LuCa: A Novel Method for Lung Cancer Delineation
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Description
2.2. Proposed Method
2.2.1. Data Pre-Processing
2.2.2. U-Net
2.2.3. Three-Dimensional Lung Cancer Delineation
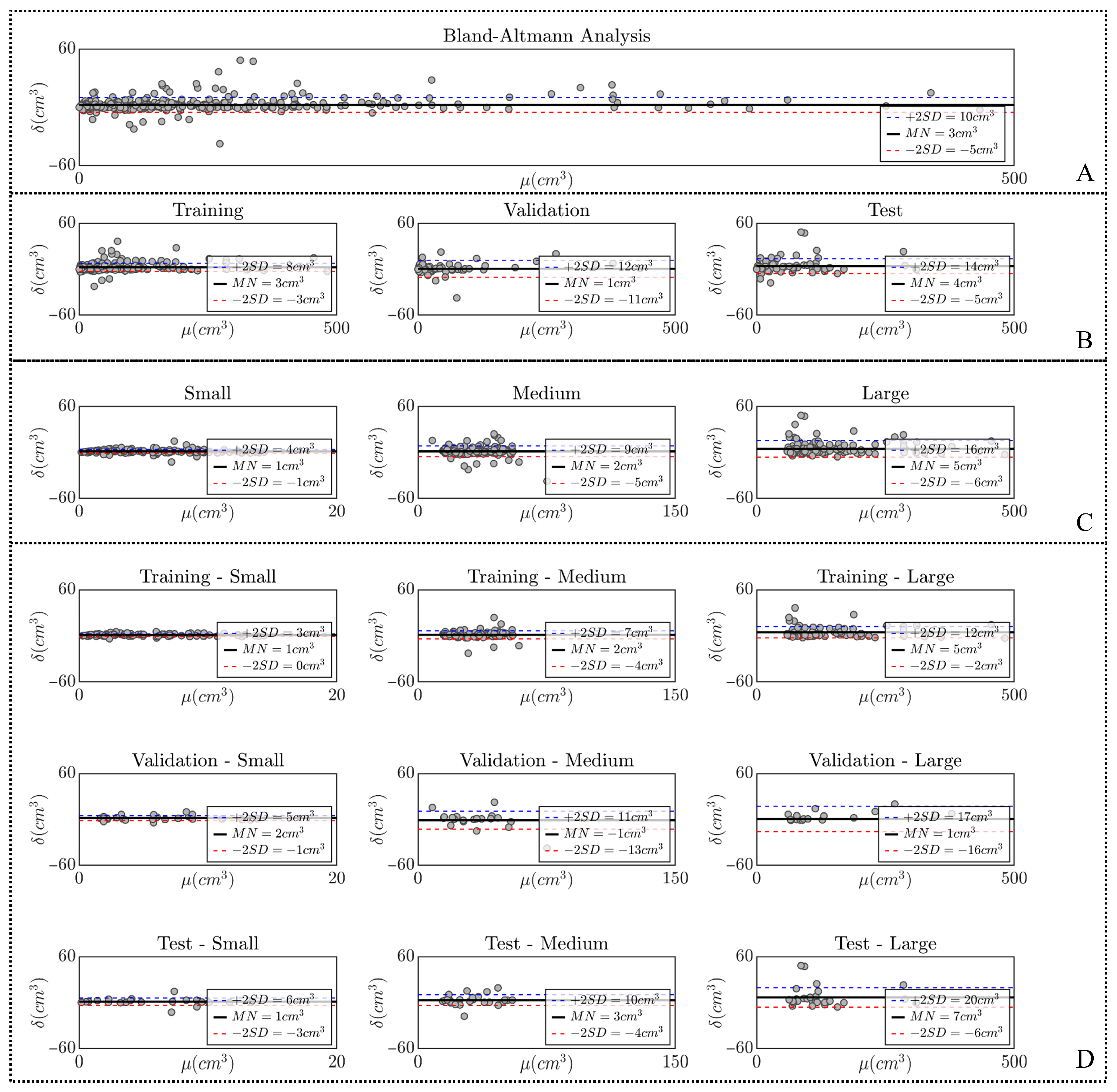
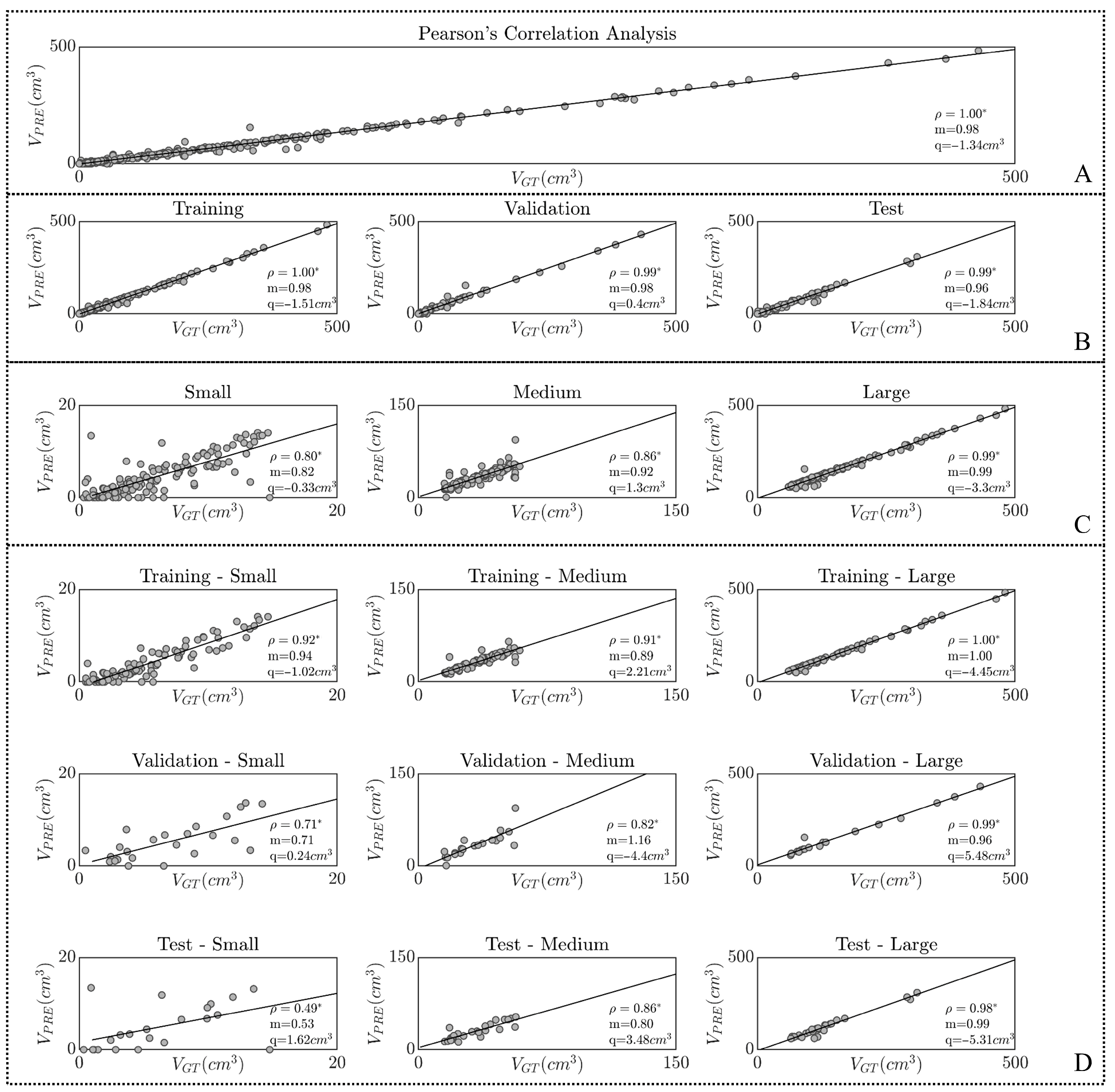
2.3. Statistical Analysis
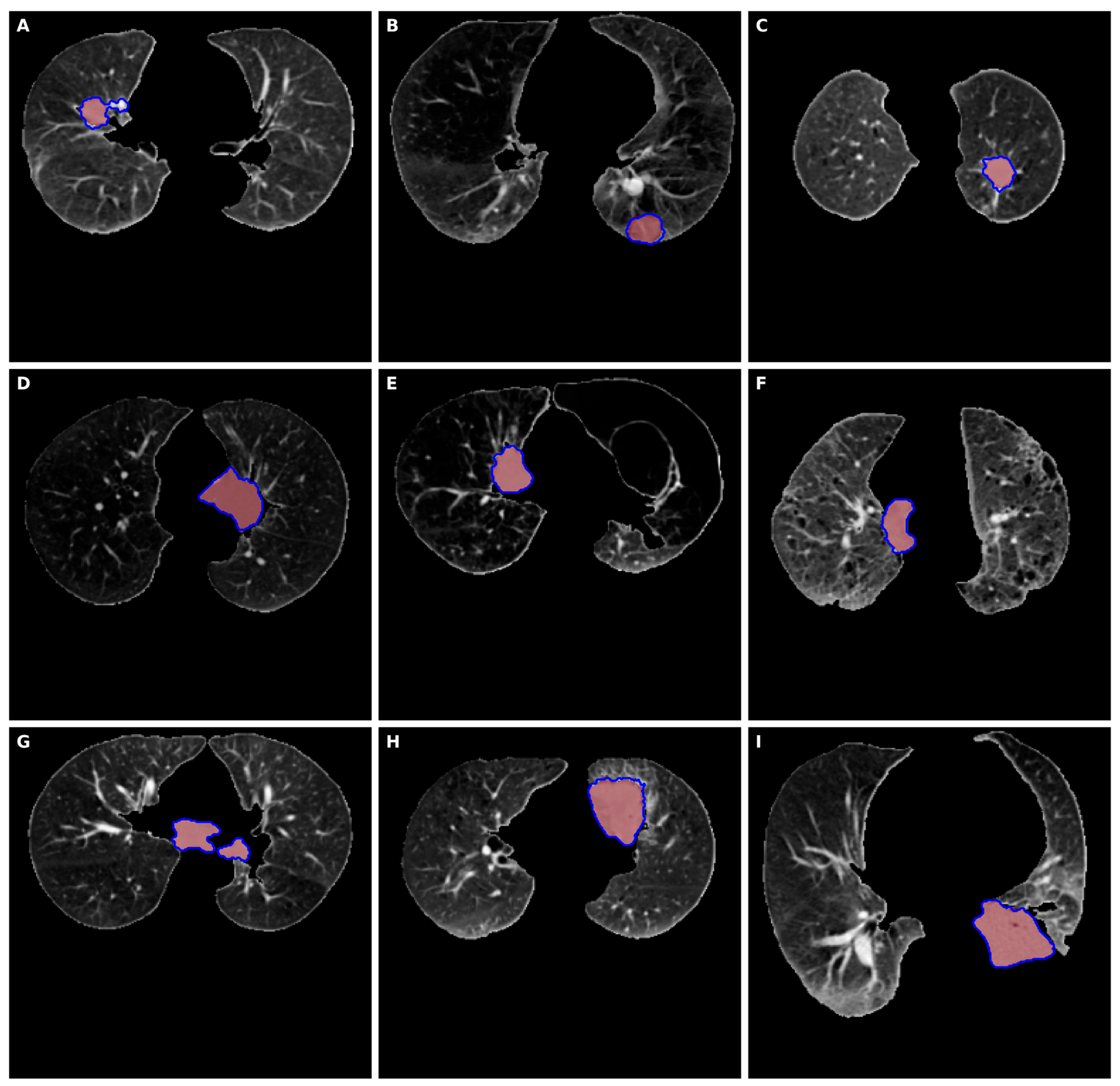
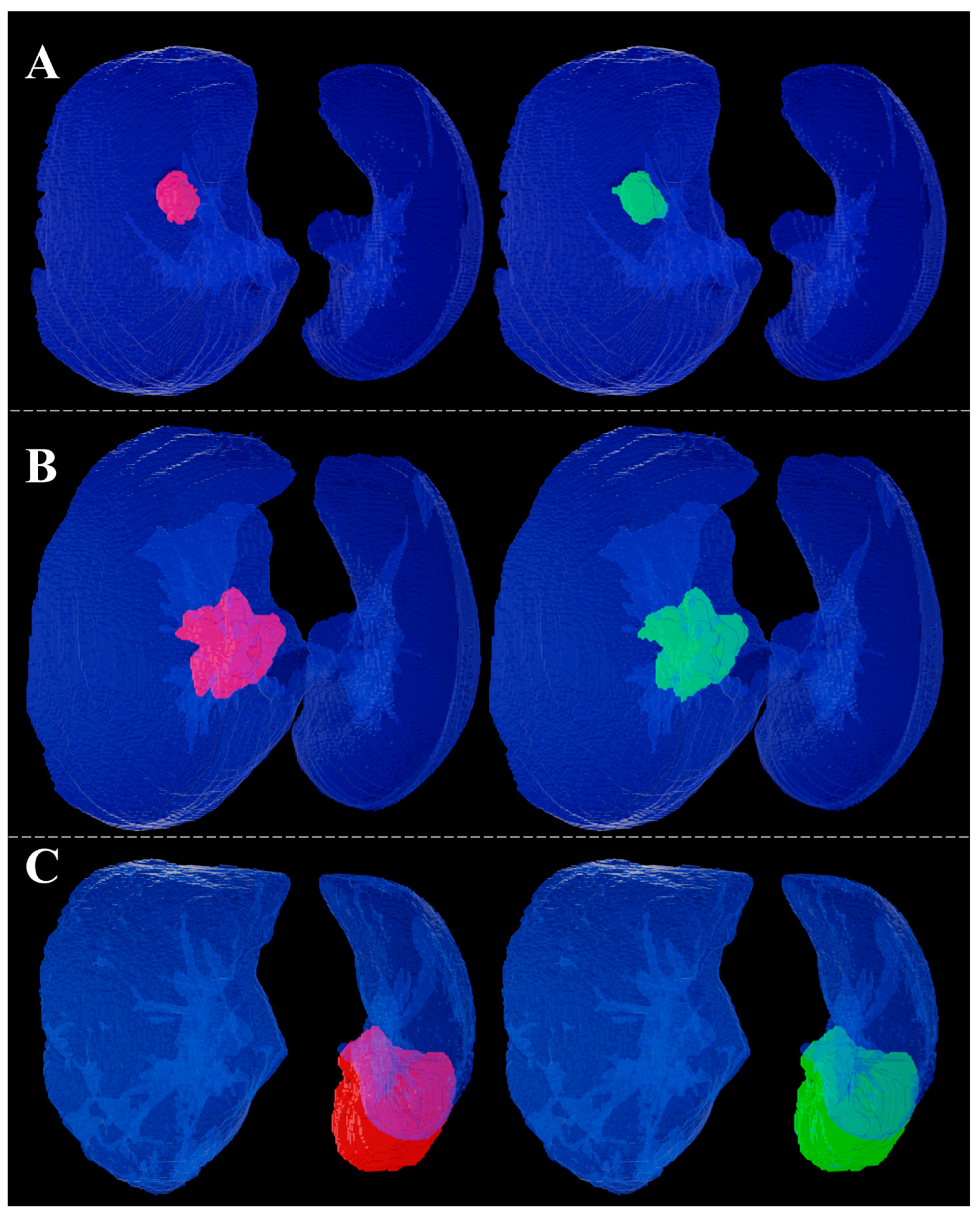
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.A.; Lam, D.C.L.; O’Toole, S.A.; Minna, J.D. Molecular Biology of Lung Cancer. J. Thorac. Dis. 2013, 5, S479–S490. [Google Scholar] [CrossRef] [PubMed]
- Thandra, K.C.; Barsouk, A.; Saginala, K.; Aluru, J.S.; Barsouk, A. Epidemiology of Lung Cancer. Contemp. Oncol. 2021, 25, 45–52. [Google Scholar] [CrossRef]
- Malhotra, J.; Malvezzi, M.; Negri, E.; La Vecchia, C.; Boffetta, P. Risk Factors for Lung Cancer Worldwide. Eur. Respir. J. 2016, 48, 889–902. [Google Scholar] [CrossRef]
- Collins, L.G.; Haines, C.; Perkel, R.; Enck, R.E. Lung Cancer: Diagnosis and Management. Am. Fam. Physician 2007, 75, 56–63. [Google Scholar]
- De Wever, W.; Coolen, J.; Verschakelen, J. Imaging Techniques in Lung Cancer. ERS J. 2011, 7, 338–346. [Google Scholar] [CrossRef]
- Panunzio, A.; Sartori, P. Lung Cancer and Radiological Imaging. Curr. Radiopharm. 2020, 13, 238–242. [Google Scholar] [CrossRef]
- The International Agency for Research on Cancer Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart (IARC WHO Classification of Tumours), 1st ed.; World Health Organization: Lyon, France, 2004.
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Song, Y.; Liu, Y.; Lin, Z.; Zhou, J.; Li, D.; Zhou, T.; Leung, M.-F. Learning from AI-Generated Annotations for Medical Image Segmentation. IEEE Trans. Consum. Electron. 2024, 71, 1473–1481. [Google Scholar] [CrossRef]
- Li, Y.; Hao, W.; Zeng, H.; Wang, L.; Xu, J.; Routray, S.; Jhaveri, R.H.; Gadekallu, T.R. Cross-Scale Texture Supplementation for Reference-Based Medical Image Super-Resolution. IEEE J. Biomed. Health Inform. 2025, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional Neural Networks: An Overview and Application in Radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Annavarapu, C.S.R.; Parisapogu, S.A.B.; Keetha, N.V.; Donta, P.K.; Rajita, G. A Bi-FPN-Based Encoder-Decoder Model for Lung Nodule Image Segmentation. Diagnostics 2023, 13, 1406. [Google Scholar] [CrossRef]
- Banu, S.F.; Sarker, M.M.K.; Abdel Nasser, M.; Rashwan, H.; Puig, D. WEU-Net: A Weight Excitation U-Net for Lung Nodule Segmentation. Appl. Sci. 2021, 101, 349–356. [Google Scholar] [CrossRef]
- Lu, D.; Chu, J.; Zhao, R.; Zhang, Y.; Tian, G. A Novel Deep Learning Network and Its Application for Pulmonary Nodule Segmentation. Comput. Intell. Neurosci. 2022, 2022, 1–6. [Google Scholar] [CrossRef]
- Kunkyab, T.; Bahrami, Z.; Zhang, H.; Liu, Z.; Hyde, D. A Deep Learning-Based Framework (Co-Retr) for Auto-Segmentation of Non-Small Cell Lung Cancer in Computed Tomography Images. J. Appl. Clin. Med. Phys. 2024, 25, e14297. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Q.; Fan, E.; Lu, N.; Chen, D.; Jiang, H.; Yu, Y. Enhancing Non-Small Cell Lung Cancer Tumor Segmentation with a Novel Two-Step Deep Learning Approach. J. Radiat. Res. Appl. Sci. 2024, 17, 100775. [Google Scholar] [CrossRef]
- Zhao, L. 3D Densely Connected Convolution Neural Networks for Pulmonary Parenchyma Segmentation from CT Images. J. Phys. Conf. Ser. 2020, 1631, 12049. [Google Scholar] [CrossRef]
- Agnes, S.A.; Anitha, J. Efficient Multiscale Fully Convolutional UNet Model for Segmentation of 3D Lung Nodule from CT Image. J. Med. Imaging 2022, 9, 052402. [Google Scholar] [CrossRef]
- Weikert, T.; Jaeger, P.F.; Yang, S.; Baumgartner, M.; Breit, H.C.; Winkel, D.J.; Sommer, G.; Stieltjes, B.; Thaiss, W.; Bremerich, J.; et al. Automated Lung Cancer Assessment on 18F-PET/CT Using Retina U-Net and Anatomical Region Segmentation. Eur. Radiol. 2023, 33, 4270–4279. [Google Scholar] [CrossRef]
- Chen, W.; Yang, F.; Zhang, X.; Xu, X.; Qiao, X. MAU-Net: Multiple Attention 3D U-Net for Lung Cancer Segmentation on CT Images. Procedia Comput. Sci. 2021, 192, 543–552. [Google Scholar] [CrossRef]
- Park, J.; Kang, S.K.; Hwang, D.; Choi, H.; Ha, S.; Seo, J.M.; Eo, J.S.; Lee, J.S. Automatic Lung Cancer Segmentation in [18F]FDG PET/CT Using a Two-Stage Deep Learning Approach. Nucl. Med. Mol. Imaging 2023, 57, 86–93. [Google Scholar] [CrossRef]
- Shirokikh, B.; Shevtsov, A.; Dalechina, A.; Krivov, E.; Kostjuchenko, V.; Golanov, A.; Gombolevskiy, V.; Morozov, S.; Belyaev, M. Accelerating 3D Medical Image Segmentation by Adaptive Small-Scale Target Localization. J. Imaging 2021, 7, 35. [Google Scholar] [CrossRef]
- Zhong, Z.; Kim, Y.; Zhou, L.; Plichta, K.; Allen, B.; Buatti, J.; Wu, X. 3D Fully Convolutional Networks for Co-Segmentation of Tumors on PET-CT Images. In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 228–231. [Google Scholar]
- Kido, S.; Kidera, S.; Hirano, Y.; Mabu, S.; Kamiya, T.; Tanaka, N.; Suzuki, Y.; Yanagawa, M.; Tomiyama, N. Segmentation of Lung Nodules on CT Images Using a Nested Three-Dimensional Fully Connected Convolutional Network. Front. Artif. Intell. 2022, 5, 782225. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, C.; Chan, H.-P.; Hadjiiski, L.M.; Chughtai, A.; Kazerooni, E.A. Hybrid U-Net-Based Deep Learning Model for Volume Segmentation of Lung Nodules in CT Images. Med. Phys. 2022, 49, 7287–7302. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, M.; Liu, Z.; Liu, Z.; Gu, D.; Zang, Y.; Dong, D.; Gevaert, O.; Tian, J. Central Focused Convolutional Neural Networks: Developing a Data-Driven Model for Lung Nodule Segmentation. Med. Image Anal. 2017, 40, 172–183. [Google Scholar] [CrossRef]
- The Cancer Imaging Archive NSCLC-Radiomics. Available online: https://www.cancerimagingarchive.net/ (accessed on 10 November 2025).
- Aggarwal, C.C. Neural Networks and Deep Learning; Springer: Cham, Switzerland, 2023. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Springer: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Geppert, J.; Asgharzadeh, A.; Brown, A.; Stinton, C.; Helm, E.J.; Jayakody, S.; Todkill, D.; Gallacher, D.; Ghiasvand, H.; Patel, M.; et al. Software Using Artificial Intelligence for Nodule and Cancer Detection in CT Lung Cancer Screening: Systematic Review of Test Accuracy Studies. Thorax 2024, 79, 1040–1049. [Google Scholar] [CrossRef] [PubMed]

| Stratification | Number | Technical Evaluation | Clinical Assessment | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DC (%) | IoU (%) | SEN (%) | PPV (%) | VGT (cm3) | VPRE (cm3) | ||||
| OVERALL | 391 | 86.8 ± 14.9 | 79.3 ± 19.6 | 93.7 ± 9.7 | 83.6 ± 18.9 | 64.2 ± 79.5 | 61.5 ± 78.3 | ||
| Data division | TRAINING | 249 | 87.4 ± 14.2 | 80.0 ± 19.0 | 94.4 ± 8.0 | 83.7 ± 18.7 | 61.9 ± 79.4 | 59.2 ± 78.2 | |
| VALIDATION | 63 | 82.6 ± 19.4 | 74.2 ± 23.8 | 90.2 ± 14.2 | 81.2 ± 23.1 | 64.6 ± 93.5 | 64.1 ± 92.6 | ||
| TEST | 79 | 88.5 ± 12.4 | 81.2 ± 17.2 | 94.1 ± 9.5 | 85.5 ± 15.7 | 71.3 ± 66.9 | 66.8 ± 65.3 | ||
| Tumor size | SMALL | 135 | 73.3 ± 17.5 | 60.7 ± 21.1 | 89.2 ± 12.0 | 66.4 ± 22.9 | 7.0 ± 3.7 | 5.6 ± 3.9 | |
| MEDIUM | 130 | 89.4 ± 10.9 | 82.1 ± 14.0 | 93.3 ± 9.6 | 87.6 ± 12.8 | 33.8 ± 13 | 32.3 ± 13.8 | ||
| LARGE | 126 | 95.4 ± 5.7 | 91.8 ± 9.0 | 97.8 ± 4.6 | 93.7 ± 8.0 | 142.7 ± 89.9 | 137.9 ± 89.6 | ||
| Subgroups | TRAIN | SMALL | 89 | 73.5 ± 17.2 | 60.9 ± 20.9 | 90.3 ± 10.4 | 65.7 ± 22.5 | 6.6 ± 3.7 | 5.2 ± 3.8 |
| MEDIUM | 85 | 91.5 ± 6.7 | 85.0 ± 10.3 | 94.6 ± 7.4 | 89.5 ± 9.2 | 34.1 ± 12.6 | 32.4 ± 12.4 | ||
| LARGE | 75 | 95.8 ± 4.6 | 92.4 ± 7.6 | 98.2 ± 1.6 | 94.0 ± 7.4 | 145.0 ± 92.3 | 140.1 ± 92.3 | ||
| VALIDATION | SMALL | 24 | 70.2 ± 18.8 | 57.1 ± 22.2 | 86.4 ± 15.8 | 64.8 ± 25.1 | 7.5 ± 4.1 | 5.9 ± 4.1 | |
| MEDIUM | 20 | 83.8 ± 20.3 | 75.6 ± 21.7 | 88.9 ± 14.1 | 85.2 ± 21.6 | 33.1 ± 14.2 | 34.0 ± 20.1 | ||
| LARGE | 19 | 95.2 ± 8.1 | 91.6 ± 11.9 | 95.7 ± 11.1 | 95.2 ± 4.7 | 160.8 ± 117.1 | 160.2 ± 113.8 | ||
| TEST | SMALL | 22 | 77.0 ± 17.4 | 65.3 ± 21 | 87.7 ± 13.0 | 73.0 ± 21.6 | 8.1 ± 3.2 | 7.0 ± 3.9 | |
| MEDIUM | 25 | 86.6 ± 10.3 | 77.7 ± 15.2 | 92.4 ± 11.2 | 83.3 ± 13.5 | 33.6 ± 13.9 | 30.3 ± 12.9 | ||
| LARGE | 32 | 94.7 ± 6.5 | 90.5 ± 10.3 | 98.0 ± 2.3 | 92.2 ± 10.5 | 126.5 ± 61.9 | 119.7 ± 62.5 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carletti, M.; Bruschi, G.; Mortada, M.J.; Burattini, L.; Sbrollini, A. LuCa: A Novel Method for Lung Cancer Delineation. Appl. Sci. 2025, 15, 12074. https://doi.org/10.3390/app152212074
Carletti M, Bruschi G, Mortada MJ, Burattini L, Sbrollini A. LuCa: A Novel Method for Lung Cancer Delineation. Applied Sciences. 2025; 15(22):12074. https://doi.org/10.3390/app152212074
Chicago/Turabian StyleCarletti, Mattia, Giulia Bruschi, MHD Jafar Mortada, Laura Burattini, and Agnese Sbrollini. 2025. "LuCa: A Novel Method for Lung Cancer Delineation" Applied Sciences 15, no. 22: 12074. https://doi.org/10.3390/app152212074
APA StyleCarletti, M., Bruschi, G., Mortada, M. J., Burattini, L., & Sbrollini, A. (2025). LuCa: A Novel Method for Lung Cancer Delineation. Applied Sciences, 15(22), 12074. https://doi.org/10.3390/app152212074










