the correlation matrix reveals that non-acylated anthocyanins are positive and closely interrelated, which indicates an increasing of their concentrations together. In contrast, coumaroyl anthocyanins tend to vary inversely with them.
3.3. Multivariate Analysis
3.3.1. Principal Compound Analysis
Principal Component Analysis (PCA) was applied to simplify our anthocyanin dataset, including all the data directly quantified and obtained by calculation parameters. To facilitate the interpretation of the results, a Varimax rotation was performed in order to minimize the number of variables influencing each factor. Three factors, accounting for 78% of the total variance, were selected since they had an eigenvalue ≥ 1. The first factor, explaining the highest percentage of variance (48.3%), is strongly associated with Glc, mostly Pe-3-glc, M-3-glc, D-3-glc, and Pe-3-glc, but very little with C-3-glc. The second factor, accounting for 22.1% of the total variance, is related with Acet and Cou anthocyanins but shows an inverse relationship with Glc. The third factor explains 7.6% of the total variance and is mostly related with C-3-glc. Figure 5 illustrates these findings using only the first two factors. The positioning of data points on these graphs helps us infer relationships between anthocyanin compounds; points that lie close together suggest related attributes.
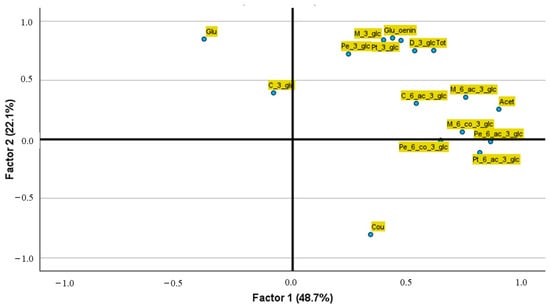
Figure 5.
Projections of anthocyanin compounds on the factor analysis in space defined by the first two factors. * Anthocyanin compounds abbreviations—D_3_glc: Delphinidin-3-glucoside; C_3_glc: Cyanidin-3-glucoside; Pt_3_glc: Petunidin-3-glucoside; Pe_3_glc: Peonidin-3-glucoside; M_3_glc: Malvidin-3-glucoside or oenin; C_6_ac_3_glc: Cyanidin-(6-acetyl)-3-glucoside; Pt_6_ac_3_glc: Petunidin-(6-acetyl)-3-glucoside; Pe_6_ac_3_glc: Peonidin-(6-acetyl)-3-glucoside; M_6_ac_3_glc: Malvidin-(6-acetyl)-3-glucoside; Pe_6_co_3_glc: Peonidin-(6-coumaroyl)-3-glucoside; M_6_co_3_glc: Malvidin-(6-coumaroyl)-3-glucoside; Glu/oenin: Glucosides without oenin; Glu: Glucosides with oenin; Acet: Acetylated Anthocyanins; Cou: Coumaroyled Anthocyanins; Tot: Total Anthocyanins.
When applying this factor analysis to differentiate cultivar or geographical origin, the visual distribution of the red wine samples showed no clear differentiation; nevertheless, there was a tendency to differentiate the aging groups, as it can be observed in Figure S1. The eldest wine samples displayed a negative score in Factor 3, with relatively positive minor results for Factor 2, while the youngest wines were mostly distributed with negative scores in Factor 2 and positive values of Factor 3.
3.3.2. Linear Discriminant Analysis
Table 4 presents the results of a Linear Discriminant Analysis (LDA) performed to classify the Canary red wines analysed based on several grouping variables such as grape cultivar, island of precedence, Tenerife DO, and wine aging. The results showed variable levels of correct classification, before and after cross-validation. Additionally, the table shows the number of discriminant functions, as well as the main anthocyanins associated with these functions, which were selected from the stepwise analysis, along with the most correlated anthocyanins influencing each function.

Table 4.
Results of the LDA to classify red wines according to the influencing factors considered.
After application of LDA to differentiate red wines according to grape cultivar using the directly determined and calculated anthocyanins, a correct classification of 75.6% was achieved, which dropped to 64.1% after cross-validation. Stepwise LDA resulted in a lower accuracy (55.6%, 50.7% after cross-validation), with Acet, M-3-glc, D-3-glc, and C-6-ac-3-glc identified as more relevant to the differentiation of red wines according to grape cultivars.
LDA applied for classifying the red wines according to the island of production showed a rate of 75.9% correct classification (69.2% after cross-validation) using all anthocyanin compounds considered. The stepwise LDA had lower correct classification (71.8%, 56.7% after cross-validation), with M-3-glc and Pt-3-glc, Tot and Glc, emerging as the most relevant variables to the differentiation according to the production island of red wines.
For red wines included under the Tenerife DO, the accuracy was lower than for other classification variables, with 66.7% correct classification (57.9% after cross-validation) using all variables. Stepwise LDA had a lower correct classification (43.9%, 40.5% after cross-validation), D-3-glc and Acet being the anthocyanins more important for separation.
Wine aging yielded the highest correct classification of red wines, achieving 86.3% correct classification (83.4% after cross-validation) using all variables. When stepwise LDA was applied, the correct classification decreased to 82.9% (80.3% after cross validation). Selected LDA variables such as Cou, Glc, and Pt-6-ac-3-glc were the more relevant variables related to aging chemical transformations.
Table 5 shows the percentage of correct classification for red wines on each grape cultivar used in their production after applying LDA. The diagonal values in the table represent the percentage of red wines correctly classified for each cultivar, while the off-diagonal values show the percentage of misclassifications. All the red wines produced from N, T, and R were correctly classified. Red wines produced from C and LN also showed a relatively high percentage of correct classification, 85.7% and 81.7%, respectively; only one (14.3%) red wine produced from Castellana cultivar was erroneously classified as LN. Further, the correct classification for the V cultivar was relatively high (76.5%); there were notable misclassifications as LN (17.6%). LP and M presented a reasonable classification rate, with a moderate accuracy (≥60%), while B and S were characterized by less appealing results. Thus, red wines from B cultivar had a rate of only 46.7% correct classification, with 20.0 and 16.7% of the red wines misclassified as red wines from LN and V, respectively. Further, it can be emphasized that 2 of the 5 (40%) red wines produced from the foreign cultivar M were erroneously classified as being from LN.

Table 5.
Percentage of correct classification (diagonal values) and misclassifications after application of LDA for differencing the red wines according to the grape cultivars used in its production.
4. Discussion
The current section, Section 4, was also divided into the same subsections: univariate analysis, correlation analysis, and multivariate analysis.
4.1. Univariate Analysis
The anthocyanin profile of red wines, as influenced by grape cultivar, geographical origin, and aging, provides valuable insights into the factors shaping pigment concentration. It is important to highlight that the international cultivars included in this study originate from commercial nurseries, where they have undergone rigorous clonal selection and are commonly grafted onto specific rootstocks to enhance uniformity and adaptability [26]. In contrast, the traditional Canary Island cultivars have been selected through generations of field cultivation by local viticulturists. These native cultivars are typically grown on their own roots, leading to greater genetic diversity and, consequently, a higher variability in wine characteristics. Additionally, the Canary Islands have a fragmented geography—comprising eight main islands—which results in significant edaphoclimatic differences across wine-producing areas.
Despite this heterogeneity, the anthocyanin concentrations observed in the red wines from autochthonous cultivars in the Canary Islands fall within the usual range reported in the literature. In general, international cultivars exhibited higher concentrations of most anthocyanins compared to local cultivars. This aligns with findings from other regions, where international grape cultivars, often selected for their higher pigment content, tend to show more intense coloration. Notably, the relatively mild and stable climate of the Canary Islands influences anthocyanin accumulation. Comparing our results obtained for the Syrah cultivar, the anthocyanin contents were lower than those values reported in red wines from tropical wine-growing regions with higher solar radiation [27], yet higher than those observed in cooler climates, such as Northern Greece [28]. This suggests that the balance between sunlight exposure and temperature plays a critical role in the production of anthocyanin. Similarly, the variability in anthocyanin composition observed across different cultivars in this study was comparable to that reported in studies conducted in France and Italy on international grape cultivars [29]. These findings highlight the importance of environmental conditions and viticultural practices in shaping the phenolic composition of wines, reinforcing the role of anthocyanin profiling as a tool for understanding terroir influence and ensuring wine authenticity.
4.1.1. Cultivar Analysis
Significant variations in the anthocyanin profile across different grape cultivars were observed, supporting that genetic factors influence pigment biosynthesis [30]. As expected, M-3-glc is the most concentrated anthocyanin in all grape cultivars, as it is the most stable non-acylated anthocyanin [6]. Its higher concentration found in foreign cultivars such as R, C, and S supports the use of these international grapes for producing red wines with intense and stable red hues. Moreover, these higher M-3-glc concentrations compared to most of the autochthonous grape cultivars are likely due to clonal selection of cultivars commonly carried out in their regions of origin, which focused on enhancing red colour. Also, Acet concentrations were generally higher in foreign cultivars (S, R, and M) and also in the autochthonous T cultivar. Our results agree with previous studies [11] where the S cultivar also presented the highest percentage of acetylated anthocyanins. These anthocyanin derivatives are known for their higher colour stability compared to their non-acetylated counterparts, as they participate in intra-molecular copigmentation processes [16]. The higher levels of acetylated anthocyanins in T cultivar might suggest that this cultivar may be better suited for producing red wines due to its capacity for maintaining colour intensity over extended storage periods than the rest of Canary traditional cultivars. Interestingly the concentration of Cou showed a distinct pattern because the V cultivar had higher concentrations compared to foreign cultivars, with significant differences with respect to R cultivar. Despite its lower Tot concentration, the V cultivar had a higher mean Cou concentration, which might provide an advantage in the production of red wines, as Cou are usually associated with colour resistance to the oxidative degradation and, therefore, could retain the colour for a longer time.
Concentrations of other minor glucosides such as D-3-glc and Pt-3-glc showed significant cultivar-specific differences. The foreign R and S cultivars, as well as the traditional C, exhibited higher concentrations, which could be due to an increased synthesis or higher colour stability in these cultivars. This could be related to a higher activity of flavonoid-3-hydrolase and flavonoid-3,5-hydrolase, which would produce different ratios between di- and trihydroxylated anthocyanins. C-3-glc presented no significant differences between cultivars and the lowest concentration, probably because it is the precursor of all the others [31]. Similarly, no significant differences between cultivars for Pe-3-glc were observed, as this anthocyanin is formed by the activity of the methyltransferase enzyme in C-3-glc [11]. The highest mean concentrations of Glc were observed in the traditional C and LN cultivars, as well as in the international R, with significant differences compared to V. This higher Glc concentration implies that these cultivars have a greater potential for producing young wines with a vivid and intense coloration. The variability in anthocyanin profiles across cultivars suggests that blending strategies could be employed to optimize wine colour and stability. For example, blending traditional cultivars rich in M-3-glc (C or LN) with those containing higher coumaryl derivatives (V) could produce wines with both immediate visual appeal and long-term colour stability.
4.1.2. Geographical Origin
Important influences of the precedence island and the DO within Tenerife Island on the anthocyanin profiles of red wines were found. Thus, grape genetics are not the only variable affecting wine colour, but there are also many other complex relationships between the vine and environmental factors influencing the anthocyanins of red wines [32].
In terms of island of precedence, the higher mean concentrations of Glc/oenin found in red wines from Tenerife and Lanzarote islands could suggest that these islands have environmental conditions that enhance the biosynthesis and stability of anthocyanin glucosides. This trend was consistent across several individual anthocyanins, such as D-3-glc, Pe-3-glc, Pt-3-glc, and M-3-glc. This higher concentrations of anthocyanins in red wines may be attributed to a higher solar radiation, particularly important in Lanzarote and South Tenerife [33], as sunlight has been proved to promote anthocyanin accumulation [34]. In contrast, lower anthocyanin concentrations in red wines from La Palma and Gran Canaria islands may result from differences in microclimates or soil composition, as a decrease in anthocyanin levels has been correlated with nitrogen deficiency [35]. The lack of significant differences in C-6-ac-3-glc and M-6-ac-3-glc between islands indicates that these compounds could be less influenced by edaphoclimatic conditions. However, the higher concentrations of Pe-6-ac-3-glc and Pt-6-ac-3-glc in red wines from Lanzarote highlight the potential for this island to produce red wines with greater anthocyanin content and diversity and associated colour stability. Interestingly, red wines from El Hierro island presented the highest mean Cou concentration, which could probably be explained by the fact that this island mostly produces traditional cultivars such as V and B, which produced red wines with high levels of Cou, as previously indicated.
The data from the five DOs within Tenerife Island further emphasize the impact of edaphoclimatic conditions on anthocyanin profiles. Red wines from Tenerife South (DO G and DO A), with higher sunlight hours, consistently exhibited higher mean D-3-glc, C-3-glc, and Glc/oenin concentrations than the rest of DOs. This suggests that these areas provide optimal conditions for the synthesis and retention of these pigments in comparison with the North DOs of the island (DO TA, DO Y, and DO O). The significant differences observed in C-3-glc within DO of Tenerife Island are interesting because it is the anthocyanin with the lowest concentration, which usually did not show significant differences for all the other variables considered (cultivar, aging, or island).
The acetylated anthocyanins followed a similar pattern to the glucoside anthocyanins. Red wines from DO G and DO A from the South of Tenerife island showed significantly higher concentrations than those from DO O, DO TA, and DO Y. This suggests that the grapes grown in DO G and DO A, which receive more sunlight hours [33], may possess environmental advantages that enhance the synthesis of these acetylated derivatives. These compounds are known for their colour stability and resistance to oxidation [16]. Furthermore, this trend appears to be present early in the biosynthesis process, as indicated by the significantly higher mean concentration observed in the simplest anthocyanin form (C-3-glc). The pattern observed for coumaroyl derivatives mirrors that of the acetylated anthocyanins, reinforcing the idea that DO G and DO A have attributes that promote the synthesis and accumulation of these coloured compounds.
4.1.3. Aging
The significant decrease in the mean concentrations of Tot and Glc with aging is consistent with anthocyanin degradation and polymerization over time [36]. In particular, the sharp decline observed in red wines aged more than 3 years suggests that the anthocyanins are progressively transformed into more complex polymeric pigments, which contribute to the colour stability of aged wines [25]. In contrast to the glucosides, the mean concentrations of Cou significantly increase with aging, supporting the assumption that these compounds are more stable during the maturation process and can be formed over time through reactions involving acylation [37]. Acetylated derivatives do not exhibit a clear trend with aging, suggesting that these compounds are less affected by the aging process compared to glucosides and coumaroyl derivatives. This variability might be due to their stability in the wine matrix components. In any case, the transformations observed lead to a loss of the bright red colour associated with young wines, resulting in the more brick-red hues typical of older wines.
4.2. Correlation Study
The highly significant and positive correlations between the major non-acylated anthocyanins—such as D-3-glc, Pt-3-glc, Pe-3-glc, and M-3-glc—support the assumption that these compounds share common biosynthetic pathways and are co-extracted during winemaking processes [31]. M-3-glc shows particularly strong correlations with other anthocyanins and the total anthocyanin content. This aligns with previous studies highlighting malvidin derivatives as predominant contributors to red wine colour and stability [38].
The significant negative correlations between Cou and Glc anthocyanin derivatives indicate a shift in anthocyanin profiles due to enzymatic acylation during wine aging. Acylation of anthocyanins is known to affect their colour stability and resistance to oxidation [16]. The negative correlation suggests that when the concentration of coumaroylated anthocyanins increases, the levels of Glc anthocyanins decrease due to their enzymatic conversion [37]. Nevertheless, the correlations between acetylated anthocyanins and their glucoside counterparts are generally moderate, indicating a less direct relationship.
The Tot concentration was strong and positively correlated with most individual anthocyanins, particularly Pt-3-glc and M-3-glc, highlighting the significant contribution of these compounds to the overall anthocyanin profile in red wines. These findings reveal the complex connections among the different anthocyanin compounds present in red wines [38].
4.3. Multivariate Analysis
4.3.1. Principal Compound Analysis
Figure 3 illustrates the separation of different anthocyanin derivatives based on their contributions to the two main factors derived from factor analysis. The arrangement of the anthocyanin compounds in the two-dimensional space highlights their differing roles and relationships in the red wines. Chemical differences among anthocyanin derivatives, produced by glucosylation, acetylation, or coumaroylation, could explain their separation through factor analysis.
Thus, Factor 1, explaining the largest proportion of variance (48.7%), predominantly separates anthocyanins based on their structural complexity, such as acetylation or glucosylation. Compounds like Pt-6-ac-3-glc, M-6-ac-3-glc, and Acet are located on the far right of the Factor 1 axis, indicating a strong and positive correlation of acetylated anthocyanins with this dimension. These compounds, characterized by their acetylated species, are likely critical for differentiating phenolic profiles of red wines by aging or specific winemaking practices which have increased their oxidation [38]. Conversely, less complex anthocyanins, such as those only from glucosides, appear closer to the centre or left side of Factor 1.
Factor 2, which explains 22.1% of the variance, differentiates anthocyanins along a different axis, more related to specific environmental and aging influences. For instance, Glc is positioned prominently at the top of the axis, while Cou is situated in the bottom-left quadrant, far from the anthocyanin cluster. This position highlights its independence from most compounds, possibly reflecting that its synthesis is mediated by hydroxycinnamic acid present in red wines [39].
A distinct clustering is observed among glucosylated anthocyanins, such as M-3-glc, Pt-3-glc, and Pe-3-glc, in the upper-right quadrant. This clustering is relevant, as it implies that monoglucosylated anthocyanins may serve as useful markers for classifying red wines based on grape cultivar or region of origin, as previously reported [11]. In contrast, acetylated and coumaroylated anthocyanins, such as M-6-ac-3-glc and Pe-6-co-3-glc, are more dispersed along Factor 1, indicating greater variability. This variability might be due to influences of winemaking and environmental factors related with wine aging [37].
In summary, acetylated anthocyanins tend to dominate the differentiation along Factor 1, while Glc and Cou anthocyanins contribute to the differentiation along Factor 2. This separation suggests that the anthocyanin profile might be a powerful tool for distinguishing red wines based on key variables like grape cultivar, geographical origin, or aging process.
4.3.2. Discriminant Analysis
Results highlight the power and limitations of LDA in classifying red wines based on their anthocyanin profiles. The relatively high classification rates of red wines by aging obtained by us suggest that the anthocyanin degradation or transformation during aging are consistent enough to effectively differentiate red wines over time. These findings agree with previous studies that demonstrated the utility of these compounds in red wine classification and authentication [11,30,40].
However, the lower accuracy for the classification of red wines according to grape cultivar and geographical origin (precedent island or Tenerife DO) highlights challenges in separating red wines based solely on anthocyanin profiles. Overlapping characteristics between closely related cultivars or within a geographically small and climatically similar region such as Tenerife Island could reduce the effectiveness of the classification. This suggests that while anthocyanins are valuable markers, additional variables may be needed for a finer differentiation.
For differentiating of red wines in basis of grape cultivar used for its elaboration, the LDA using all variables achieved a correct classification of 75.6%, which dropped to 64.1% after cross-validation. There are red wines produced from some grape cultivars that were well-classified (100%) within their group, which contrasts with that observed in other red wines elaborated from other cultivars. A moderate ability to distinguish cultivars was found, suggesting overlap of the phenolic profiles among different grape cultivars. Other factors could be having an influence on anthocyanin concentrations, such as winemaking techniques or terroir factors. Therefore, while anthocyanin profiles are valuable for differentiating certain grape cultivars, they may not be sufficient for distinguishing closely related cultivars. Interestingly, stepwise LDA, which uses a reduced set of variables, often yielded lower accuracy compared to using all the variables. This could imply that phenolic compounds work synergistically in red wine classification and removing less significant variables might inadvertently weaken the model’s robustness. However, stepwise LDA provides valuable insights into the most discriminatory compounds, such as M-3-glc and acetylated anthocyanins, which can guide further targeted research.
5. Conclusions
Anthocyanin levels in wines from the Canary Islands made from international grape cultivars are comparable to those reported in other regions of the world. Red wines produced from international grape cultivars generally exhibited higher anthocyanin concentrations than those from autochthonous cultivars varieties, particularly for malvidin-3-glucoside. Some autochthonous cultivars, however, showed higher contents of coumaryl anthocyanin derivatives, which are associated with an enhanced pigment stability over time. These findings highlight the potential for strategic blending to optimize wine color properties and stability. Furthermore, cultivar diversity highlights the importance of preserving and promoting the region’s native cultivars. Further research into the genetic factors influencing these profiles would provide insights into the potential of each cultivar for winemaking and quality improvement.
Anthocyanin concentrations and profiles in red wines are also influenced by geographical origin. Thus, the wines from different islands, as well as different Denominations of Origin within Tenerife Island, exhibited significant variations in the anthocyanin profile. Sunlight exposure, temperature, and other edaphoclimatic conditions appear to play a key role in pigment biosynthesis. Therefore, the red wines from southern Tenerife and Lanzarote showed the highest concentrations of glucoside anthocyanins. Given the observed influence of terroir, viticultural practices such as canopy management, irrigation, and harvest timing could be adjusted to enhance anthocyanin biosynthesis under specific environmental conditions. Aging of red wines was the factor with the highest impact on anthocyanin composition. A marked decline in glucoside anthocyanins and an increase in coumaroyl derivatives was clearly observed in aging, which could be associated with the improvement of red wine color stability.
Multivariate analyses (principal component and linear discriminant analyses) demonstrated that anthocyanin profiles tend to classify red wines according to grape cultivar, island of origin, Denomination of Origin, and aging. Notably, the highest classification accuracy was achieved for aging differentiation. The use of multivariate statistical techniques applied to the anthocyanin composition could have potential as a cost-effective tool for verifying the red wine authenticity. This could aid in the prevention of fraud and the protection of regional wine identities.
In future research, it could also be interesting to examine the impact of winemaking techniques, including maceration time and fermentation parameters, on anthocyanin stability. Additionally, exploring the influence of microbial activity during fermentation on pigment transformations, as well as assessing the consistency of classification patterns across multiple vintages, would provide valuable insights into potential interannual variability.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/app15041755/s1—Figure S1: Factor 2 and 3 scores of wine samples described by aging; Table S1: Retention times and abbreviations of the compounds quantified; Table S2: Anthocyanins (mg/l) according to the DO from Tenerife Island; Table S3: Results of anthocyanin concentrations (mg/l) according to ageing.
Author Contributions
Conceptualization, C.D.-R. and J.H.-R.; methodology, C.D.-R.; software, J.H.-R.; validation, C.D.-R.; formal analysis, J.H.-R.; investigation, J.H.-R.; resources, C.D.-R.; data curation, C.D.-R.; writing—original draft preparation, J.H.-R.; writing—review and editing, C.D.-R.; visualization, J.H.-R.; supervision, C.D.-R.; project administration, J.H.-R.; funding acquisition, C.D.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data are unavailable due to the winemakers’ and cellar privacy.
Acknowledgments
We acknowledge Canary Island wineries for their support with the samples.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Villegas-Aguilar, M.d.C.; Sánchez-Marzo, N.; Fernández-Ochoa, Á.; Del Río, C.; Montaner, J.; Micol, V.; Herranz-López, M.; Barrajón-Catalán, E.; Arráez-Román, D.; Cádiz-Gurrea, M.d.l.L. Evaluation of Bioactive Effects of Five Plant Extracts with Different Phenolic Compositions against Different Therapeutic Targets. Antioxidants 2024, 13, 217. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, Y.; Zhang, T.; Chen, X. Fingerprint and difference analysis of flavonoids of Hippophae plants grown on the Tibetan plateau. J. Food Compos. Anal. 2024, 128, 106010. [Google Scholar] [CrossRef]
- Alimoglu, G.; Guzelmeric, E.; Yuksel, P.I.; Celik, C.; Deniz, I.; Yesilada, E. Monofloral and polyfloral bee pollens: Comparative evaluation of their phenolics and bioactivity profiles. LWT 2021, 142, 110973. [Google Scholar] [CrossRef]
- Blasi, F.; Ianni, F.; Cossignani, L. Phenolic profiling for geographical and varietal authentication of extra virgin olive oil. Trends Food Sci. Technol. 2024, 147, 104444. [Google Scholar] [CrossRef]
- Lucini, L.; Rocchetti, G.; Kane, D.; Trevisan, M. Phenolic fingerprint allows discriminating processed tomato products and tracing different processing sites. Food Control 2017, 73, 696–703. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, C.; Huang, X.; Hu, D. Anthocyanin stability and degradation in plants. Plant Signal. Behav. 2021, 16, 1987767. [Google Scholar] [CrossRef]
- Sacchi, K.L.; Bisson, L.F.; Adams, D.O. A review of the effect of winemaking techniques on phenolic extraction in red wines. Am. J. Enol. Vitic. 2005, 56, 197–206. [Google Scholar] [CrossRef]
- Castellarin, S.D.; Pfeiffer, A.; Sivilotti, P.; Degan, M.; Peterlunger, E.; Di Gaspero, G. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007, 30, 1381–1399. [Google Scholar] [CrossRef]
- Kobayashi, S.; Ishimaru, M.; Ding, C.K.; Yakushiji, H.; Goto, N. Comparison of UDP-glucose: Flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci. 2001, 160, 543–550. [Google Scholar] [CrossRef]
- Azuma, A.; Yakushiji, H.; Koshita, Y.; Kobayashi, S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 2012, 236, 1067–1080. [Google Scholar] [CrossRef]
- Ortega-Regules, A.; Romero-Cascales, I.; López-Roca, J.M.; Ros-García, J.M.; Gómez-Plaza, E. Anthocyanin fingerprint of grapes: Environmental and genetic variations. J. Sci. Food Agric. 2006, 86, 1460–1467. [Google Scholar] [CrossRef]
- Shi, T.; Su, Y.; Lan, Y.; Duan, C.; Yu, K. The molecular basis of flavonoid biosynthesis response to water, light, and temperature in grape berries. Front. Plant Sci. 2024, 15, 1441893. [Google Scholar] [CrossRef]
- de Rosas, I.; Deis, L.; Baldo, Y.; Cavagnaro, J.B.; Cavagnaro, P.F. High temperature alters anthocyanin concentration and composition in grape berries of Malbec, Merlot, and Pinot Noir in a cultivar-dependent manner. Plants 2022, 11, 926. [Google Scholar] [CrossRef]
- Rienth, M.; Vigneron, N.; Darriet, P.; Sweetman, C.; Burbidge, C.; Bonghi, C.; Walker, R.P.; Famiani, F.; Castellarin, S.D. Grape berry secondary metabolites and their modulation by abiotic factors in a climate change scenario—A review. Front. Plant Sci. 2021, 12, 643258. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef]
- Cheynier, V.; Duenas-Paton, M.; Salas, E.; Maury, C.; Souquet, J.; Sarni-Manchado, P.; Fulcrand, H. Structure and properties of wine pigments and tannins. Am. J. Enol. Vitic. 2006, 57, 298–305. [Google Scholar] [CrossRef]
- Koljančić, N.; Furdíková, K.; de Araújo Gomes, A.; Špánik, I. Wine authentication: Current progress and state of the art. Trends Food Sci. Technol. 2024, 150, 104598. [Google Scholar] [CrossRef]
- Garcia-Viñola, V.; Ruiz-de-Villa, C.; Gombau, J.; Poblet, M.; Bordons, A.; Reguant, C.; Rozès, N. Simultaneous Analysis of Organic Acids, Glycerol and Phenolic Acids in Wines Using Gas Chromatography-Mass Spectrometry. Foods 2024, 13, 186. [Google Scholar] [CrossRef]
- Bambina, P.; Spinella, A.; Lo Papa, G.; Chillura Martino, D.F.; Lo Meo, P.; Cinquanta, L.; Conte, P. 1H-NMR Spectroscopy Coupled with Chemometrics to Classify Wines According to Different Grape Varieties and Different Terroirs. Agriculture 2024, 14, 749. [Google Scholar] [CrossRef]
- Ianeselli, A.; Longo, E.; Poggesi, S.; Montali, M.; Boselli, E. A Complete Analysis Pipeline for the Processing, Alignment and Quantification of HPLC–UV Wine Chromatograms. Chromatographia 2024, 87, 159–166. [Google Scholar] [CrossRef]
- Merkytė, V.; Longo, E.; Windisch, G.; Boselli, E. Phenolic compounds as markers of wine quality and authenticity. Foods 2020, 9, 1785. [Google Scholar] [CrossRef]
- Ibern-Gómez, M.; Andrés-Lacueva, C.; Lamuela-Raventós, R.M.; Waterhouse, A.L. Rapid HPLC analysis of phenolic compounds in red wines. Am. J. Enol. Vitic. 2002, 53, 218–221. [Google Scholar] [CrossRef]
- Baiano, A.; Terracone, C. Varietal differences among the phenolic profiles and antioxidant activities of seven table grape cultivars grown in the south of Italy based on chemometrics. J. Agric. Food Chem. 2011, 59, 9815–9826. [Google Scholar] [CrossRef]
- Ginjom, I.; D’Arcy, B.; Caffin, N.; Gidley, M. Phenolic compound profiles in selected Queensland red wines at all stages of the wine-making process. Food Chem. 2011, 125, 823–834. [Google Scholar] [CrossRef]
- Vivar-Quintana, A.M.; Santos-Buelga, C.; Rivas-Gonzalo, J.C. Anthocyanin-derived pigments and colour of red wines. Anal. Chim. Acta 2002, 458, 147–155. [Google Scholar] [CrossRef]
- Theocharis, S.; Gkrimpizis, T.; Karadimou, C.; Alatzas, A.; Koundouras, S.; Taskos, D. Modulating ‘Xinomavro’(Vitis vinifera L.) Vine Growth and Berry Composition: A Comparative Analysis of Rootstock Effects. Horticulturae 2024, 10, 490. [Google Scholar] [CrossRef]
- de Oliveira, J.B.; Egipto, R.; Laureano, O.; de Castro, R.; Pereira, G.E.; Ricardo-da-Silva, J.M. Chemical composition and sensory profile of Syrah wines from semiarid tropical Brazil–Rootstock and harvest season effects. LWT 2019, 114, 108415. [Google Scholar] [CrossRef]
- Stavridou, K.; Soufleros, E.H.; Bouloumpasi, E.; Dagkli, V. The phenolic potential of wines from French grape varieties Cabernet Sauvignon, Merlot and Syrah cultivated in the region of Thessaloniki (Northern Greece) and its evolution during aging. Food Nutr. Sci. 2016, 7, 122–137. [Google Scholar] [CrossRef]
- Ponder, A.; Frąckowiak, M.; Kruk, M.; Hallmann, E. Estimation of Chemical Compounds in Selected Italian and French Wines Produced through Organic and Conventional Methods. Appl. Sci. 2024, 14, 2466. [Google Scholar] [CrossRef]
- Díaz-Fernández, Á.; Díaz-Losada, E.; Moreno, D.; Sánchez, M.E.V. Anthocyanin profile of Galician endangered varieties. A tool for varietal selection. Food Res. Int. 2022, 154, 110983. [Google Scholar] [CrossRef]
- Núñez, V.; Monagas, M.; Gomez-Cordovés, M.C.; Bartolomé, B. Vitis vinifera L. cv. Graciano grapes characterized by its anthocyanin profile. Postharvest Biol. Technol. 2004, 31, 69–79. [Google Scholar] [CrossRef]
- LaFountain, A.M.; Yuan, Y. Repressors of anthocyanin biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef]
- Spanish Meteorological Agency (AEMET). Available online: https://www.aemet.es/es/serviciosclimaticos/datosclimatologicos/valoresclimatologicos?k=coo (accessed on 12 February 2024).
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef]
- An, J.; Qu, F.; Yao, J.; Wang, X.; You, C.; Wang, X.; Hao, Y. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef]
- Prat-García, S.; Oliveira, J.; del Alamo-Sanza, M.; de Freitas, V.; Nevares, I.; Mateus, N. Characterization of anthocyanins and anthocyanin-derivatives in red wines during ageing in custom oxygenation oak wood barrels. Molecules 2020, 26, 64. [Google Scholar] [CrossRef]
- He, F.; Liang, N.; Mu, L.; Pan, Q.; Wang, J.; Reeves, M.J.; Duan, C. Anthocyanins and their variation in red wines I. Monomeric anthocyanins and their color expression. Molecules 2012, 17, 1571–1601. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Liang, N.; Mu, L.; Pan, Q.; Wang, J.; Reeves, M.J.; Duan, C. Anthocyanins and their variation in red wines II. Anthocyanin derived pigments and their color evolution. Molecules 2012, 17, 1483–1519. [Google Scholar] [CrossRef]
- Yang, P.; Basílio, N.; Liao, X.; Xu, Z.; Dangles, O.; Pina, F. Influence of Acylation by Hydroxycinnamic Acids on the Reversible and Irreversible Processes of Anthocyanins in Acidic to Basic Aqueous Solution. J. Agric. Food Chem. 2024, 72, 25955–25971. [Google Scholar] [CrossRef]
- Cosme, F.; Milheiro, J.; Pires, J.; Guerra-Gomes, F.I.; Filipe-Ribeiro, L.; Nunes, F.M. Authentication of Douro DO monovarietal red wines based on anthocyanin profile: Comparison of partial least squares–discriminant analysis, decision trees and artificial neural networks. Food Control 2021, 125, 107979. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).