Figure 1.
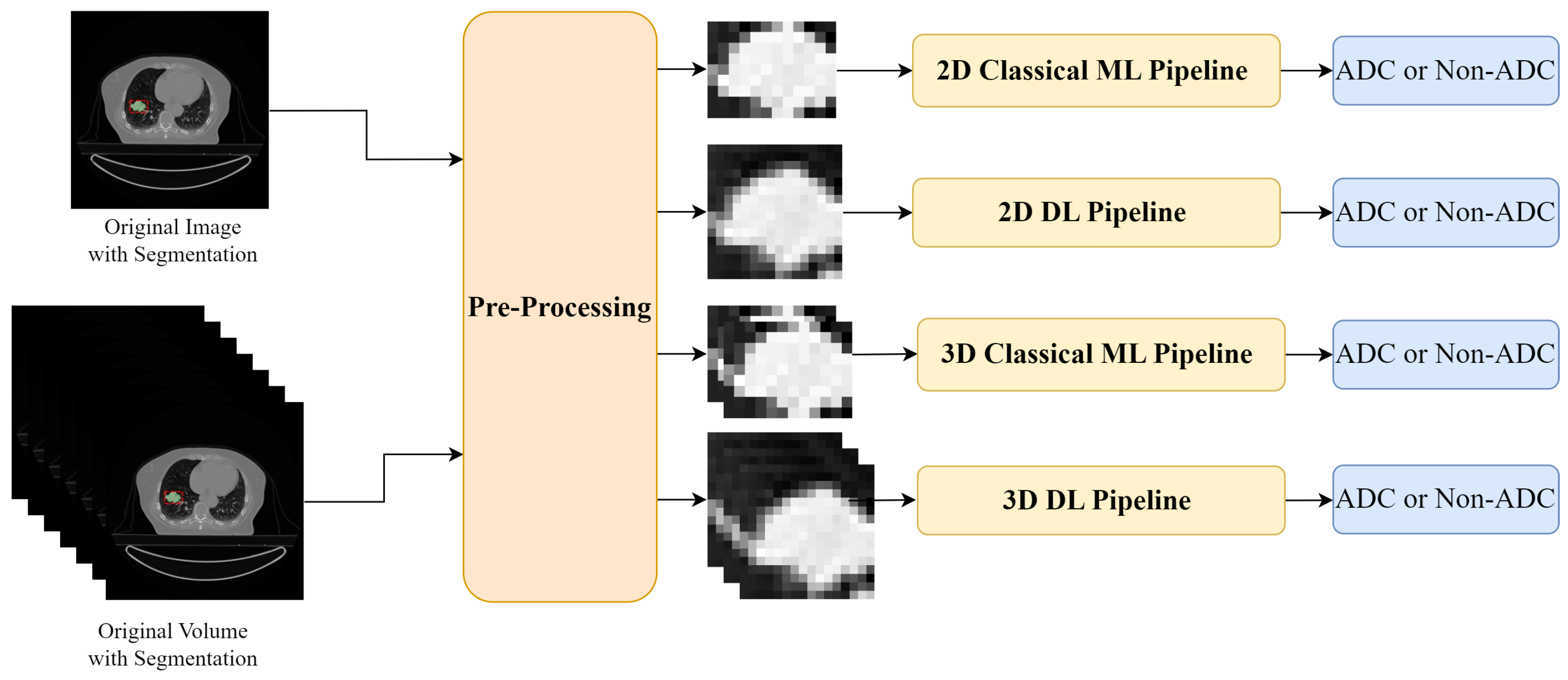
Experimental Setup: two input forms were used—a 2D slice and a 3D CT scan. The inputs were pre-processed, firstly intensity normalization was applied and then, according to each pipeline, the ROI was defined: for the ML approach, the tumours were cropped according to the original segmentation masks; while for the DL approach, a fixed-size bounding box centred on the tumour mask was selected. The tumour regions were then used as input to the Classical ML and DL pipelines for binary classification in adenocarcinoma (ADC) and non-adenocarcinoma (Non-ADC).
Figure 1.
Experimental Setup: two input forms were used—a 2D slice and a 3D CT scan. The inputs were pre-processed, firstly intensity normalization was applied and then, according to each pipeline, the ROI was defined: for the ML approach, the tumours were cropped according to the original segmentation masks; while for the DL approach, a fixed-size bounding box centred on the tumour mask was selected. The tumour regions were then used as input to the Classical ML and DL pipelines for binary classification in adenocarcinoma (ADC) and non-adenocarcinoma (Non-ADC).
Figure 2.
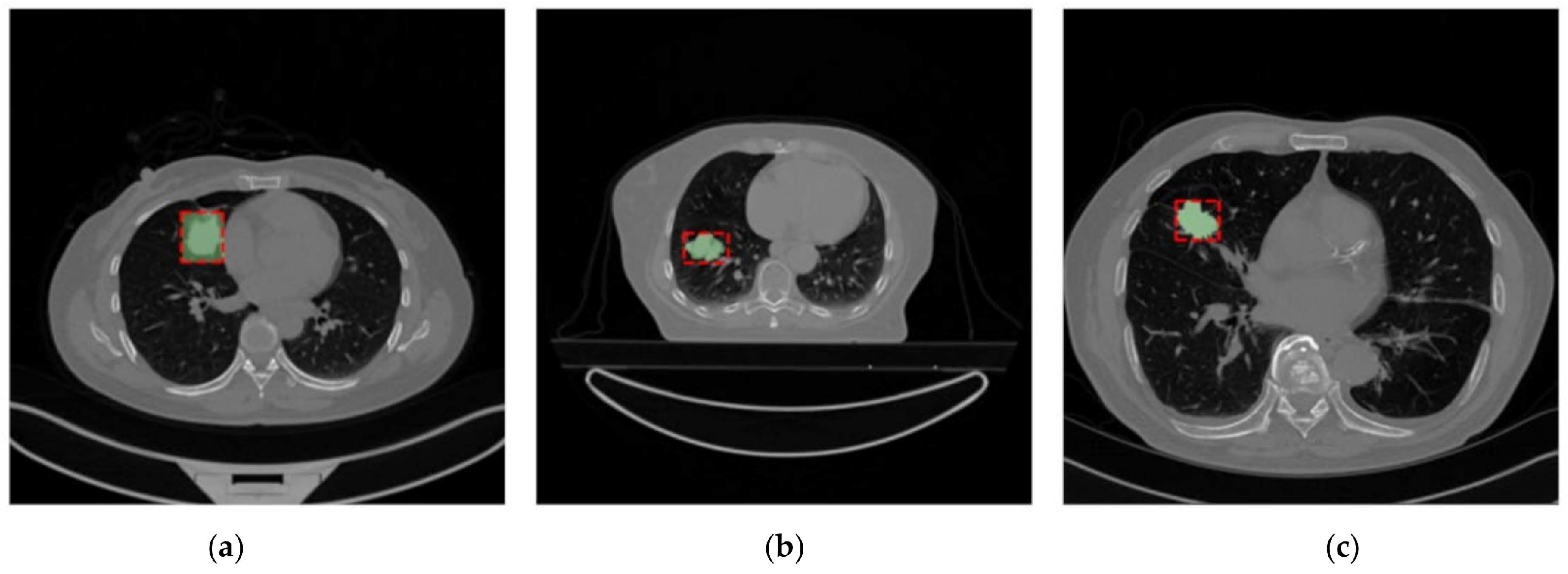
Example of images from the Lung-PET-CT-Dx Dataset (a), the NSCLC-Radiomics Dataset (b), and the NSCLC-Radiogenomics (c) Dataset. The segmentations present in the dataset are shown in green and the bounding box used after the dataset uniformization is presented in red.
Figure 2.
Example of images from the Lung-PET-CT-Dx Dataset (a), the NSCLC-Radiomics Dataset (b), and the NSCLC-Radiogenomics (c) Dataset. The segmentations present in the dataset are shown in green and the bounding box used after the dataset uniformization is presented in red.
Figure 3.
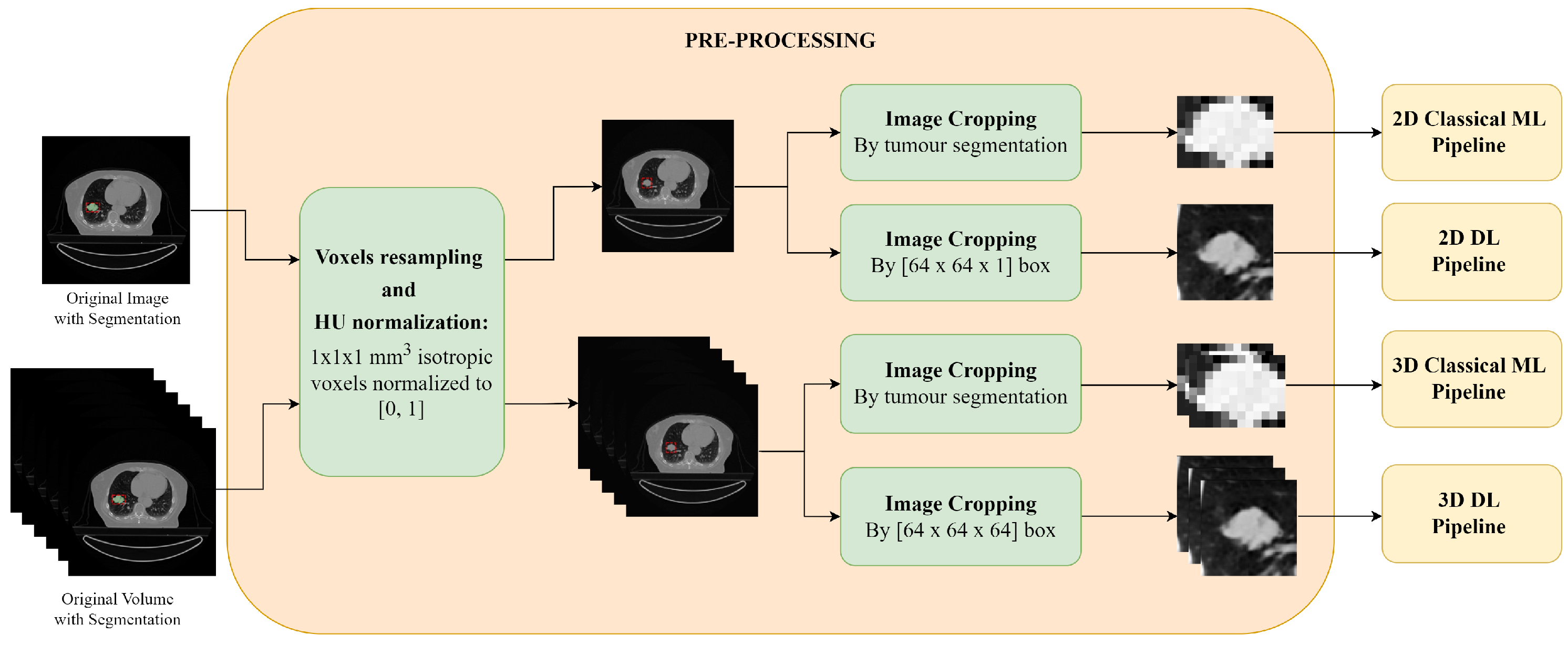
Pre-processing of the 2D slices and the 3D CT scans: first a voxel resampling and discretization is applied, followed by image cropping. For the images and volumes used for the DL pipeline, image resampling and normalization are also applied.
Figure 3.
Pre-processing of the 2D slices and the 3D CT scans: first a voxel resampling and discretization is applied, followed by image cropping. For the images and volumes used for the DL pipeline, image resampling and normalization are also applied.
Figure 4.
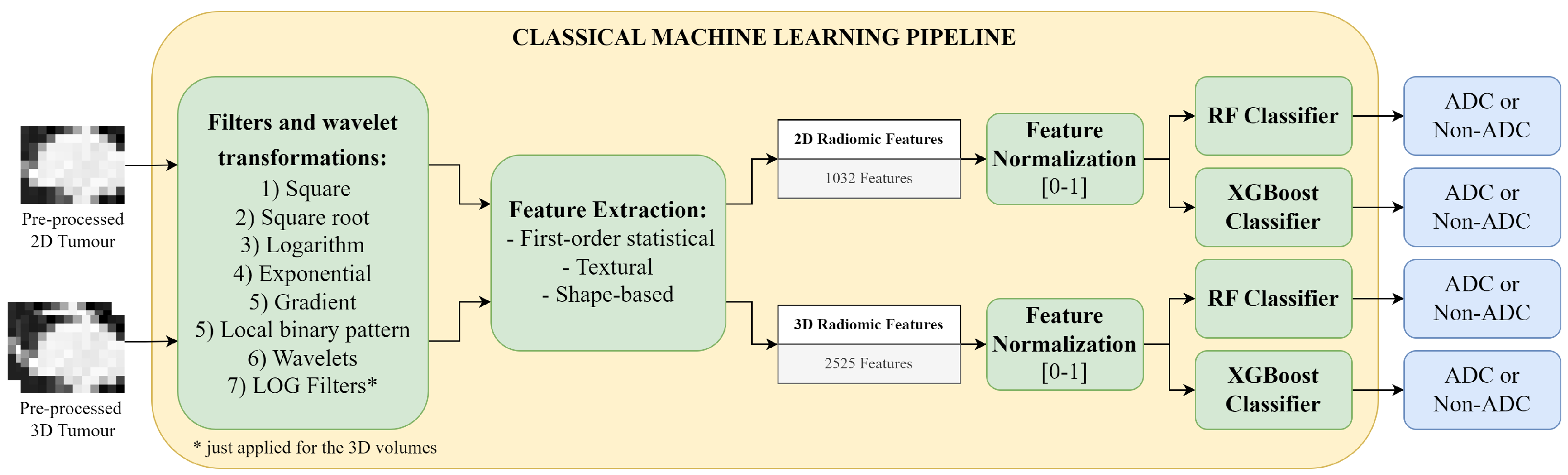
Classical Machine Learning Pipeline: first several filters are applied to the 2D tumour image and the 3D tumour volume, for radiomics feature extraction; then the features are normalized and used as input to the Random Forest (RF) or the eXtreme Gradient Boosting (XGBoost) classifier for the binary classification between adenocarcinoma (ADC) and non-adenocarcinoma (Non-ADC).
Figure 4.
Classical Machine Learning Pipeline: first several filters are applied to the 2D tumour image and the 3D tumour volume, for radiomics feature extraction; then the features are normalized and used as input to the Random Forest (RF) or the eXtreme Gradient Boosting (XGBoost) classifier for the binary classification between adenocarcinoma (ADC) and non-adenocarcinoma (Non-ADC).
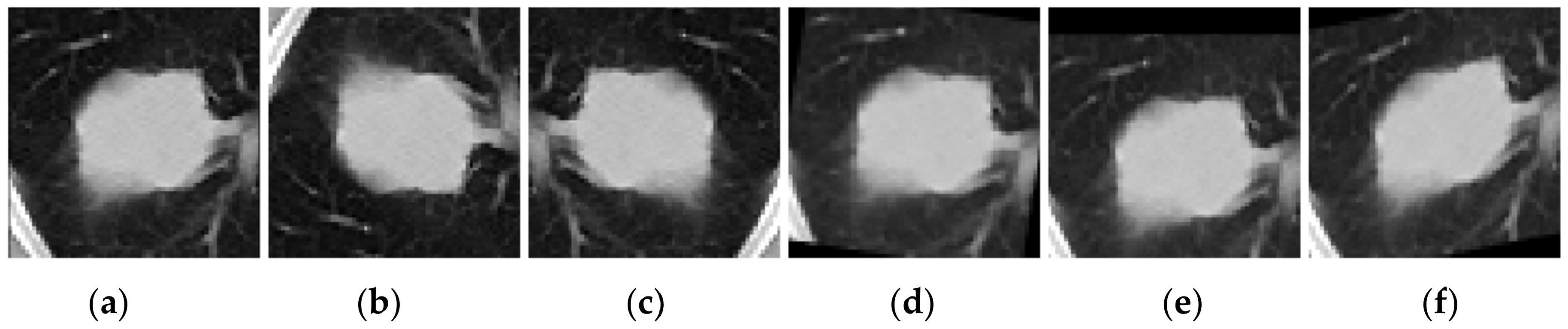
Figure 5.
Samples resulting from data augmentation transformations applied in the deep learning approach, respectively: (a) original image, (b) vertical flip, (c) horizontal flip, (d) rotation of 10°, (e) shift of 10%, and (f) shear of 10%.
Figure 5.
Samples resulting from data augmentation transformations applied in the deep learning approach, respectively: (a) original image, (b) vertical flip, (c) horizontal flip, (d) rotation of 10°, (e) shift of 10%, and (f) shear of 10%.
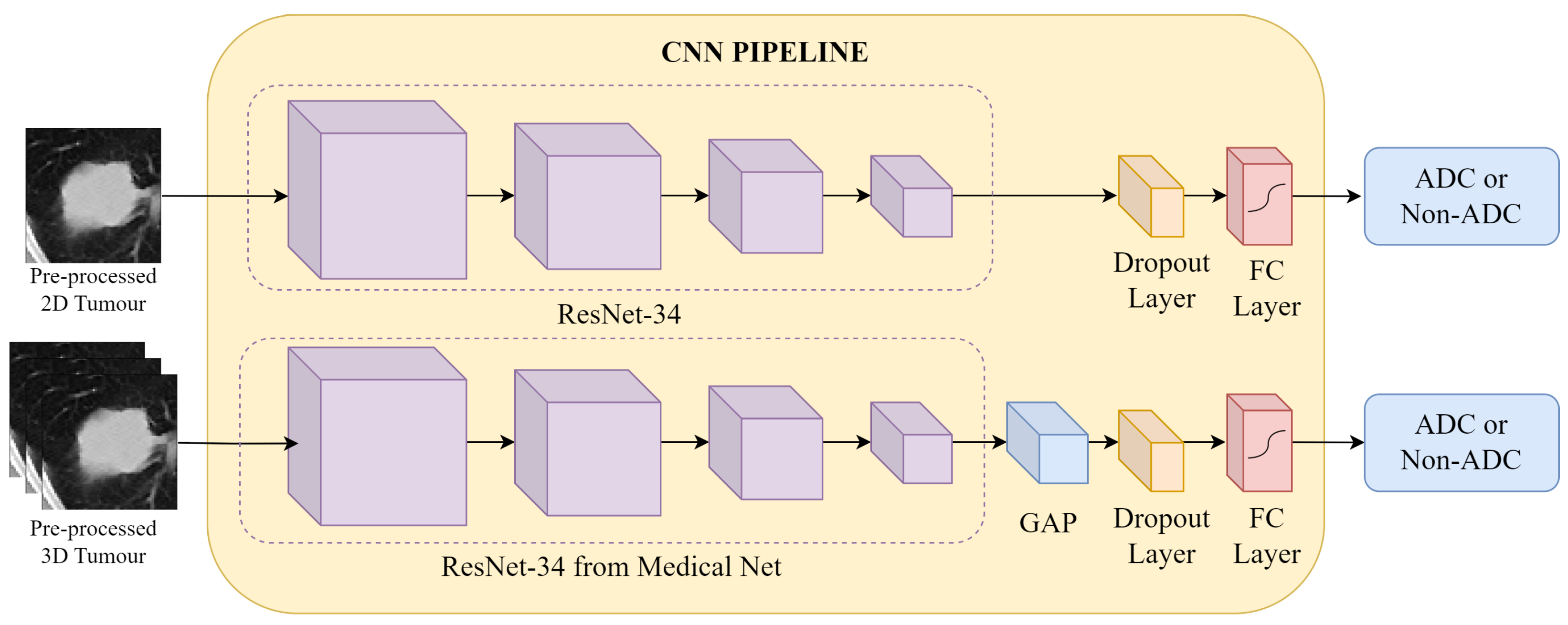
Figure 6.
CNN Pipeline: the pre-processed CT patches are fed into the respective 2D and 3D networks. In both approaches, feature extraction is conducted by a ResNet-34 architecture, followed by classification at a fully connected (FC) layer into the binary labels adenocarcinoma (ADC) and non-adenocarcinoma (Non-ADC).
Figure 6.
CNN Pipeline: the pre-processed CT patches are fed into the respective 2D and 3D networks. In both approaches, feature extraction is conducted by a ResNet-34 architecture, followed by classification at a fully connected (FC) layer into the binary labels adenocarcinoma (ADC) and non-adenocarcinoma (Non-ADC).
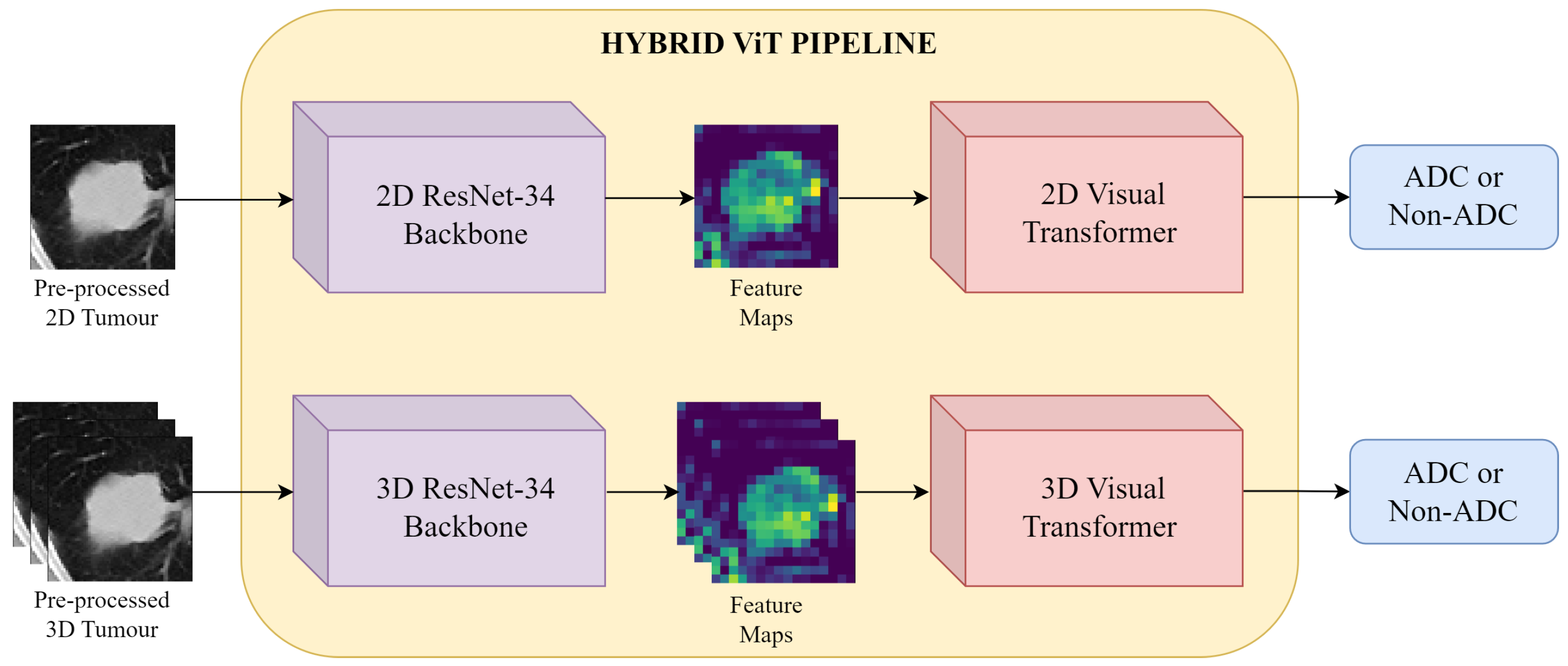
Figure 7.
Hybrid ViT Pipeline: the pre-processed CT patches are first input into the ResNet-34 backbone for feature extraction. The resulting feature maps are then fed to a ViT encoder, and finally binary classification is performed.
Figure 7.
Hybrid ViT Pipeline: the pre-processed CT patches are first input into the ResNet-34 backbone for feature extraction. The resulting feature maps are then fed to a ViT encoder, and finally binary classification is performed.
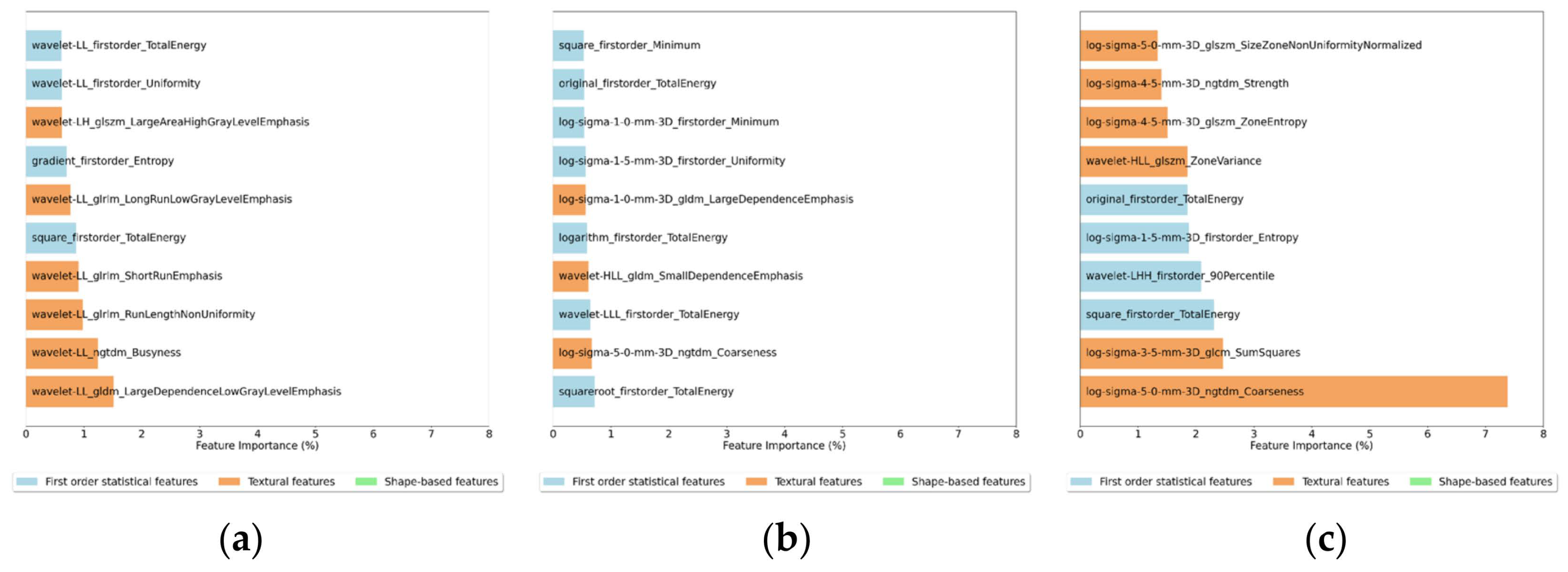
Figure 8.
Feature importance for the 10 most important features for the (a) 2D Random Forest, (b) 3D Random Forest and (c) 3D XGBoost Classifiers.
Figure 8.
Feature importance for the 10 most important features for the (a) 2D Random Forest, (b) 3D Random Forest and (c) 3D XGBoost Classifiers.
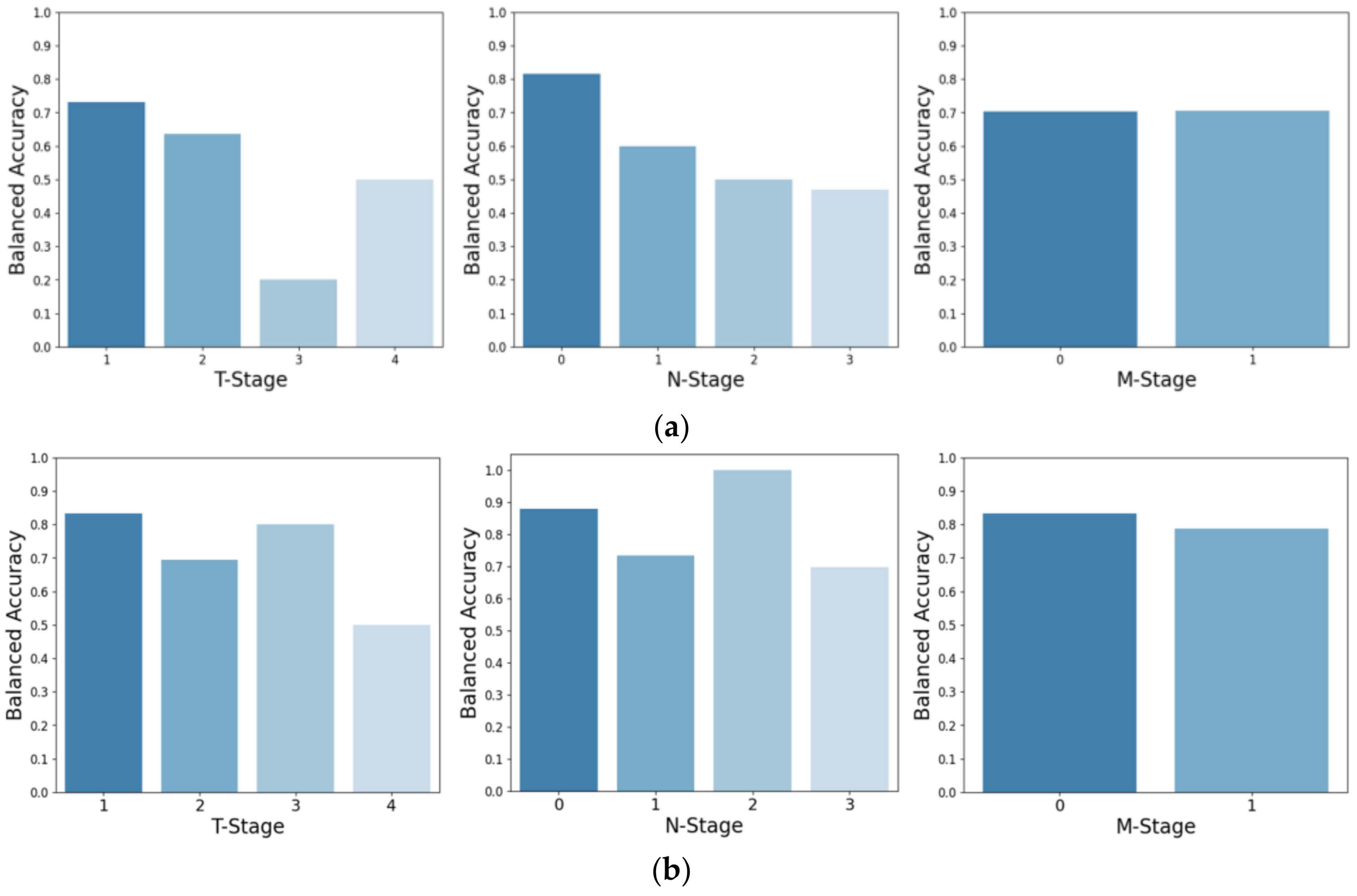
Figure 9.
Balanced accuracy based on tumour size (T-Stage), on the extent of tumour spreading (N-Stage), and metastasis state (M-Stage). The top row (a) represents the balanced accuracy for the Lung-PET-CT-Dx test set classified by the ResNet-34, while the bottom row (b) shows the corresponding results when classified by the Hybrid ViT.
Figure 9.
Balanced accuracy based on tumour size (T-Stage), on the extent of tumour spreading (N-Stage), and metastasis state (M-Stage). The top row (a) represents the balanced accuracy for the Lung-PET-CT-Dx test set classified by the ResNet-34, while the bottom row (b) shows the corresponding results when classified by the Hybrid ViT.
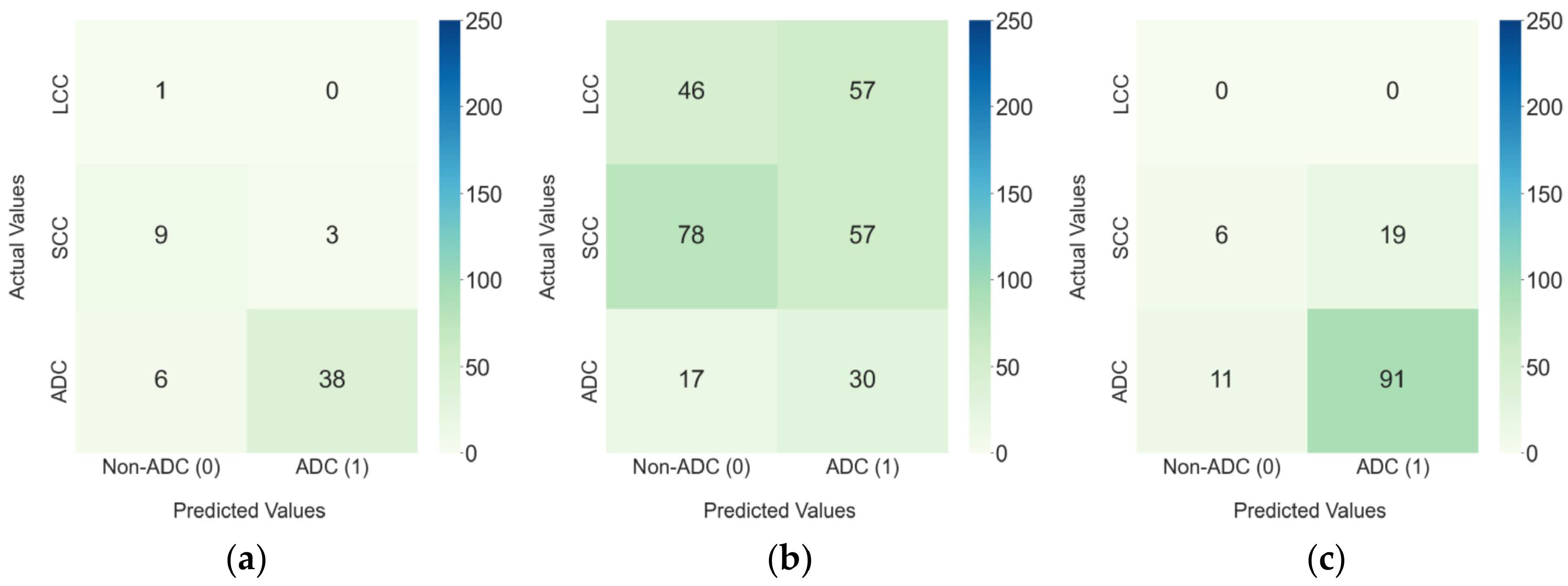
Figure 10.
Confusion matrixes for the 3D Hybrid ViT model in (a) the Lung-PET-CT-Dx internal test set and the external test sets: (b) NSCLC-Radiomics and (c) NSCLC-Radiogenomics.
Figure 10.
Confusion matrixes for the 3D Hybrid ViT model in (a) the Lung-PET-CT-Dx internal test set and the external test sets: (b) NSCLC-Radiomics and (c) NSCLC-Radiogenomics.
Table 1.
Characterization of the final training, validation, and testing subsets, grouped by class, for the experiments performed in this work.
Table 1.
Characterization of the final training, validation, and testing subsets, grouped by class, for the experiments performed in this work.
| Dataset | Class | Total | Train | Validation | Test |
|---|
| Lung-PET-CT-Dx | ADC | 221 | 146 | 31 | 44 |
| Non-ADC | 64 | 38 | 13 | 13 |
| Total | 285 | 184 | 44 | 57 |
| NSCLC-Radiomics | ADC | 47 | - | - | 47 |
| Non-ADC | 238 | - | - | 238 |
| Total | 285 | - | - | 285 |
| NSCLC-Radiogenomics | ADC | 102 | - | - | 102 |
| Non-ADC | 25 | - | - | 25 |
| Total | 127 | - | - | 127 |
Table 2.
Hyper-parameter values for the RF classifier.
Table 2.
Hyper-parameter values for the RF classifier.
| Hyper-Parameter | Values |
|---|
| NEstimators | {50, 100, 150, 200, 250, 300, 400, 500, 750, 1000} |
| MaxDepth | {None, 1, 10, 50, 100, 150, 200, 300} |
| MinSamplesSplit | {2, 10, 15, 20, 25, 30, 50, 75, 100} |
| MinSamplesLeaf | {1, 2, 4, 6, 8, 10} |
| Criterion | {gini, entropy, log_loss} |
Table 3.
Hyper-parameter values for the XGBoost classifier.
Table 3.
Hyper-parameter values for the XGBoost classifier.
| Hyper-Parameter | Values |
|---|
| NEstimators | {50, 100, 150, 200, 250, 300, 400, 500, 750, 1000} |
| MaxDepth | {None, 1, 10, 50, 100, 150, 200, 300} |
| MinChildWeight | {1, 2, 4, 6} |
| {0.001, 0.01, 0.1} |
| {0, 0.5, 1, 2} |
| {0, 0.1, 0.2, 0.3} |
Table 4.
Range of values defined for the parameters of the data augmentation transformations.
Table 4.
Range of values defined for the parameters of the data augmentation transformations.
| Transformations | Range |
|---|
| Flips | - |
| Rotation | [−20, 20]° |
| Shift | [−15, 15]% |
| Shear | [−20, 20]% |
Table 5.
Hyper-parameter values for the CNN.
Table 5.
Hyper-parameter values for the CNN.
| Hyper-Parameter | Values |
|---|
| Dropout | {0, 0.5} |
| Learning Rate | {0.000005, 0.000001, 0.00001} |
| Weight Decay | {0, 0.001, 0.01} |
Table 6.
Hyper-parameter values for the Hybrid ViT.
Table 6.
Hyper-parameter values for the Hybrid ViT.
| Hyper-Parameter | Values |
|---|
| ViT Layers | {6, 8, 10} |
| Learning Rate | {0.00001, 0.00005, 0.0001} |
| Weight Decay | {0, 0.001, 0.01} |
Table 7.
Test results (AUC, balanced accuracy, precision, recall and specificity) of Random Forest and ResNet-34 on three different test sets (Lung-PET-CT-Dx, NSCLC-Radiomics, and NSCLC-Radiogenomics) for 2D and 3D input types. The highest values within each metric are highlighted in bold for each model type and test set.
Table 7.
Test results (AUC, balanced accuracy, precision, recall and specificity) of Random Forest and ResNet-34 on three different test sets (Lung-PET-CT-Dx, NSCLC-Radiomics, and NSCLC-Radiogenomics) for 2D and 3D input types. The highest values within each metric are highlighted in bold for each model type and test set.
| Classifier | Input | Test Set | AUC | Balanced Accuracy | Precision | Recall | Specificity |
|---|
| Random Forest | 2D | Lung-PET-CT-Dx | 0.806 | 0.712 | 0.867 | 0.886 | 0.538 |
| NSCLC-Radiomics | 0.543 | 0.497 | 0.164 | 0.936 | 0.059 |
| NSCLC-Radiogenomics | 0.659 | 0.531 | 0.814 | 0.941 | 0.120 |
| 3D | Lung-PET-CT-Dx | 0.836 | 0.762 | 0.889 | 0.909 | 0.615 |
| NSCLC-Radiomics | 0.560 | 0.509 | 0.168 | 0.830 | 0.189 |
| NSCLC-Radiogenomics | 0.676 | 0.555 | 0.821 | 0.990 | 0.120 |
| ResNet-34 | 2D | Lung-PET-CT-Dx | 0.783 | 0.660 | 0.861 | 0.705 | 0.615 |
| NSCLC-Radiomics | 0.577 | 0.598 | 0.222 | 0.638 | 0.559 |
| NSCLC-Radiogenomics | 0.495 | 0.527 | 0.814 | 0.814 | 0.240 |
| 3D | Lung-PET-CT-Dx | 0.841 | 0.700 | 0.955 | 0.477 | 0.923 |
| NSCLC-Radiomics | 0.576 | 0.550 | 0.219 | 0.340 | 0.761 |
| NSCLC-Radiogenomics | 0.589 | 0.555 | 0.839 | 0.510 | 0.600 |
Table 8.
Test results (AUC, balanced accuracy, precision, recall and specificity) of XGBoost and Hybrid ViT on three different test sets (Lung-PET-CT-Dx, NSCLC-Radiomics, and NSCLC-Radiogenomics) for 3D input type. The highest values within each metric are highlighted in bold for each test set.
Table 8.
Test results (AUC, balanced accuracy, precision, recall and specificity) of XGBoost and Hybrid ViT on three different test sets (Lung-PET-CT-Dx, NSCLC-Radiomics, and NSCLC-Radiogenomics) for 3D input type. The highest values within each metric are highlighted in bold for each test set.
| Input | Classifier | Test Set | AUC | Balanced Accuracy | Precision | Recall | Specificity |
|---|
| 3D | XGBoost | Lung-PET-CT-Dx | 0.853 | 0.801 | 0.909 | 0.909 | 0.692 |
| NSCLC-Radiomics | 0.570 | 0.533 | 0.176 | 0.872 | 0.193 |
| NSCLC-Radiogenomics | 0.745 | 0.550 | 0.820 | 0.980 | 0.120 |
| Hybrid ViT | Lung-PET-CT-Dx | 0.869 | 0.816 | 0.927 | 0.864 | 0.769 |
| NSCLC-Radiomics | 0.617 | 0.580 | 0.208 | 0.638 | 0.521 |
| NSCLC-Radiogenomics | 0.613 | 0.567 | 0.827 | 0.892 | 0.240 |
Table 9.
Cumulative feature importance for each type of feature (first-order statistical, textural and shape-based features) in the (a) 2D Random Forest, (b) 3D Random Forest and (c) 3D XGBoost Classifiers.
Table 9.
Cumulative feature importance for each type of feature (first-order statistical, textural and shape-based features) in the (a) 2D Random Forest, (b) 3D Random Forest and (c) 3D XGBoost Classifiers.
| Model | Type of Features |
|---|
| | First Order Statistical | Textural | Shape-Based |
| 2D Random Forest | 34.6 % | 64.7 % | 0.6 % |
| 3D Random Forest | 28.3 % | 70.7 % | 0.9 % |
| 3D XGBoost | 24.8 % | 73.8 % | 1.4 % |