4.1. Results from Experiment 1
Table 5 presents the descriptive statistics of optimal solutions and statistical test results of the GAs for a population size of 20 (
). The median values of the optimal solutions are illustrated in
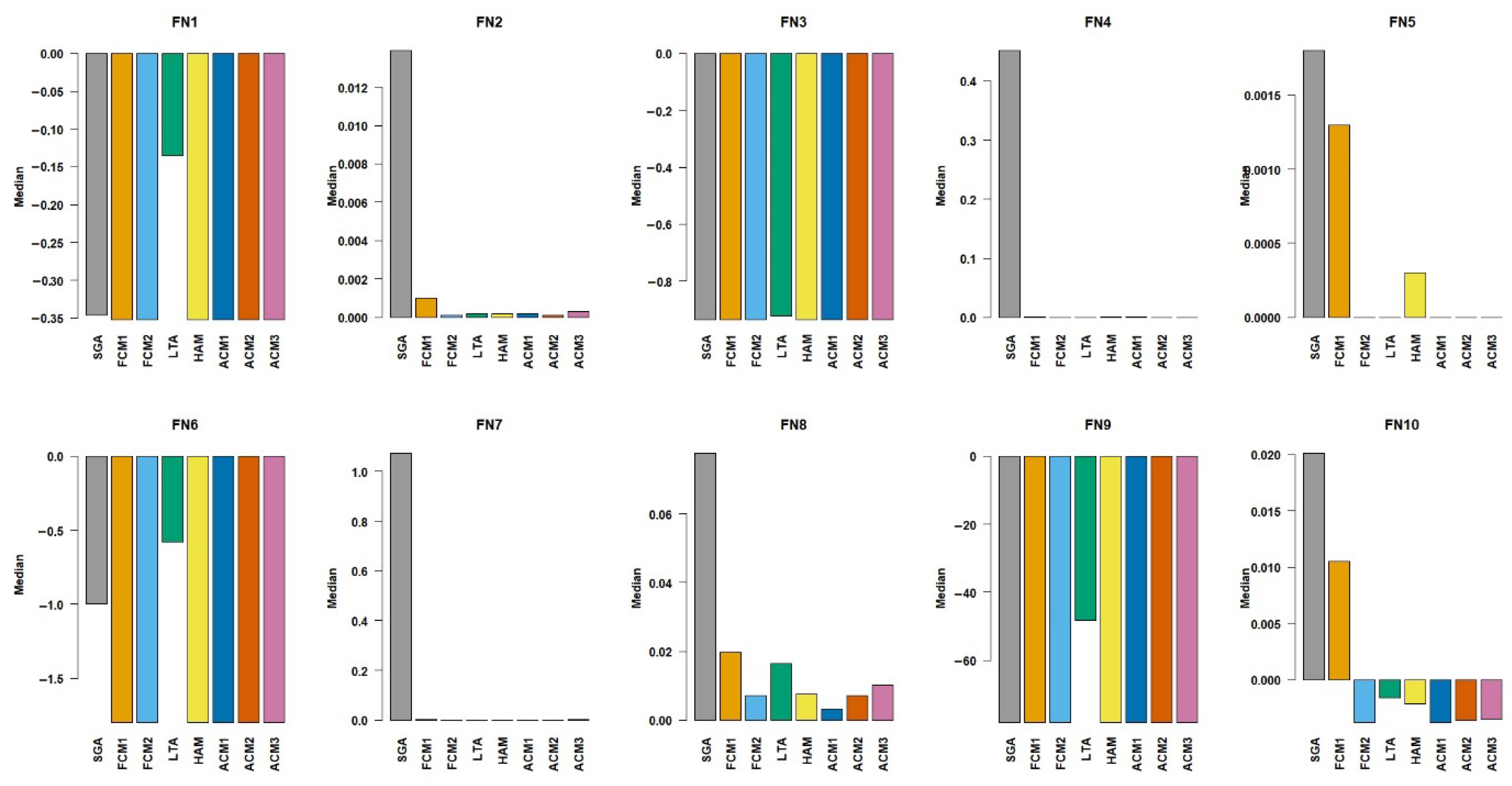
Figure 3. The results show that FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 converged to the global optimum value of approximately −0.3524 for the test function, FN1. SGA deviated from the optimum slightly, with a median value of −0.3463 and produced a relatively larger standard deviation of 0.0776, which hints that it is less stable than the other methods. Despite the FN1’s less complex structure, LTA underperformed, with a median value of −0.1356, having difficulty converging to the global optimum. For FN2, the methods FCM2, HAM, ACM1, ACM2, and ACM3 had near-zero errors and were in close approximation of the known global minimum. SGA and FCM1 produced relatively higher median values, implying less efficiency when handling the non-separable structure of FN2. In the tests with FN3, the methods FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 returned results that were identical and close to the global optimum, −1. LTA showed a visible performance gap, with a median value of −0.9234 compared to the global optimum, indicating a bit lesser capability in handling steep gradient changes. For FN4, SGA recorded a substantially high median value of 0.4513 and a large variance, reflecting poor performance and instability. On the other hand, the methods FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 could find solutions with negligible deviations from the global optimum, proving superior in global search and local refinement in a multimodal structure. For the FN5, the methods FCM2, LTA, HAM, ACM1, ACM2, and ACM3 found the global optimum. SGA’s median value of 0.0018 was slightly above the optimum, suggesting minor inefficiencies, while FCM1 also showed small but visible deviations. As for the FN6, the FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 located the global optimum of approximately −1.8013. Meanwhile, LTA did not perform well with the median value of −0.5813, and SGA underperformed with −1.000, struggling to navigate the rugged search space. For FN7, the methods FCM2, HAM, ACM1, ACM2, and ACM3 deviated very slightly from the global optimum, expressing fine exploitation abilities. SGA, however, produced a higher median of 1.0723 and a large standard deviation of 1.2864, showing less performance. On FN8, the best-performing methods were FCM2, ACM1, and ACM2, producing median values that were close to the global optimum of 0. SGA, FCM1, and HAM had slightly higher median deviations, facing some difficulties in fine-tuning solutions along the curved valley towards the optimum. For FN9, except LTA, all the methods managed to locate the global optimum −78.3323, whereas LTA performed significantly worse because of premature convergences to locate the deepest minima. For FN10, the methods FCM2, HAM, ACM1, ACM2, and ACM3 found the global optimum. SGA and LTA deviated a bit from the optimum, hinting at minor inefficiencies in convergence.
The NCGOs and ranks of the compared methods are given in
Table 6 for
. Based on the NCGO results in
Table 6, the methods FCM2, HAM, ACM1, ACM2, and ACM3 displayed a highly satisfactory performance, often attaining the maximum possible NCGO values for some functions such as FN1, FN6, and FN9. They maintained high rankings in both unimodal and multimodal problems, as well as the difficult non-separable cases like FN2, FN4, and FN8. Among these methods, the FCM2’s performance stood out, as it achieved top positions frequently. Throughout the test functions, FCM2 also achieved the lowest rank sum of 20.5. This was an unexpected superiority from the perspectives of this study, but it is worth mentioning that Hassanat et al. [
25] had also reported that FCM2 configuration performs exceptionally better with small population sizes. On the FCM2’s performance in small population sizes (e.g.,
), the fixed and relatively high mutation rate (
) has a critical part. Genetic diversity tends to diminish rapidly in small populations and this may lead to premature convergence. Thus, the high mutation rate in FCM2 can be a countermeasure since it maintains the diversity. The balanced crossover rate of (
) acts as complementary by ensuring steady exploitation without overwhelming the search with convergence pressure. As a result, the fixed but balanced setup of FCM2 seems to be effective especially under limited population diversity.
ACM1, ACM2, and ACM3 also performed very well with rank sums of 33, 35, and 36, respectively. HAM had a rank sum of 41, showing moderate performance due to a zero-mutation rate in early generations. SGA and LTA returned weaker results, frequently producing zero NCGOs for complex multimodal functions such as FN3, FN6, and FN9. Although the SGA achieved moderate success in FN1 and FN5, its performance remained consistently lower than that of the leading methods. LTA occasionally attained mid-tier rankings, as in FN2, FN4, and FN7, but these instances can be considered as exceptions rather than a consistent pattern. Overall, the integration of NCGO-based rankings and rank sums clearly confirms that FCM2 is the most successful and surprisingly dominant method for the given benchmark set with , followed closely by ACM2, ACM1, and ACM3.
Table 7 presents the descriptive statistics and statistical comparisons of the optimal solutions obtained from different parameter tunings and control methods for a population size of 50 (
). As also illustrated in
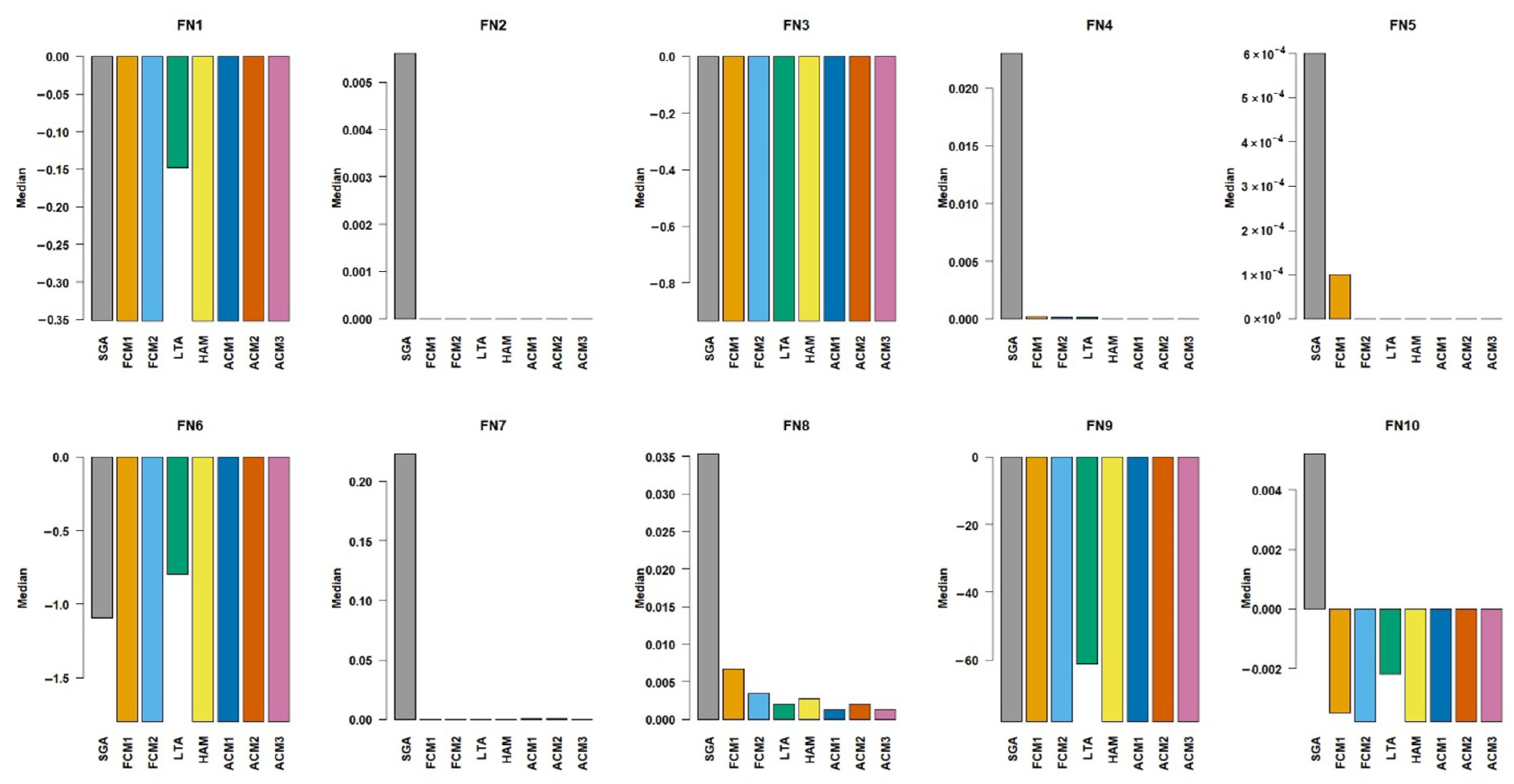
Figure 4, the methods FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 successfully converged to the global optimum of approximately −0.3524 for the FN1 test function. Meanwhile, SGA produced a median of −0.3506 close to the global optimum, LTA performed considerably worse, with a median of −0.1481, suggesting it struggled to locate the optimum despite the simplicity of the FN1 function. On FN2, all the methods except SGA have been very successful by achieving a median of exactly 0, which is the global optimum. SGA yielded a larger median of 0.0056 for FN2. Regarding the function FN3, the medians of all methods have been found as very close to the global optimum of −1, while LTA had a slightly higher median of −0.9347. In FN4, however, the methods HAM, ACM1, ACM2, and ACM3 had the medians equal to the global optimum; there were no significant differences with FCM1, FCM2, and LTA, also implying their capability in multimodal optimisation. For this function, even though SGA’s median is a bit higher than the optimum it found the best results close to the global optimum. For FN5, the methods FCM2, LTA, HAM, ACM1, ACM2, and ACM3 reached the global optimum. SGA produced a slightly higher median of 0.0006. On FN6, the methods FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 perfectly found the optimum value. SGA’s median of −1.0929 indicates weaker performance, while LTA’s result of −0.7994 reflects significant difficulty in navigating the rugged search space. For FN7, the methods FCM1, FCM2, LTA, HAM, ACM1, ACM2, and ACM3 achieved medians very close to zero, showing effective exploitation ability. For FN8, although ACM1 and ACM3 gave the media results very close to the global optimum, LTA, HAM, and ACM2 also approached the global optimum, pointing to their efficient fine-tuning capability. SGA and FCM1 performed slightly worse, suggesting they were less precise in handling the curved search space of FN8. On FN9, all methods but SGA and LTA perfectly achieved the global optimum. SGA’s median was quite close, but it was less stable due to larger standard deviations of solutions, whereas LTA’s median indicates premature convergence and a significant amount of divergence. Lastly, for FN10, FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 did not differ in median performance. SGA and LTA exhibited small but consistent deviations, reflecting slight inefficiencies in convergence accuracy.
Table 8 presents a summary of the performance of methods across test functions, focusing on NCGOs and their corresponding ranks. The table also includes the rank sums for overall performance. As seen in the table, FCM2 emerged as the best-performing method with the lowest rank sum and perfect NCGO scores for six functions of FN2, FN4, FN5, FN6, FN9, and FN10, confirming its superiority. Among the stronger performers, ACM1 and ACM3 came closest to FCM2, while HAM and ACM2 showed good results, particularly on FN3, FN4, FN7, and FN8. FCM1 performed well on some functions, such as FN1, FN2, FN6, and FN9, but poorly on others, indicating problem-specific sensitivity. SGA was the weakest with the highest rank sum of 76.5, and failing entirely on FN3, FN7, and FN8 with zero NCGOs. LTA matched FCM1 in rank but was highly inconsistent, with zero NCGOs for FN1, FN3, FN6, and FN9.
Table 9 lists the descriptive statistics for the optimal solutions found for a larger population size of
. As shown in
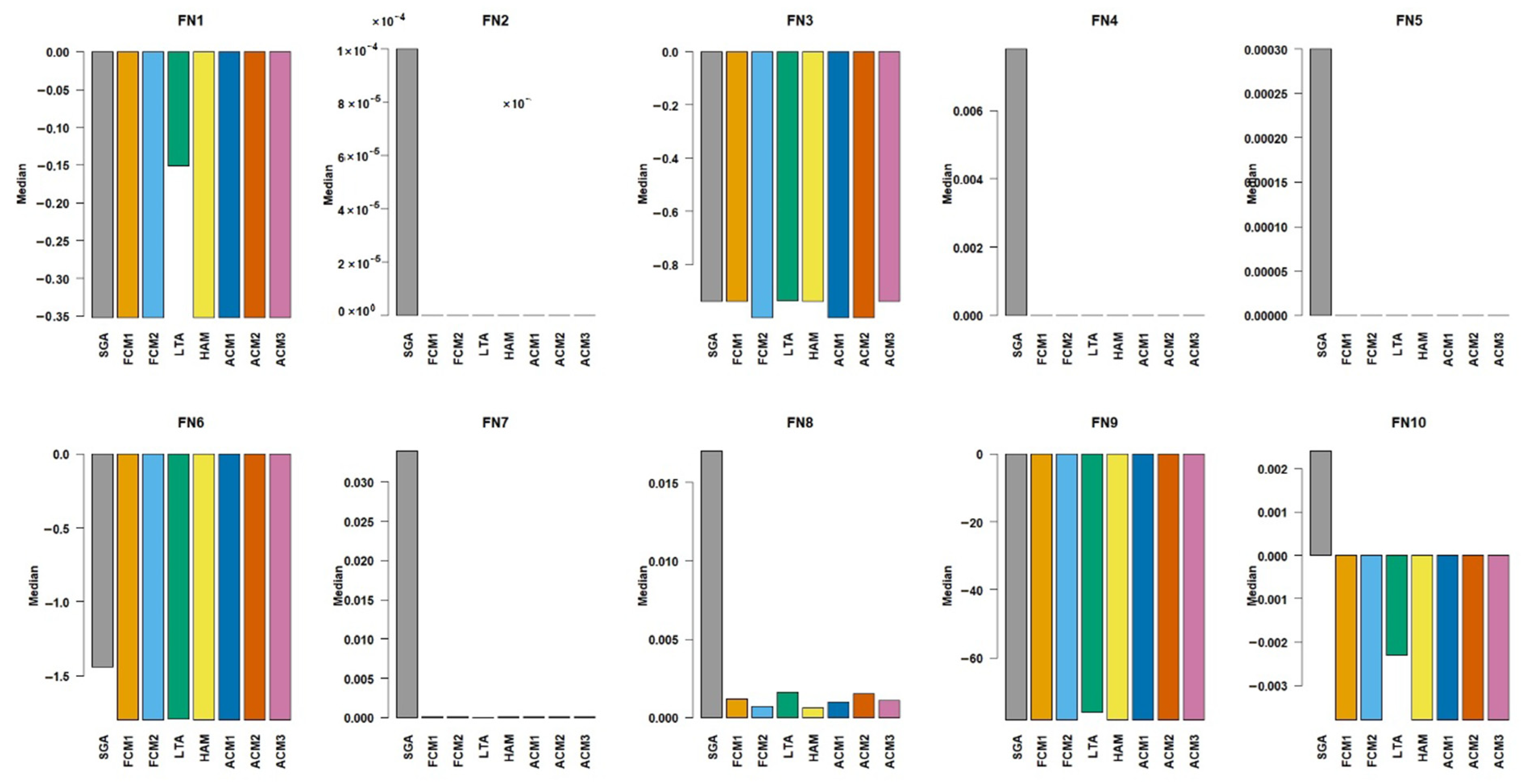
Table 9 and as the median values in
Figure 5 display, the methods FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 achieved excellent results across almost all of the functions. For FN1, FN2, FN4, FN5, FN6, FN9, and FN10, these methods either matched up or were only negligibly different from the global optimum, as indicated by their median values. Their standard deviations were often zero or close to it, showing a remarkable stability and reliability in finding the optimal solution in larger populations.
SGA method, in general, performed worse than the other methods. For example, in FN1, FN2, and FN5, SGA’s median was statistically different from the more successful methods, and its standard deviation was notably higher, suggesting less consistent convergence. However, when considered on its own, it showed a little improvement for the larger population size compared to that of the previous test, where .
LTA showed a mixed performance. It struggled heavily with certain functions, such as FN1 and FN9, where its median values fell far from the global optimum. For FN9, the standard deviation was 4.4622, indicating a huge variability in its results. However, it performed well on functions like FN4 and FN7, achieving pristine medians of 0.0000 and 0.0000, respectively, statistically on par with the best methods. We may therefore conclude that the LTA’s effectiveness is highly dependent on the characteristics of the optimisation problem, such as population size.
According to the results in
Table 10, FCM2 is the best-performing method with the lowest rank sum of 30.0. It achieved an impressive NCGO score of 30 on many functions, including FN2, FN4, FN5, FN6, FN7, FN9, and FN10, an indicator of its ability to converge to the global optimum across a variety of problem types. There was a slight dip in performance on FN3 and FN8, but it still managed to reserve the ranks of 1 and 2, respectively. The methods ACM1, HAM, and ACM2 also performed very well, with rank sum values of 35.5, 36.0, and 36.5, respectively. These scores indicate high performance, nearly matching that of the FCM2. The trio achieved NCGO scores of 30 with most functions, showing significant consistency. Compared to the FCM2’s performance, the most visible differences were observed for FN3 and FN8, where their NCGO values were a bit lower, which suggests a minor but not statistically significant difference in their fine-tuning capabilities. In the third group, the methods FCM1 and ACM3 showed a decent overall performance with rank sums of 45.0 and 42.0, respectively. FCM1 consistently located the global optimum on functions like FN1, FN2, FN5, FN7, FN9, and FN10. However, its performance fell dramatically on FN3 and FN8, where its NCGO values decreased to 10 and 14, respectively. This inconsistency suggests that although FCM1 is highly effective, it may struggle with certain complex function landscapes. ACM3 followed a similar pattern, with satisfactory performance on most functions while scoring a lower NCGO on FN3 and FN8. The LTA and SGA methods were the weakest performers. LTA has a quite large rank sum of 58.5, and its performance is highly inconsistent. It totally failed on functions like FN1, FN3, and FN9, demonstrating a clear weakness to specific types of optimisation problems. In contrast, though, the LTA’s performances on FN2, FN4, FN5, and FN7 were perfect, which suggests a significant dependency on the function’s characteristics. SGA showed the worst performance, with a very large rank sum of 76.5. Its NCGO values were consistently low across the methods, and it failed to find the optimum in any run for FN3, confirming its ineffectiveness for more challenging problems, even with an increased population size.
4.2. Results from Experiment 2
Experiment 2 begins with the settings,
,
, and,
. According to the results presented in
Table 11 and
Figure 6, the SGA performed the worst in general. For the FN4 function, SGA with a median of 0.0491 performed significantly worse than all the other methods. The remaining methods showed no statistically significant difference in median performance, ranging from 0.0013 to 0.0022. On the FN7 function, the SGA again stands out with a significantly higher median value. The methods HAM and ACM1 were statistically similar and the best-performing group. The methods FCM1, FCM2, LTA, ACM2, and ACM3 performed similarly to each other. For the FN8 function, the analysis of median values again shows that SGA performed significantly worse than all other methods. The methods FCM2, LTA, HAM, ACM1, ACM2, and ACM3 gave statistically similar results.
According to the NCGOs and ranks given in
Table 12, the best method overall was the ACM2, followed by the HAM and FCM2.
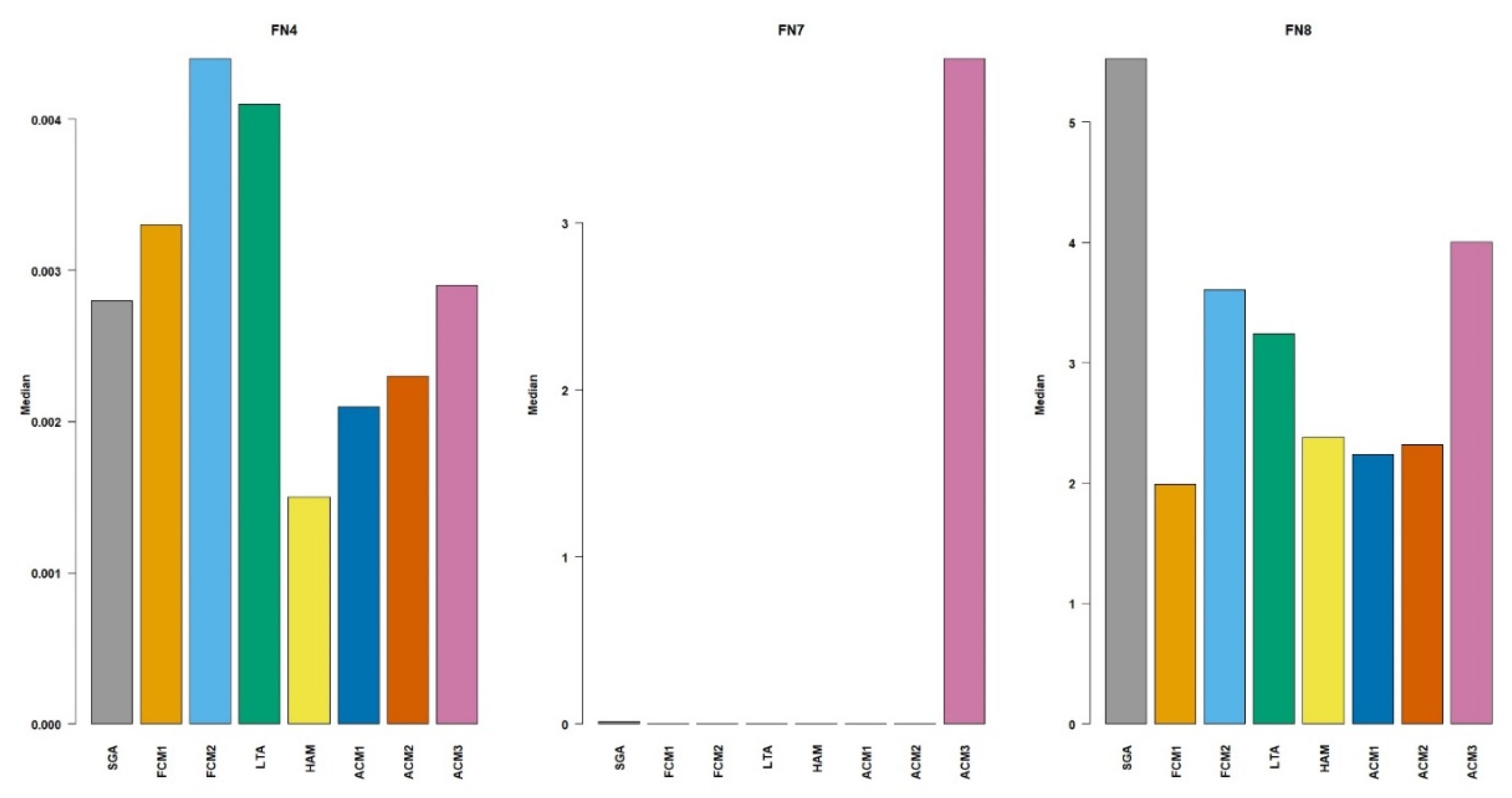
The results for the configuration of
and
on the test functions are shown in
Table 13 and plotted in
Figure 7. For the FN4 benchmark function, SGA performed statistically worse than all other methods, with a median value of 0.0177. The best performance was achieved by HAM and ACM2, with a median value of 0.0004 for both. These are followed by the methods FCM2, ACM1, and ACM3. On FN7, the performance results were more distinct. SGA and ACM3 performed significantly worse than the other methods. FCM1, with a median value of 0.0000, was the single best-performing method. The methods FCM2, ACM1, and ACM2 formed a statistically similar group, representing the second-best performers. LTA and HAM had an intermediate performance. For FN8, most of the methods exhibited a similar level of performance. Although ACM2 had the lowest median value, the methods FCM1, FCM2, HAM, ACM1, ACM2, and ACM3 belonged to a single, statistically similar group. This group collectively represents the best performance. In contrast, SGA and LTA performed worse than the methods.
According to the NCGO ranking results shown in
Table 14, the best performance was scored by the ACM2, followed by the FCM2 and HAM, with the rank values of 7, 8, and 9, respectively.
The results for the configuration of
and
on the test functions are shown in
Table 15 and
Table 16, and they are plotted in
Figure 8. For FN4, HAM demonstrated a statistically superior performance, outperforming the rest. A large group of methods, including SGA, FCM1, ACM1, ACM2, and ACM3, performed similarly to this top-tier method, with no significant differences among them. However, FCM2 and LTA showed a statistically worse performance compared to the best one. This indicates that while HAM was the standout performer, a large number of methods were also highly effective. The performance on FN7 was clearly divided into three distinct levels. The methods FCM1, HAM, ACM1, and ACM2 achieved the best results with the lowest median values, performing in a statistically similar manner. A second group, consisting of SGA, FCM2, and LTA, showed significantly worse results than the top performers. Finally, ACM3 had a median so high that it was statistically worse than all other methods. On the FN8 function, FCM1 was statistically the best, surpassing all others. The group of HAM, ACM1, and ACM2 performed worse than FCM1 but were still more effective than the rest. FCM2 and LTA showed an intermediate level of performance, while SGA had the worst performance of all, with a median value that was statistically higher than every other method.
According to the NCGOs in
Table 16, the ACM2 was the best method, followed by the HAM and ACM1, on the test function FN4, while the HAM was the best, followed by FCM1 and FCM2, on the test function FN7. On the other hand, the ACM1 and ACM2 also performed well on this function, as seen in
Figure 8. Finally, on the test function FN8 (Rosenbrock), all of the methods had the same rank score. Picheny et al. [
52] reported that, even though the valley of the Rosenbrock function is easy to find, convergence to the global minimum is very difficult for the larger dimensions with it. The zero NCGO values observed for the Rosenbrock function in
Table 16 also suggested a substantial difficulty for all of the compared algorithms in locating the global optimum. Although the Rosenbrock function has a narrow valley guiding toward the global minimum, its curvature becomes highly non-linear and poorly conditioned in higher dimensions. Consequently, standard crossover and mutation operators struggle to produce offspring that remain within the valley. The problem could be tackled with brute force by increasing the maximum number of generations to at least 2000 to locate the global minimum of this function when its dimension is higher than 15. However, GAs without a hybrid local search property tend to oscillate or converge prematurely; thus, increasing the number of generations would not be the most robust approach. The integration of gradient-based or adaptive search mechanisms that can handle the poorly conditioned curvatures may offer a better solution.
In summary, Experiment 2 underlined the strength of the ACM2 algorithm in higher-dimensional optimisation problems. For problem dimensions of and , ACM2 had lower median values across the benchmark functions FN4 and FN8, indicating more accurate convergence toward optimality. ACM2 also displayed superior overall NCGO rankings than other methods, including HAM and FCM2. Although several methods demonstrated competitive performances on individual test functions, the ACM2′s lower median solutions and better NCGOs have verified its superiority for exploration and exploitation in high-dimensional search spaces. It was therefore sufficiently evident that ACM2 exhibits significant robustness and reliability, making it well-suited for challenging high-dimensional optimisation tasks.
4.3. Results from Experiment 3
The performance of the methods on the boost converter implementation is compared in
Table 17 where the best, the worst, median, mean, and standard deviations of best fitness values across the generations are presented.
The Kruskal–Wallis rank sum test for the best fitness values across generations indicated statistically significant differences between the algorithms (, , ). According to the post hoc analysis, ACM1 and ACM2 performed comparably to FCM1 and FCM2, while LTA was slightly weaker. HAM and ACM3 were in the same group, with lower performances despite their occasional good results. SGA performed the worst overall.
Regarding the best solutions, ACM3 achieved the smallest value of 0.0001, and ACM2 became the second with 0.0002, while FCM1, HAM, and ACM1 followed them with 0.0005. According to the standard deviations, FCM1 (0.8993), FCM2 (1.2284), and ACM2 (1.9466) displayed the highest robustness. HAM (2.4651) and SGA (2.3444) showed more variability, meaning less stable outcomes. ACM1 (2.1493) and ACM3 (2.2077) displayed relatively high deviations, showing that although they offered excellent solutions on occasion, they are susceptible to producing less stable results.
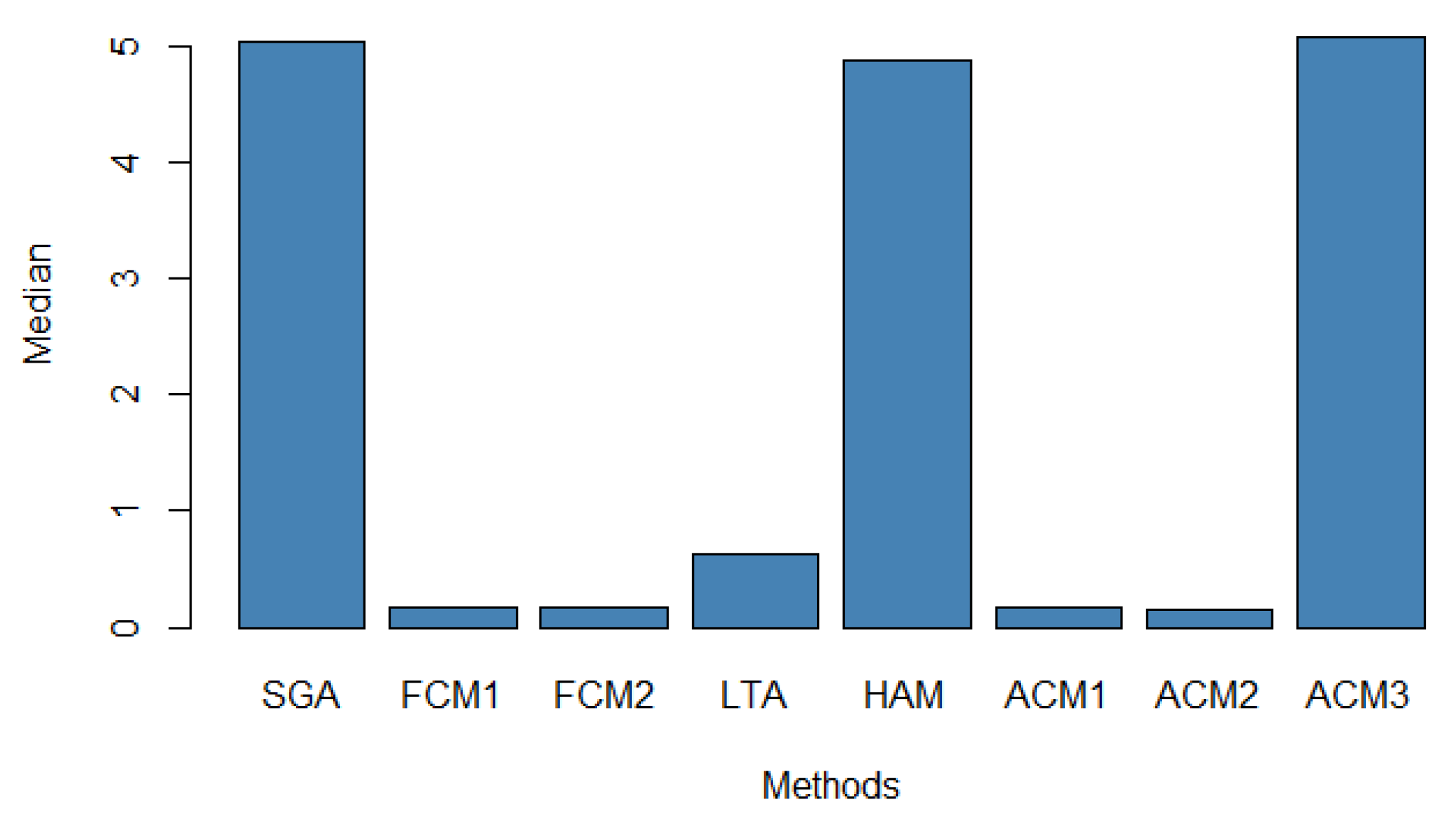
The median best fitness values are illustrated in
Figure 9, where ACM2 achieved the lowest median. The ACM1, FCM1, and FCM2 were very close to ACM2. In contrast, SGA, HAM, and ACM3 showed much higher median values. The LTA was in between. In summary, the algorithms were ranked as ACM2 > FCM1 > FCM2 > ACM1 > LTA > HAM > ACM3 > SGA.
Overall, the ACM3 might offer the best possible outcome, but it severely lacks robustness and stability. Therefore, the ACM2 is the best candidate if the most rewarding but also balanced outcome is sought. The difference between their best is only 0.0001, and ACM2 is much more robust and stable, having the lowest median value among the methods. The FCM1 might have a better mean value than the ACM2, but given the stochastic nature of the runs, median values are more reliable.