Two Non-Learning Systems for Profile-Extraction in Images Acquired from a near Infrared Camera, Underwater Environment, and Low-Light Condition
Abstract
1. Introduction
2. Materials and Methods
2.1. Instruments Used
2.2. Frameworks of the Algorithms
| Algorithm 1. Atanh filter. |
|
| (5) |
|
| for each iteration j = 1: a10 do |
| lhj = 2j(lh−1) + 1; |
| lgj = 2j(lg−1) + 1; |
| hj(1: lhj) = 0; |
| gj(1: lgj) = 0; |
| for each iteration n1 = 1:lh do |
| hj(2j(n1−1) + 1) = h(n1); |
| end for |
| for each iteration n1 = 1:lg do |
| gj(2j(n1−1) + 1) = g(n1); |
| end for |
| a(:, :, j + 1) = conv(hj, hj, a(:, :, j)); |
| dx(:, :, j + 1) = conv(delta, gj, a(:, :, j)); |
| dy(:, :, j + 1) = conv(gj, delta, a(:, :, j)); |
| x = dx(:, :, j + 1); |
| y = dy(:, :, j + 1); |
| end for |
| Here, the coefficients from 2 to j + 1 are decomposed. |
|
| Algorithm 2. Sech filter. |
|
|
| for each iteration j2 = 1:J do |
| lhj2 = 2j2(lh2−1) + 1; |
| lgj2 = 2j2(lg2−1) + 1; |
| hj2(1: lhj2) = 0; |
| gj2(1: lgj2) = 0; |
| for each iteration n2 = 1:lh2 do |
| hj2(2j2(n2−1) + 1) = h2(n2); |
| end for |
| for each iteration n2 = 1:lg2 do |
| gj2(2j2(n2−1) + 1) = g2(n2); |
| end for |
| a2(:, :, j2 + 1) = conv(hj2, hj2, a2(:, :, j2)); |
| dx2(:, :, j2 + 1) = conv(delta2, gj2, a2(:, :, j2)); |
| dy2(:, :, j2 + 1) = conv(gj2, delta2, a2(:, :, j2)); |
| x2 = dx2(:, :, j2 + 1); |
| y2 = dy2(:, :, j2 + 1); |
| end for |
| Here, the coefficients from 2 to j2 + 1 are decomposed. The obtained image is shown using the output value of convolution. |
| Algorithm 3. Image fusion. |
|
3. Results
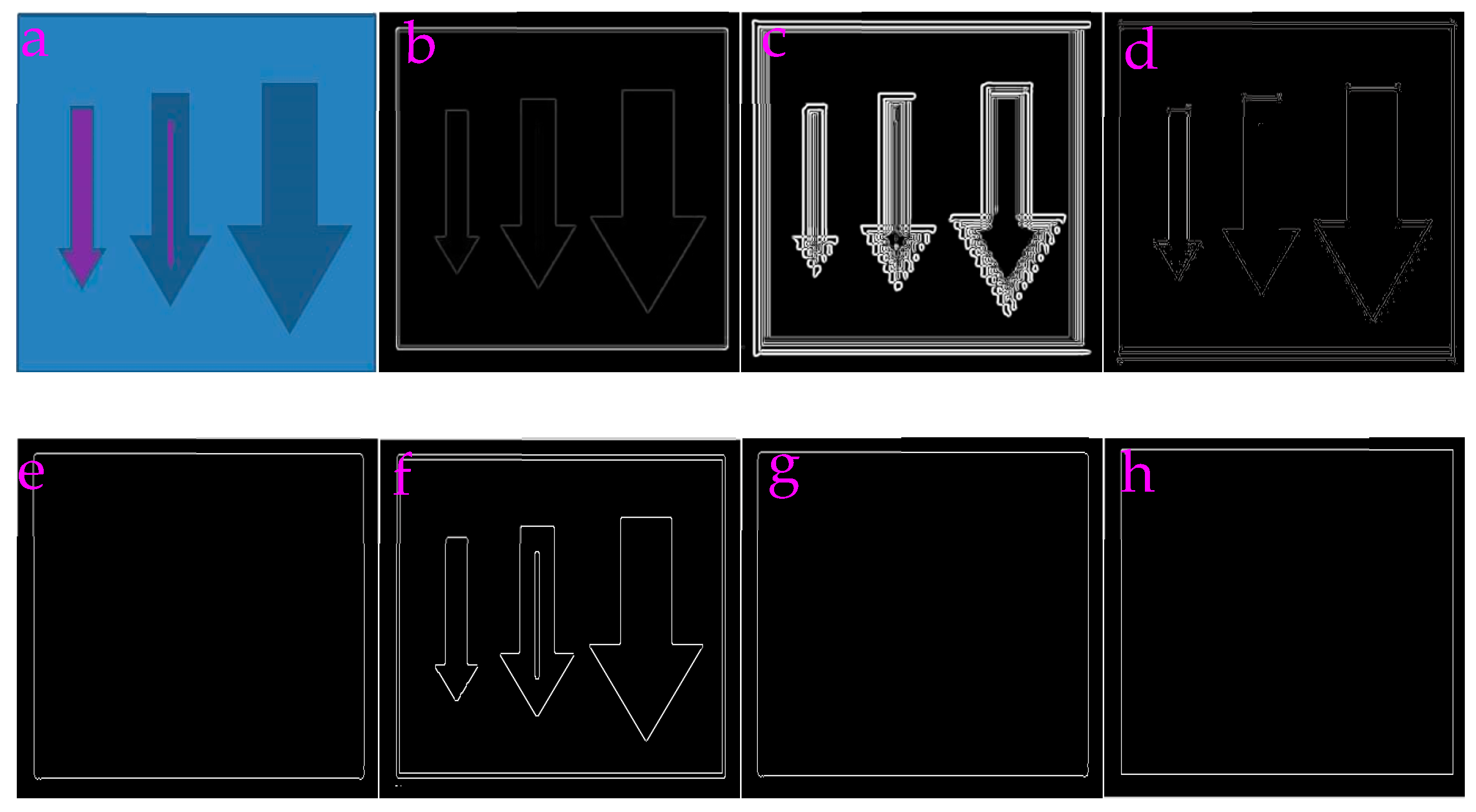
3.1. Capability of Profile-Extraction
3.2. Profile-Extraction of the Images Acquired from near Infrared Camera
3.3. Application for Extracting the Profiles in Underwater Images
3.4. Enhancing Low-Light Images
3.5. Application for Image Fusion
3.6. Application for Detecting the Edge
3.7. Application for Detecting the Array
4. Discussion
4.1. Features of Our Filters
4.2. Potential Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, C.; Zheng, Y.; Long-Iyer, K.; Wright, E.C.; Li, Y.; Tian, L. Fluorescence Imaging of Neural Activity, Neurochemical Dynamics, and Drug-Specific Receptor Conformation with Genetically Encoded Sensors. Annu. Rev. Neurosci. 2022, 45, 273–294. [Google Scholar] [CrossRef]
- Wang, Q.; Li, X.; Qian, B.; Hu, K.; Liu, B. Fluorescence imaging in the surgical management of liver cancers: Current status and future perspectives. Asian J. Surg. 2022, 45, 1375–1382. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, T.; Wang, X.; Li, J.; Zhao, H. Real-time near-infrared fluorescence imaging mediated by blue dye in breast cancer patients. J. Surg. Oncol. 2020, 121, 964–966. [Google Scholar] [CrossRef]
- Marsden, M.; Weaver, S.S.; Marcu, L.; Campbell, M.J. Intraoperative Mapping of Parathyroid Glands Using Fluorescence Lifetime Imaging. J. Surg. Res. 2021, 265, 42–48. [Google Scholar] [CrossRef]
- Huh, W.K.; Johnson, J.L.; Elliott, E.; Boone, J.D.; Leath, C.A., 3rd; Kovar, J.L.; Kim, K.H. Fluorescence Imaging of the Ureter in Minimally Invasive Pelvic Surgery. J. Minim. Invasive Gynecol. 2021, 28, 332–341.e14. [Google Scholar] [CrossRef]
- Paraboschi, I.; De Coppi, P.; Stoyanov, D.; Anderson, J.; Giuliani, S. Fluorescence imaging in pediatric surgery: State-of-the-art and future perspectives. J. Pediatr. Surg. 2021, 56, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Lauwerends, L.J.; van Driel, P.B.A.A.; Baatenburg, d.J.R.J.; Hardillo, J.A.U.; Koljenovic, S.; Puppels, G.; Mezzanotte, L.; Lowik, C.W.G.M.; Rosenthal, E.L.; Vahrmeijer, A.L.; et al. Real-time fluorescence imaging in intraoperative decision making for cancer surgery. Lancet Oncol. 2021, 22, e186–e195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, K.; Chi, C.; Hu, Z.; Tian, J. Intraoperative fluorescence molecular imaging accelerates the coming of precision surgery in China. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 2531–2543. [Google Scholar] [CrossRef] [PubMed]
- de Wit, J.G.; Vonk, J.; Voskuil, F.J.; de Visscher, S.A.H.J.; Schepman, K.P.; Hooghiemstra, W.T.R.; Linssen, M.D.; Elias, S.G.; Halmos, G.B.; Plaat, B.E.C.; et al. EGFR-targeted fluorescence molecular imaging for intraoperative margin assessment in oral cancer patients: A phase II trial. Nat. Commun. 2023, 14, 4952. [Google Scholar] [CrossRef]
- Hao, H.; Wang, X.; Qin, Y.; Ma, Z.; Yan, P.; Liu, C.; Chen, G.; Yang, X. Ex vivo near-infrared targeted imaging of human bladder carcinoma by ICG-anti-CD47. Front. Oncol. 2023, 13, 1083553. [Google Scholar] [CrossRef]
- Sun, Y.; Zhong, X.; Dennis, A.M. Minimizing near-infrared autofluorescence in preclinical imaging with diet and wavelength selection. J. Biomed. Opt. 2023, 28, 094805. [Google Scholar] [CrossRef] [PubMed]
- George, M.B.; Lew, B.; Blair, S.; Zhu, Z.; Liang, Z.; Srivastava, I.; Chang, A.; Choi, H.; Kim, K.; Nie, S.; et al. Bioinspired color-near infrared endoscopic imaging system for molecular guided cancer surgery. J. Biomed. Opt. 2023, 28, 056002. [Google Scholar] [CrossRef]
- Okusanya, O.T.; Holt, D.; Heitjan, D.; Deshpande, C.; Venegas, O.; Jiang, J.; Judy, R.; DeJesus, E.; Madajewski, B.; Oh, K.; et al. Intraoperative near-infrared imaging can identify pulmonary nodules. Ann. Thorac. Surg. 2014, 98, 1223–1230. [Google Scholar] [CrossRef]
- Keating, J.J.; Runge, J.J.; Singhal, S.; Nims, S.; Venegas, O.; Durham, A.C.; Swain, G.; Nie, S.; Low, P.S.; Holt, D.E. Intraoperative near-infrared fluorescence imaging targeting folate receptors identifies lung cancer in a large-animal model. Cancer 2017, 123, 1051–1060. [Google Scholar] [CrossRef]
- Cheng, S.; Jin, Z.; Wu, X.; Liang, J. Transmission map and background light guided enhancement of unpaired underwater image. Neurocomputing 2025, 621, 129270. [Google Scholar] [CrossRef]
- Fu, C.; Liu, R.; Fan, X.; Chen, P.; Fu, H.; Yuan, W.; Zhu, M.; Luo, Z. Rethinking general underwater object detection: Datasets, Challenges, and solutions. Neurocomputing 2023, 517, 243–256. [Google Scholar] [CrossRef]
- Saleh, A.; Sheaves, M.; Jerry, D.; Azghadi, M.R. Adaptive deep learning framework for robust unsupervised underwater image enhancement. Expert Sys. Appl. 2025, 268, 126314. [Google Scholar] [CrossRef]
- Ye, B.; Jin, S.; Li, B.; Yan, S.; Zhang, D. Dual Histogram Equalization Algorithm Based on Adaptive Image Correction. Appl. Sci. 2023, 13, 10649. [Google Scholar] [CrossRef]
- Zarie, M.; Parsayan, A.; Hajghassem, H. Image contrast enhancement using triple clipped dynamic histogram equalization based on standard deviation. IET Image Process. 2019, 13, 1081–1089. [Google Scholar] [CrossRef]
- Rao, B.S. Dynamic Histogram Equalization for contrast enhancement for digital images. Appl. Softw. Comput. 2020, 89, 106114. [Google Scholar] [CrossRef]
- Paul, A.; Sutradhar, T.; Bhattacharya, P.; Maity, S.P. Infrared images enhancement using fuzzy dissimilarity histogram equalization. Optik 2021, 247, 167887. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, Z.; Jiang, D.; Tong, X.; Tao, B.; Jiang, G.; Kong, J.; Yun, J.; Liu, Y.; Liu, X.; et al. Low-Illumination Image Enhancement Algorithm Based on Improved Multi-Scale Retinex and ABC Algorithm Optimization. Front. Bioeng. Biotechnol. 2022, 10, 865820. [Google Scholar] [CrossRef]
- Liu, D.; Chang, F.; Zhang, H.; Liu, L. Level set method with Retinex-corrected saliency embedded for image segmentation. IET Image Process. 2021, 15, 1530–1541. [Google Scholar] [CrossRef]
- Wen, C.; Nie, T.; Li, M.; Wang, X.; Huang, L. Image Restoration via Low-Illumination to Normal-Illumination Networks Based on Retinex Theory. Sensors 2023, 23, 8442. [Google Scholar] [CrossRef] [PubMed]
- Chao, K.; Song, W.; Shao, S.; Liu, D.; Liu, X.; Zhao, X. CUI-Net: A correcting uneven illumination net for low-light image enhancement. Sci. Rep. 2023, 13, 12894. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Chen, X.; Ren, K.; Miao, X.; Chen, Z.; Jin, Y. Low-light image enhancement via adaptive frequency decomposition network. Sci. Rep. 2023, 13, 14107. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wen, C.; Liu, W.; He, W. A depth iterative illumination estimation network for low-light image enhancement based on retinex theory. Sci. Rep. 2023, 13, 19709. [Google Scholar] [CrossRef]
- Latke, V.; Narawade, V. Detection of dental periapical lesions using retinex based image enhancement and lightweight deep learning model. Image Vis. Comput. 2024, 146, 105016. [Google Scholar] [CrossRef]
- Liu, C.; Wang, Z.; Birch, P.; Wang, X. Efficient Retinex-based framework for low-light image enhancement without additional networks. IEEE Trans. Circuits Syst. Video Technol. 2025, 35, 4896–4909. [Google Scholar] [CrossRef]
- Liu, R.; Ma, L.; Zhang, J.; Fan, X.; Luo, Z. Retinex-inspired unrolling with cooperative prior architecture search for low-light image enhancement. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021. [Google Scholar]
- Liu, J. Retinex-based Lightweight Network for Low Light Image Enhancement. In Proceedings of the 2024 IEEE 6th International Conference on Power, Intelligent Computing and Systems (ICPICS), Shenyang, China, 26–28 July 2024; pp. 109–115. [Google Scholar]
- Lin, Y.H.; Yu, C.M.; Wu, C.Y. Towards the Design and Implementation of an Image-Based Navigation System of an Autonomous Underwater Vehicle Combining a Color Recognition Technique and a Fuzzy Logic Controller. Sensors 2021, 21, 4053. [Google Scholar] [CrossRef]
- Tang, Y.; Qiu, J.; Gao, M. Fuzzy Medical Computer Vision Image Restoration and Visual Application. Comput. Math. Methods Med. 2022, 2022, 6454550. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, J.; Huang, S.; Fang, Y.; Lin, P.; Que, Y. Multimodal Medical Image Fusion Based on Fuzzy Discrimination with Structural Patch Decomposition. IEEE J. Biomed. Health Inform. 2019, 23, 1647–1660. [Google Scholar] [CrossRef] [PubMed]
- Alphonse, A.S.; Benifa, J.V.B.; Muaad, A.Y.; Chola, C.; Heyat, M.B.B.; Murshed, B.A.H.; Samee, N.A.; Alabdulhafith, M.; Al-antari, M.A. A Hybrid Stacked Restricted Boltzmann Machine with Sobel Directional Patterns for Melanoma Prediction in Colored Skin Images. Diagnostics 2023, 13, 1104. [Google Scholar] [CrossRef]
- Sharifrazi, D.; Alizadehsani, R.; Roshanzamir, M.; Joloudari, J.H.; Shoeibi, A.; Jafari, M.; Hussain, S.; Sani, Z.A.; Hasanzadeh, F.; Khozeimeh, F.; et al. Fusion of convolution neural network, support vector machine and Sobel filter for accurate detection of COVID-19 patients using X-ray images. Biomed. Signal Process. Control. 2021, 68, 102622. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Ma, Y. SAR minimum entropy autofocusing based on Prewitt operator. PLoS ONE 2023, 18, e0276051. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yan, S.; Lu, N.; Yang, D.; Lv, H.; Wang, S.; Zhu, X.; Zhao, Y.; Wang, Y.; Ma, Z.; et al. Automated retinal boundary segmentation of optical coherence tomography images using an improved Canny operator. Sci. Rep. 2022, 12, 1412. [Google Scholar] [CrossRef]
- Haq, I.; Anwar, S.; Shah, K.; Khan, M.T.; Sah, S.A. Fuzzy Logic Based Edge Detection in Smooth and Noisy Clinical Images. PLoS ONE 2015, 10, e0138712. [Google Scholar] [CrossRef]
- Jia, M.; Xu, J.; Yang, R.; Li, Z.; Zhang, L.; Wu, Y. Three filters for the enhancement of the images acquired from fluorescence microscope and weak-light-sources and the image compression. Heliyon 2023, 9, e20191. [Google Scholar] [CrossRef]
- Chaudhuri, S.; Chatterjee, S.; Katz, N.; Nelson, M.; Goldbaum, M. Detection of blood vessels in retinal images using two-dimensional matched filters. IEEE Trans. Med. Imaging 1989, 8, 263–269. [Google Scholar] [CrossRef]
- Sun, Y.; Jin, Y.; Chen, X.; Xu, Y.; Yan, X.; Liu, Z. Unsupervised detail and color restorer for Retinex-based low-light image enhancement. Eng. Appl. Artif. Intell. 2025, 153, 110867. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, R.; Geng, X.; Li, Z.; Wu, Y. Two filters for acquiring the profiles from images obtained from weak-light background, fluorescence microscope, transmission electron microscope, and near-infrared camera. Sensors 2023, 23, 6207. [Google Scholar] [CrossRef]
- Okusanya, O.T.; DeJesus, E.M.; Jiang, J.X.; Judy, R.P.; Venegas, O.G.; Deshpande, C.G.; Heitjan, D.F.; Nie, S.; Low, P.S.; Singhal, S. Intraoperative molecular imaging can identify lung adenocarcinomas during pulmonary resection. J. Thorac. Cardiovasc. Surg. 2015, 150, 28–35. [Google Scholar] [CrossRef]
- Predina, J.D.; Okusanya, O.; Newton, A.D.; Low, P.; Singhal, S. Standardization and Optimization of Intraoperative Molecular Imaging for Identifying Primary Pulmonary Adenocarcinomas. Mol. Imaging Biol. 2018, 20, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.D.; Predina, J.D.; Frenzel-Sulyok, L.G.; Low, P.S.; Singhal, S.; Roses, R.E. Intraoperative Molecular Imaging Utilizing a Folate Receptor-Targeted Near-Infrared Probe Can Identify Macroscopic Gastric Adenocarcinomas. Mol. Imaging Biol. 2021, 23, 11–17. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Lin, Y.; Alam, M.Z.; Wu, Y. Synthesizing Ag+: MgS, Ag+: Nb2S5, Sm3+: Y2S3, Sm3+: Er2S3, and Sm3+: ZrS2 Compound Nanoparticles for Multicolor Fluorescence Imaging of Biotissues. ACS Omega 2020, 5, 32868–32876. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ou, P.; Fronczek, F.R.; Song, J.; Lin, Y.; Wen, H.-M.; Xu, J. Simultaneous Enhancement of Near-Infrared Emission and Dye Photodegradation in a Racemic Aspartic Acid Compound via MetalIon Modification. ACS Omega 2019, 4, 19136–19144. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lin, Y.; Xu, J. Synthesis of Ag-Ho, Ag-Sm, Ag-Zn, Ag-Cu, Ag-Cs, Ag-Zr, Ag-Er, Ag-Y and Ag-Co metal organic nanoparticles for UV-Vis-NIR wide-range bio-tissue imaging. Photochem. Photobiol. Sci. 2019, 18, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ou, P.; Song, J.; Zhang, L.; Lin, Y.; Song, P.; Xu, J. Synthesis of praseodymium-and molybdenum- sulfide nanoparticles for dye-photodegradation and near-infrared deep-tissue imaging. Mater. Res. Express 2020, 7, 036203. [Google Scholar] [CrossRef]
- Fang, Z.; Wu, Q.; Huang, D.; Guan, D. An Improved DCP-Based Image Defogging Algorithm Combined with Adaptive Fusion Strategy. Math. Probl. Eng. 2021, 2021, 1436255. [Google Scholar] [CrossRef]
- Li, D.; Zhou, J.; Wang, S.; Zhang, D.; Zhang, W.; Alwadai, R.; Alenezi, F.; Tiwari, P.; Shi, T. Adaptive weighted multiscale retinex for underwater image enhancement. Eng. Appl. Artif. Intell. 2023, 123, 106457. [Google Scholar] [CrossRef]
- Zhang, W.; Dong, L.; Xu, W. Retinex-inspired color correction and detail preserved fusion for underwater image enhancement. Comput. Electron. Agric. 2022, 192, 106585. [Google Scholar] [CrossRef]
- Zhang, X.; Stramski, D.; Reynolds, R.A.; Blocker, E.R. Light scattering by pure water and seawater: The depolarization ratio and its variation with salinity. Appl. Opt. 2019, 58, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Galaktionov, I.; Nikitin, A.; Sheldakova, J.; Toporovsky, V.; Kudryashov, A. Focusing of a laser beam passed through a moderately scattering medium using phase-only spatial light modulator. Photonics 2022, 9, 296. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, X.; Xiong, Y.; Gray, D.J.; He, M.-X. Variability of relationship between the volume scattering function at 180° and the backscattering coefficient for aquatic particles. Appl. Opt. 2020, 59, C31–C34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, L.; Xiong, Y.; Huot, Y.; Gray, D. Experimental Estimates of Optical Backscattering Associated With Submicron Particles in Clear Oceanic Waters. Geophys. Res. Lett. 2020, 47, e2020GL087100. [Google Scholar] [CrossRef]
- Zhao, Y.; Poulin, C.; Mckee, D.; Hu, L.; Agagliate, J.; Yang, P.; Zhang, X. A closure study of ocean inherent optical properties using flow cytometry measurements. J. Quant. Spectrosc. Radiat. Transf. 2020, 241, 106730. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, X.; Xiong, Y.; He, M.-X. Calibration of the LISST-VSF to derive the volume scattering functions in clear waters. Opt. Express 2019, 27, A1188–A1206. [Google Scholar] [CrossRef]
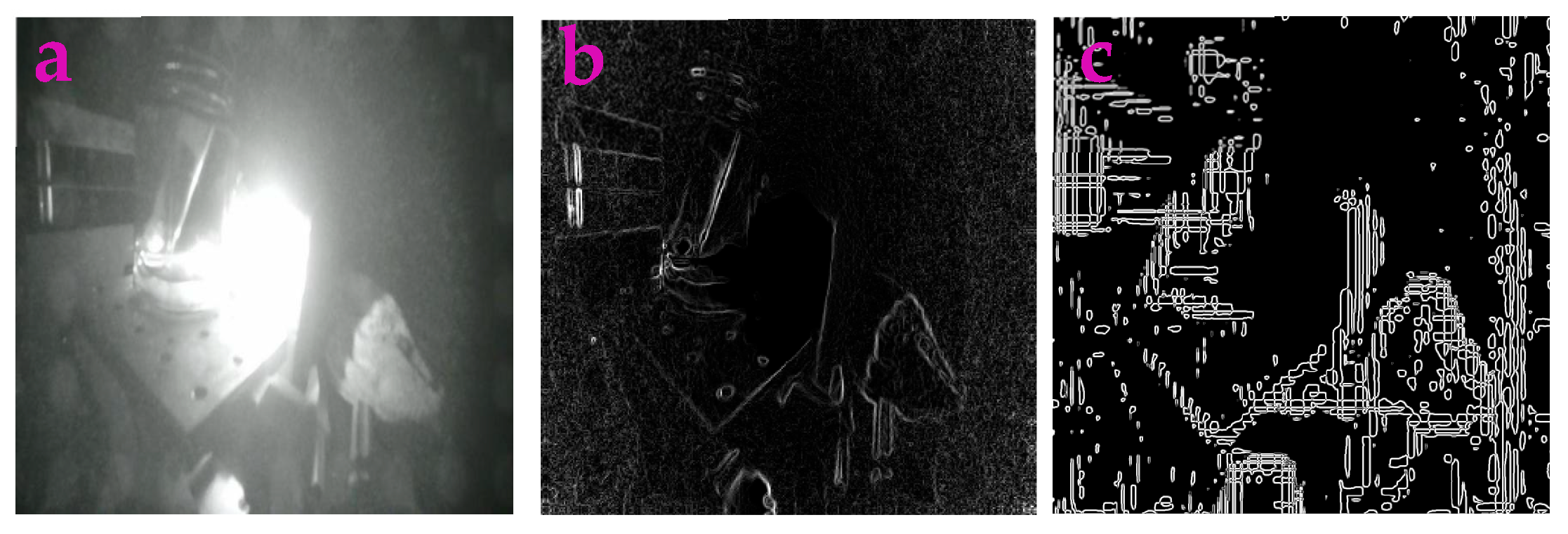
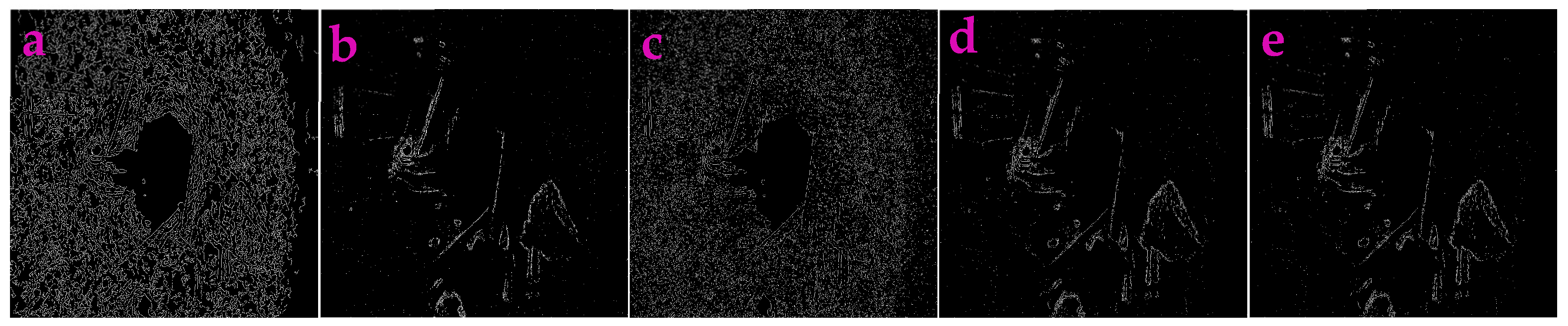
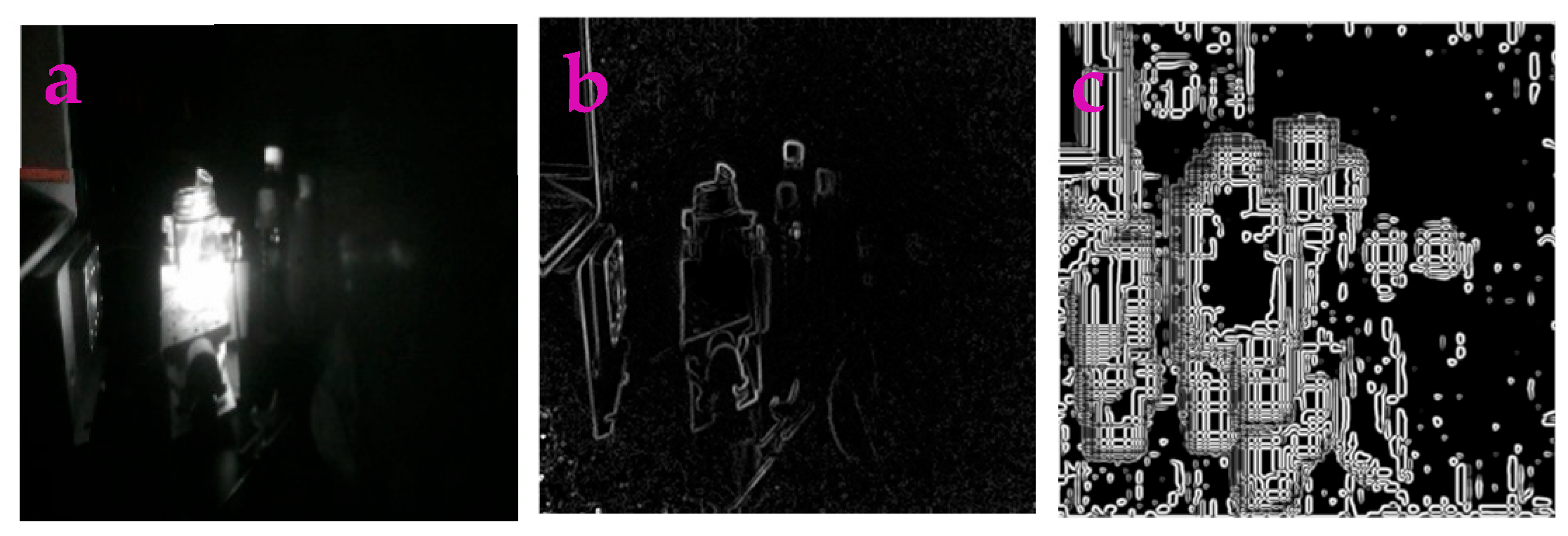
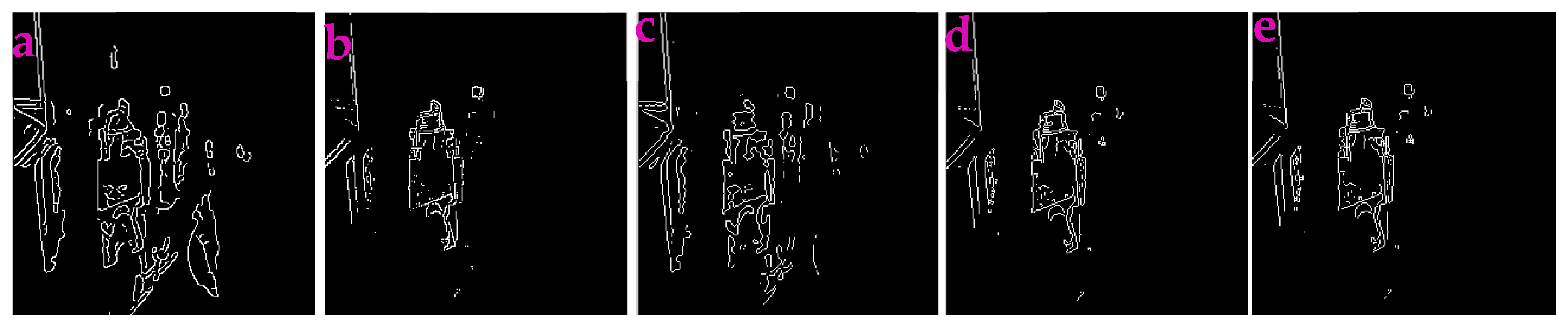



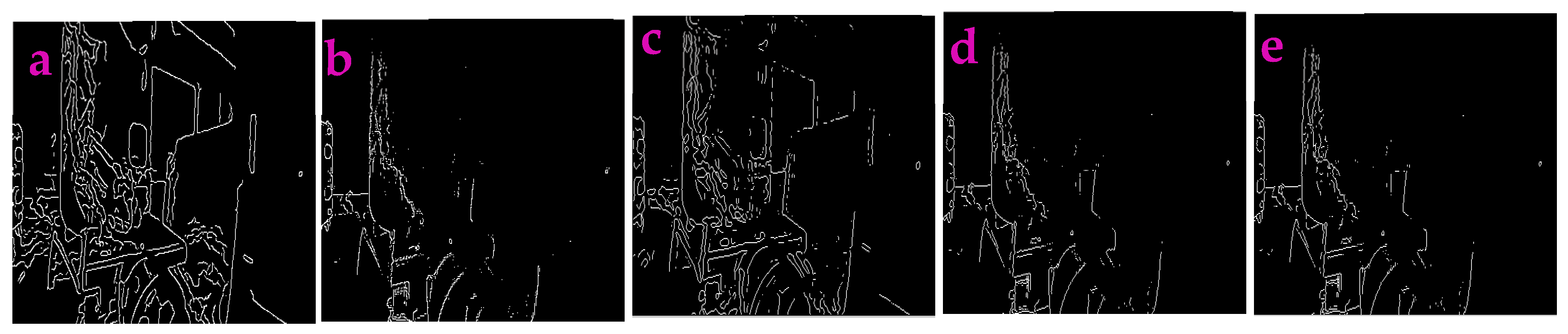

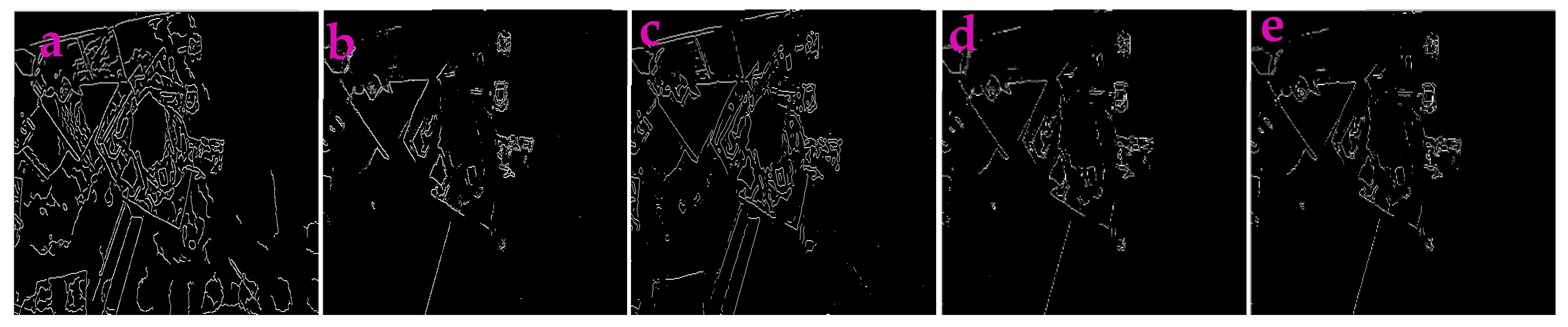

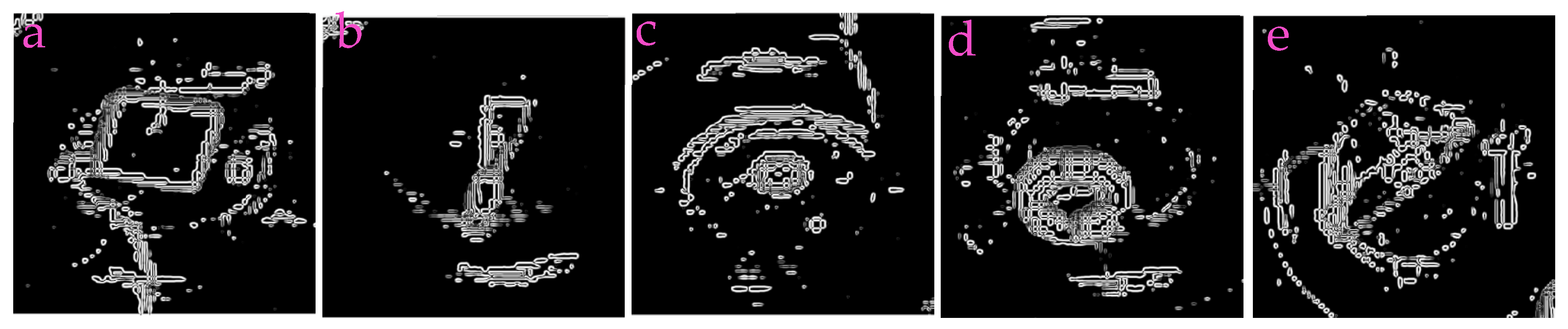
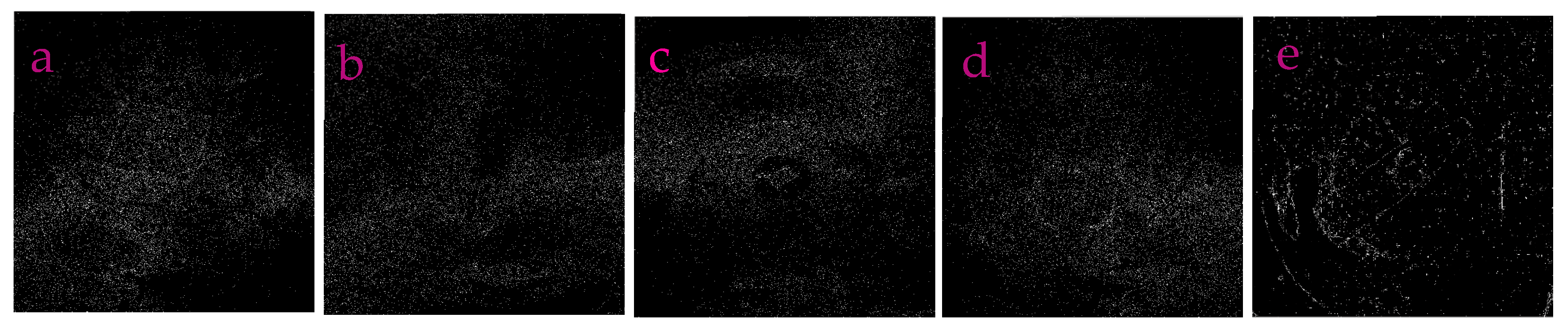


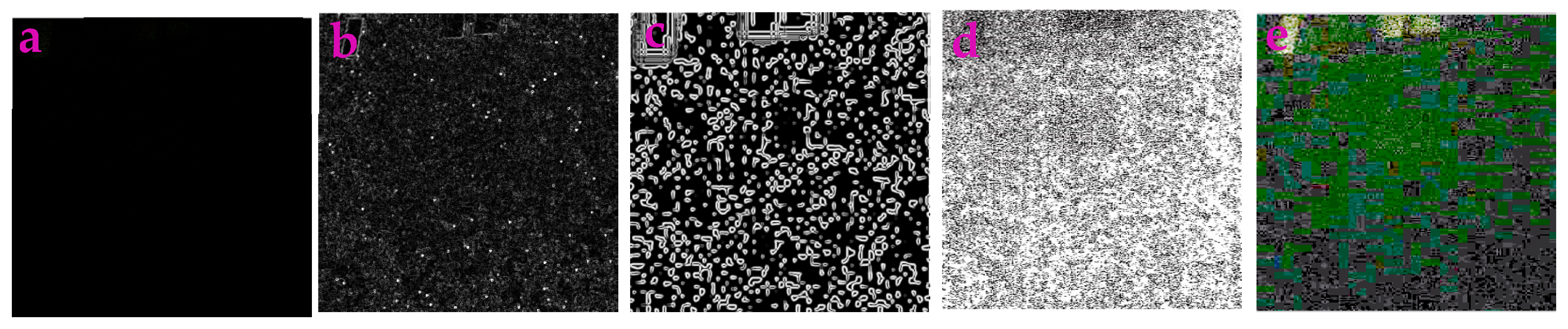

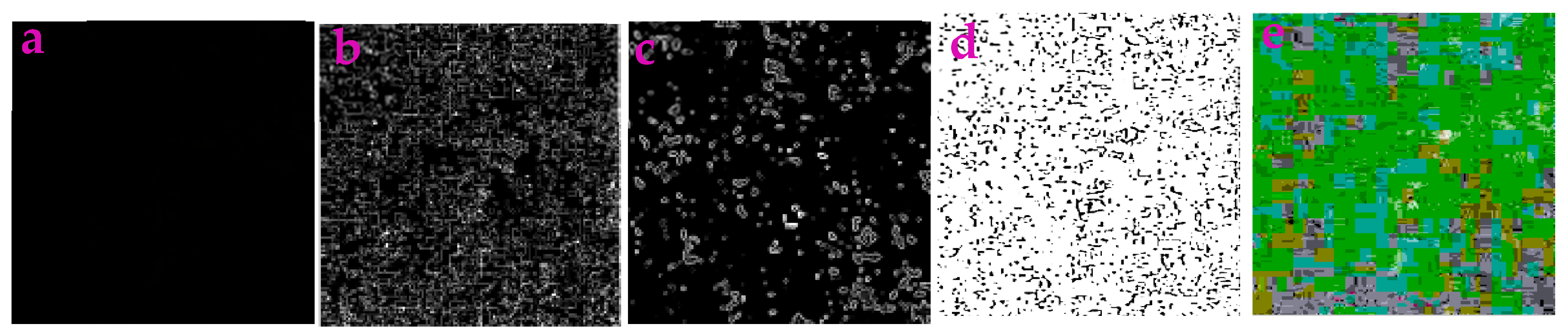



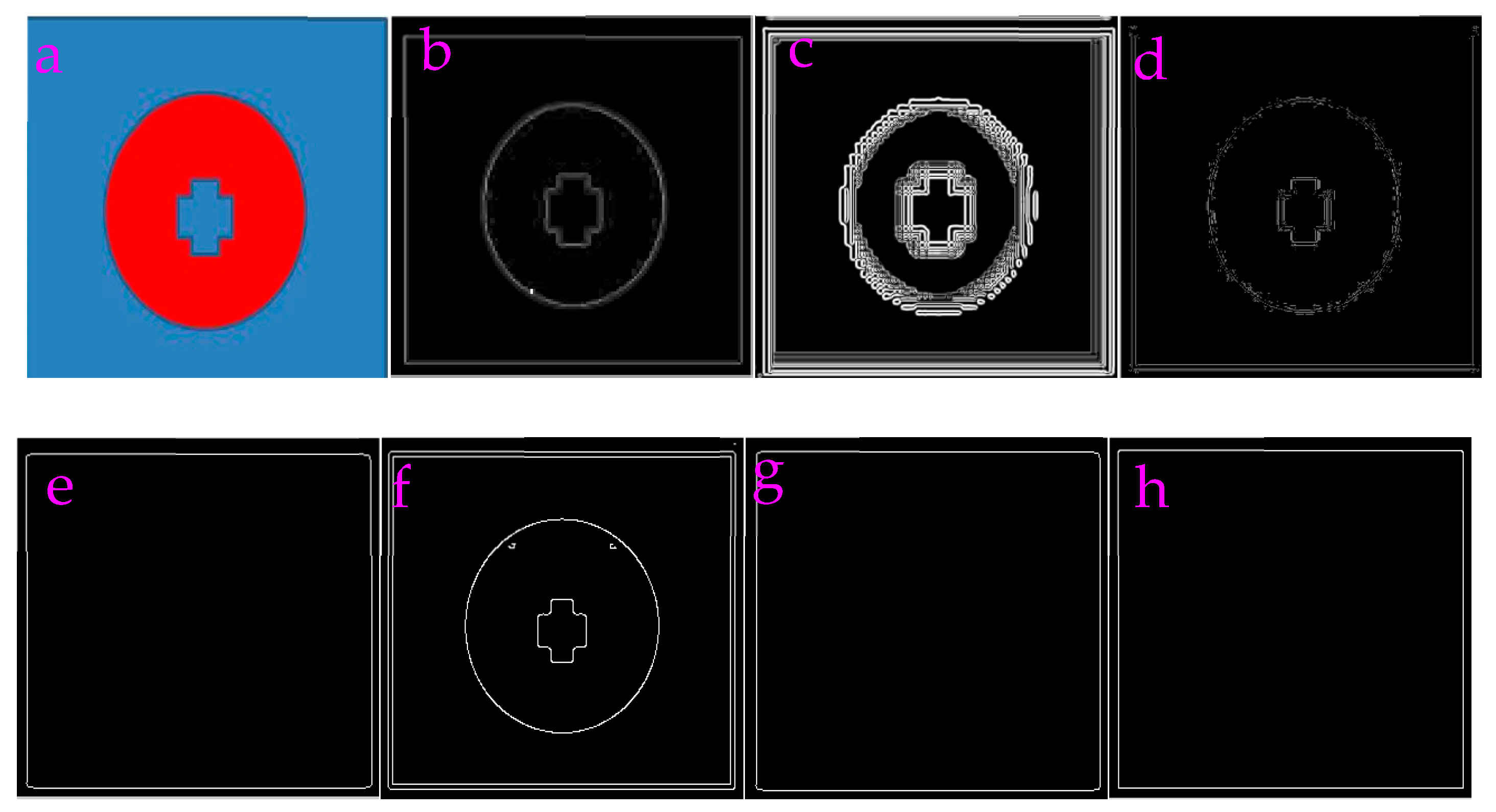



| Filters/Operators | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 65.4637 | 0.996 s | 728.9 MB | 0.3140 | 24.3794 | 0.9302 |
| Sech | 11.4179 | 1.009 s | 602.3 MB | 0.7784 | 24.8438 | 0.8359 |
| Canny | 0 | 0.199 s | 520.3 MB | 0 | 24.0654 | 1 |
| Roberts | 0 | 0.172 s | 510.3 MB | 0 | 24.0654 | 1 |
| Log | 0 | 0.253 s | 510.4 MB | 0 | 24.0654 | 1 |
| Sobel | 0 | 0.135 s | 487.1 MB | 0 | 24.0654 | 1 |
| Prewitt | 0 | 0.143 s | 503.6 MB | 0 | 24.0654 | 1 |
| Filters/Operators | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 9.9236 | 1.097 s | 785.4 MB | 2.9209 | 28.8381 | 0.5104 |
| Sech | 32.0996 | 1.049 s | 643.0 MB | 3.0511 | 28.9682 | 0.4953 |
| Canny | 0 | 0.178 s | 515.5 MB | 0.0019 | 25.9191 | 0.9996 |
| Roberts | 0 | 0.135 s | 516.2 MB | 6.7269 × 10−5 | 25.9173 | 1 |
| Log | 0 | 0.149 s | 515.9 MB | 0.0094 | 25.9266 | 0.9978 |
| Sobel | 0 | 0.140 s | 516.5 MB | 5.4751 × 10−5 | 25.9172 | 1 |
| Prewitt | 0 | 0.134 s | 540.3 MB | 5.1612 × 10−5 | 25.9172 | 1 |
| Filters/Operators | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 9.2901 | 1.128 s | 781.6 MB | 1.006 | 26.9232 | 0.7932 |
| Sech | 13.4065 | 1.019 s | 652.4 MB | 0.4059 | 24.4713 | 0.9108 |
| Canny | 0 | 0.171 s | 529.1 MB | 0 | 24.0654 | 1 |
| Roberts | 0 | 0.132 s | 540.4 MB | 0 | 24.0654 | 1 |
| Log | 0 | 0.163 s | 537.9 MB | 0 | 24.0654 | 1 |
| Sobel | 0 | 0.139 s | 523.1 MB | 0 | 24.0654 | 1 |
| Prewitt | 0 | 0.135 s | 527.6 MB | 0 | 24.0654 | 1 |
| Filters/Operators | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 9.7894 | 1.104 s | 612.1 MB | 0.0349 | 24.1003 | 0.9920 |
| Sech | 18.4000 | 1.120 s | 484.2 MB | 1.4380 | 25.5034 | 0.7181 |
| Canny | 0 | 0.169 s | 370.8 MB | 0 | 24.0654 | 1 |
| Roberts | 0 | 0.132 s | 365.1 MB | 0 | 24.0654 | 1 |
| Log | 0 | 0.139 s | 374.8 MB | 0 | 24.0654 | 1 |
| Sobel | 0 | 0.138 s | 376.8 MB | 0 | 24.0654 | 1 |
| Prewitt | 0 | 0.134 s | 387.9 MB | 0 | 24.0654 | 1 |
| Filters/Operators | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 9.7894 | 1.121 s | 658.9 MB | 0.0427 | 24.1081 | 0.9902 |
| Sech | 18.4000 | 1.046 s | 525.1 MB | 0.9717 | 25.0371 | 0.7998 |
| Canny | 0 | 0.160 s | 397.7 MB | 0 | 24.0654 | 1 |
| Roberts | 0 | 0.130 s | 397.6 MB | 4.2977 × 10−7 | 24.0654 | 1 |
| Log | 0 | 0.139 s | 397.4 MB | 0 | 24.0654 | 1 |
| Sobel | 0 | 0.152 s | 397.2 MB | 0 | 24.0654 | 1 |
| Prewitt | 0 | 0.138 s | 396.9 MB | 4.2977 × 10−7 | 24.0654 | 1 |
| Parameters | Water26 | Water27 | Water30 | Water32 | Water38 |
|---|---|---|---|---|---|
| ME | 15.9886 | 3.3147 | 3.1197 | 19.5336 | 7.8664 |
| Running time | 4.229 s | 1.918 s | 2.163 s | 2.006 s | 1.697 s |
| Memory usage | 771.7 MB | 771.8 MB | 771.9 MB | 772.2 MB | 772.7 MB |
| SNR | 0.2271 | 0.0584 | 0.0878 | 0.1203 | 0.9862 |
| PSNR | 24.2925 | 24.1238 | 24.1533 | 24.1857 | 25.0570 |
| NMSE | 0.9490 | 0.9866 | 0.9800 | 0.9727 | 0.7969 |
| Parameter | Water26 | Water27 | Water30 | Water32 | Water38 |
|---|---|---|---|---|---|
| ME | 6.5804 | 3.4154 | 5.2177 | 6.1686 | 6.5603 |
| Running time | 1.291 s | 1.282 s | 1.178 s | 1.138 s | 1.192 s |
| Memory usage | 533.2 MB | 532.9 MB | 533.7 MB | 533.8 MB | 529.0 MB |
| SNR | 0.9715 | 0.4698 | 0.7234 | 0.8979 | 0.9384 |
| PSNR | 25.0369 | 24.5352 | 24.789 | 24.9633 | 25.0092 |
| NMSE | 0.7996 | 0.8975 | 0.8466 | 0.8132 | 0.8057 |
| Parameters | Water26 | Water27 | Water30 | Water32 | Water38 |
|---|---|---|---|---|---|
| ME | 0 | 0 | 0 | 0 | 0 |
| Running Time | 0.9 s | 0.809 s | 0.852 s | 0.836 s | 0.619 s |
| Memory usage | 418.8 MB | 406.6 MB | 406.8 MB | 406.9 MB | 387.9 MB |
| SNR | 0 | 0 | 0 | 0 | 6.1607 × 10−5 |
| PSNR | 24.0654 | 24.0654 | 24.0655 | 24.0654 | 24.0709 |
| NMSE | 1 | 1 | 1 | 1 | 1 |
| Filters | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 65.3793 | 1.004 s | 539.6 MB | 3.1006 | 31.5750 | 0.4897 |
| Sech | 17.4831 | 0.994 s | 461.7 MB | 1.6328 | 30.1071 | 0.6866 |
| matched | 0 | 0.271 s | 356.3 MB | 7.8876 | 36.3619 | 0.1626 |
| Retinex | 2.4510 | 0.321 s | 362.8 MB | 0.4822 | 28.9475 | 0.8949 |
| Filters | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 11.6951 | 1.050 s | 585.1 MB | 9.2541 | 39.8031 | 0.1187 |
| Sech | 18.2741 | 0.759 s | 494.9 MB | 3.4882 | 34.0373 | 0.4479 |
| matched | 0 | 0.338 s | 372.8 MB | 10.4385 | 40.9875 | 0.0904 |
| Retinex | 5.9403 | 0.455 s | 366.3 MB | 48.7877 | 79.3368 | 1.3220 × 10−5 |
| Filters | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 15.0668 | 0.970 s | 574.4 MB | 11.446 | 52.9 | 0.0717 |
| Sech | 15.7241 | 0.787 s | 496.4 MB | 4.4845 | 45.9386 | 0.3561 |
| matched | 0 | 0.224 s | 365.0 MB | 19.9880 | 61.4421 | 0.0100 |
| Retinex | 14.3534 | 0.240 s | 364.3 MB | 23.7531 | 65.2071 | 0.0042 |
| Filters | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 17.3630 | 1.182 s | 580.9 MB | 8.1247 | 46.7810 | 0.1540 |
| Sech | 25.7916 | 0.770 s | 494.8 MB | 6.4217 | 45.0781 | 0.2279 |
| matched | 0 | 0.285 s | 367.8 MB | 10.1747 | 48.8310 | 0.0961 |
| Retinex | 4.4329 | 0.394 s | 364.3 MB | 54.1691 | 92.8255 | 3.8290 × 10−6 |
| Filters | ME | Running Time | Memory Usage | SNR | PSNR | NMSE |
|---|---|---|---|---|---|---|
| Atanh | 14.8250 | 1.021 s | 571.5 MB | 10.6666 | 53.1607 | 0.0858 |
| Sech | 10.0528 | 0.771 s | 495.4 MB | 3.7103 | 46.2044 | 0.4256 |
| matched | 0 | 0.242 s | 365.1 MB | 18.6418 | 61.1359 | 0.0137 |
| Retinex | 4.8891 | 0.249 s | 363.0 MB | 37.5639 | 80.0580 | 1.7523 × 10−4 |
| Parameters | m = 2 | m = 22 | m = 42 | m = 62 |
|---|---|---|---|---|
| NMSE | 0.0262 | 0.0253 | 0.0145 | 0.0098 |
| SNR | 15.8119 | 15.9655 | 18.3778 | 20.1049 |
| PSNR | 39.8773 | 40.0309 | 42.4432 | 44.1703 |
| Parameters | n = 2 | n = 6 | n = 8 | n = 12 |
|---|---|---|---|---|
| NMSE | 0.0070 | 0.0028 | 0.0023 | 0.0179 |
| SNR | 21.5188 | 25.5952 | 26.3806 | 17.4633 |
| PSNR | 45.5842 | 49.6606 | 50.4460 | 41.5287 |
| Images | Atanh | Sech |
|---|---|---|
| Leaf1 | 1.440 s | 1.109 s |
| Leaf2 | 1.300 s | 1.109 s |
| Leaf3 | 1.333 s | 1.102 s |
| Leaf4 | 1.286 s | 1.101 s |
| Leaf5 | 1.316 s | 1.092 s |
| Water26 | 1.268 s | 1.092 s |
| Water27 | 1.254 s | 1.123 s |
| Water30 | 1.310 s | 1.200 s |
| Water32 | 1.269 s | 1.103 s |
| Water38 | 0.982 s | 1.022 s |
| Shape2 | 0.969 s | 1.001 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, T.; Xu, J.; Li, Z.; Wu, Y. Two Non-Learning Systems for Profile-Extraction in Images Acquired from a near Infrared Camera, Underwater Environment, and Low-Light Condition. Appl. Sci. 2025, 15, 11289. https://doi.org/10.3390/app152011289
Sun T, Xu J, Li Z, Wu Y. Two Non-Learning Systems for Profile-Extraction in Images Acquired from a near Infrared Camera, Underwater Environment, and Low-Light Condition. Applied Sciences. 2025; 15(20):11289. https://doi.org/10.3390/app152011289
Chicago/Turabian StyleSun, Tianyu, Jingmei Xu, Zongan Li, and Ye Wu. 2025. "Two Non-Learning Systems for Profile-Extraction in Images Acquired from a near Infrared Camera, Underwater Environment, and Low-Light Condition" Applied Sciences 15, no. 20: 11289. https://doi.org/10.3390/app152011289
APA StyleSun, T., Xu, J., Li, Z., & Wu, Y. (2025). Two Non-Learning Systems for Profile-Extraction in Images Acquired from a near Infrared Camera, Underwater Environment, and Low-Light Condition. Applied Sciences, 15(20), 11289. https://doi.org/10.3390/app152011289







