Deterministic Modeling of Muller’s Ratchet Effect in Populations Evolving in an Environment of Finite Capacity
Abstract
1. Introduction
2. Methods
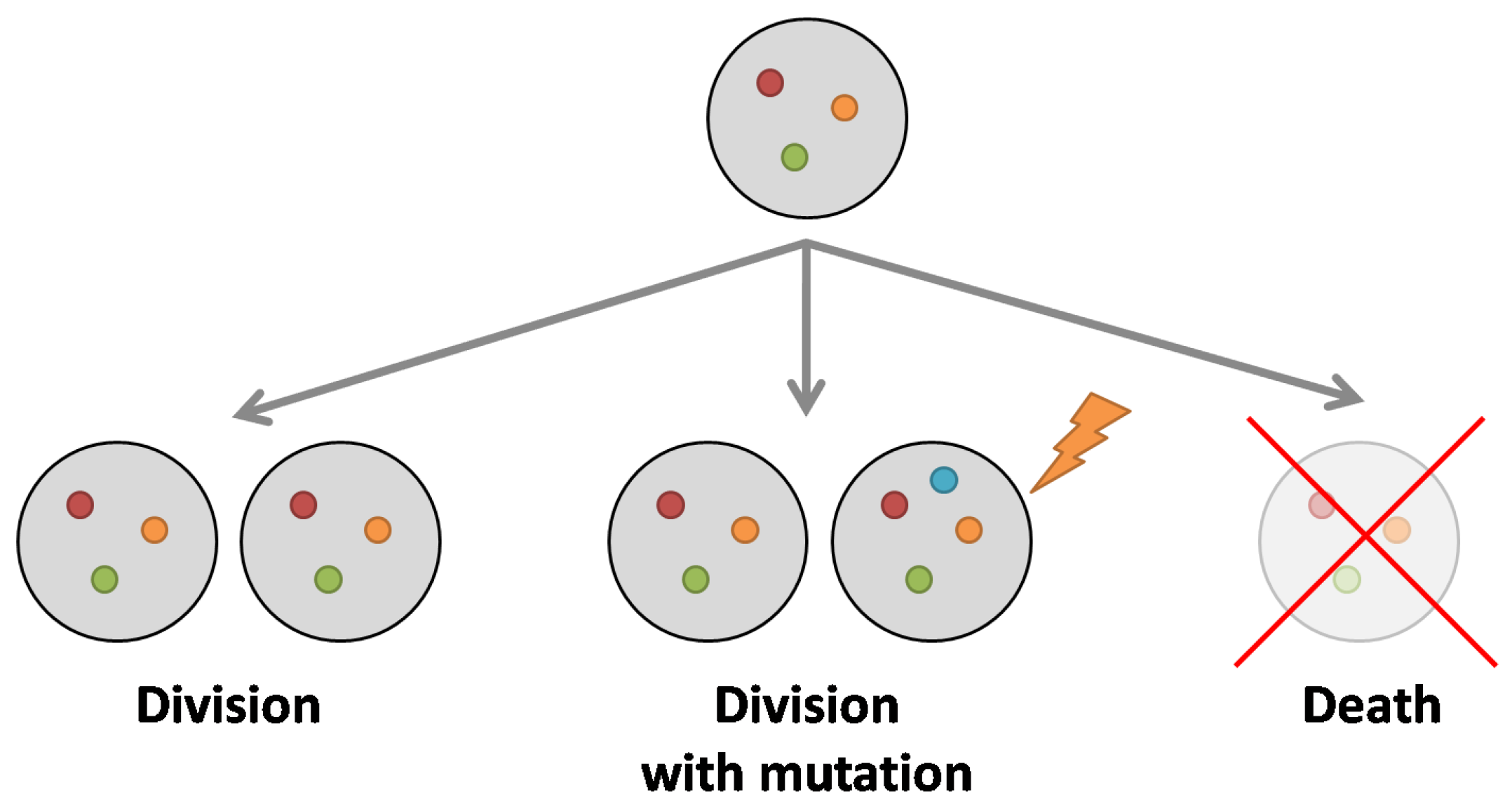
- Evolution is asexual, with events of births (replications), deaths and mutations.
- The population comprises individuals, species, organisms or cells of different reproductive fitness.
- The population is under pressure from an environment of finite capacity, where the size of the population modifies the rate of the death process.
- Random, weakly deleterious mutations take place during birth processes. They modify reproductive fitness of individuals.
2.1. Evolution of Population with Mutation and Selection in an Environment of Finite Capacity
2.2. Deterministic Model of Propagation of Weakly Deleterious Mutations in Population Evolution
2.3. Deterministic Balance Equations
2.4. Evolution of the Population Size
2.5. Equations for Evolution of Frequencies of Classes
2.6. Traveling Waves of Mutations
2.7. Stationary Mutation Front
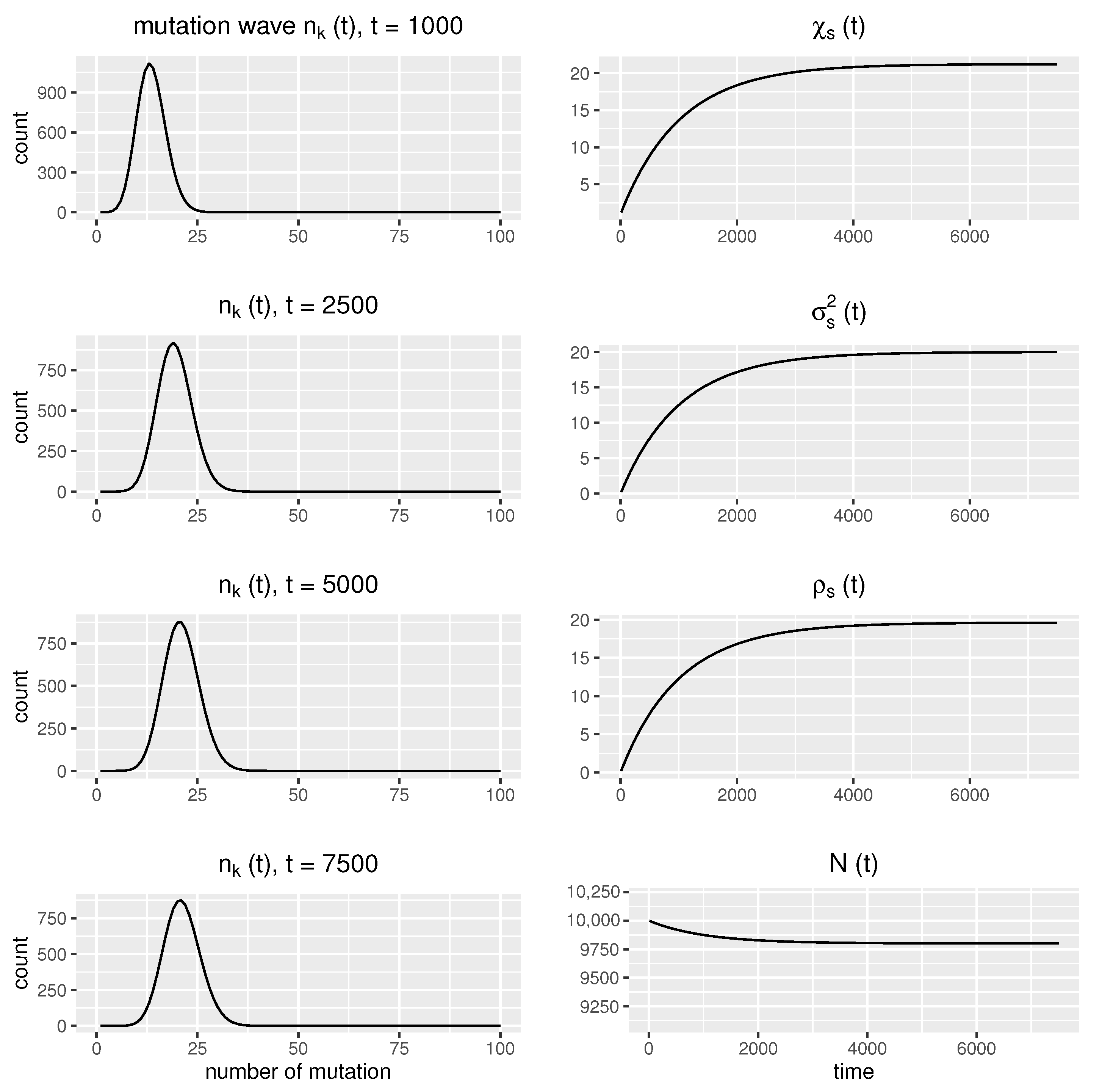
2.8. Deterministic Modeling Versus Stochastic Simulations

3. Results
3.1. Modified Deterministic Models
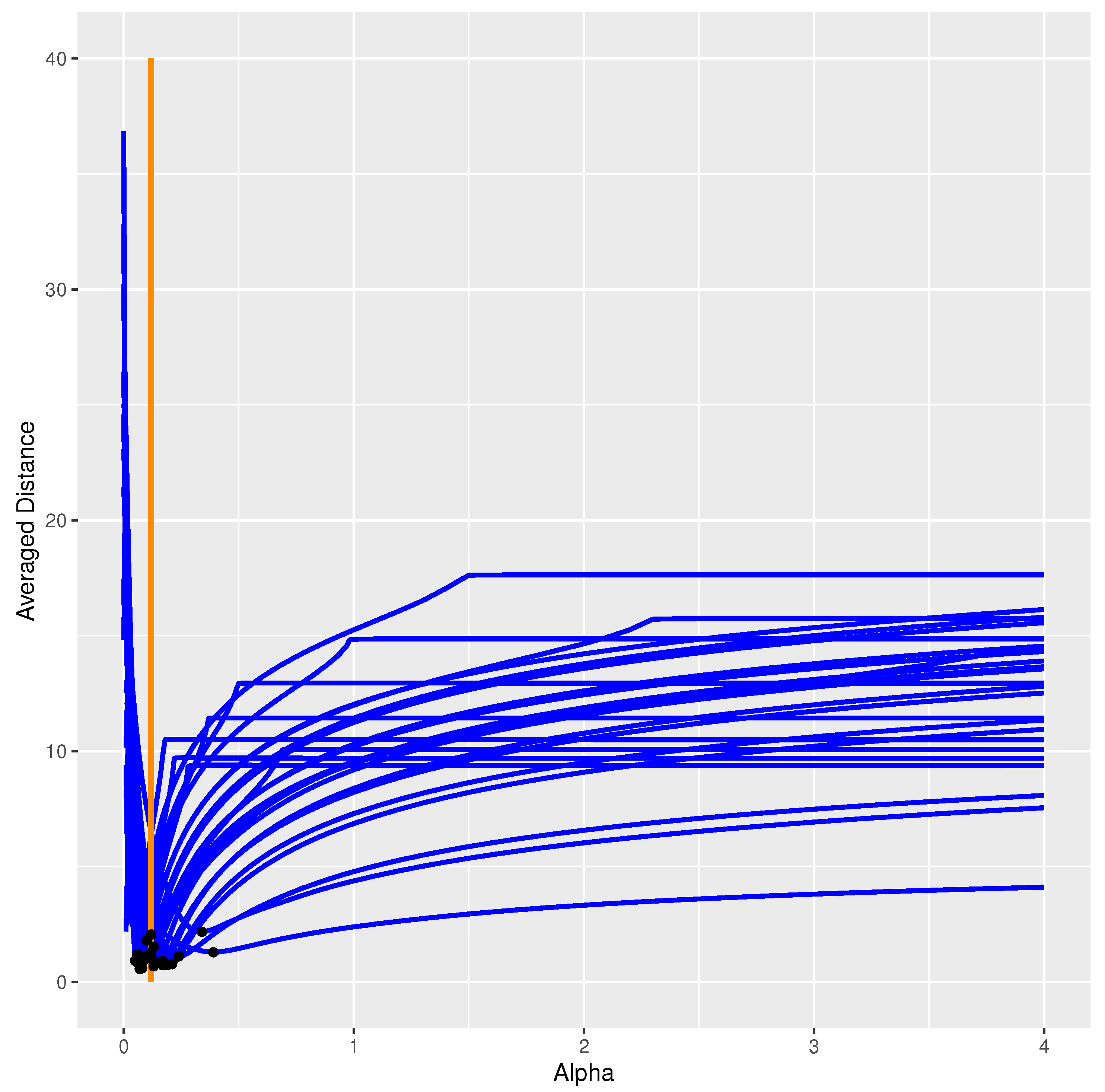
3.2. Cutoff Modification
3.3. Exponential Modification
3.4. Characterization and Comparison of Two Modifications
3.4.1. Stationary Mutation Fronts Versus Advancing Mutation Waves
- The least-loaded class is .
- The stationarity condition is
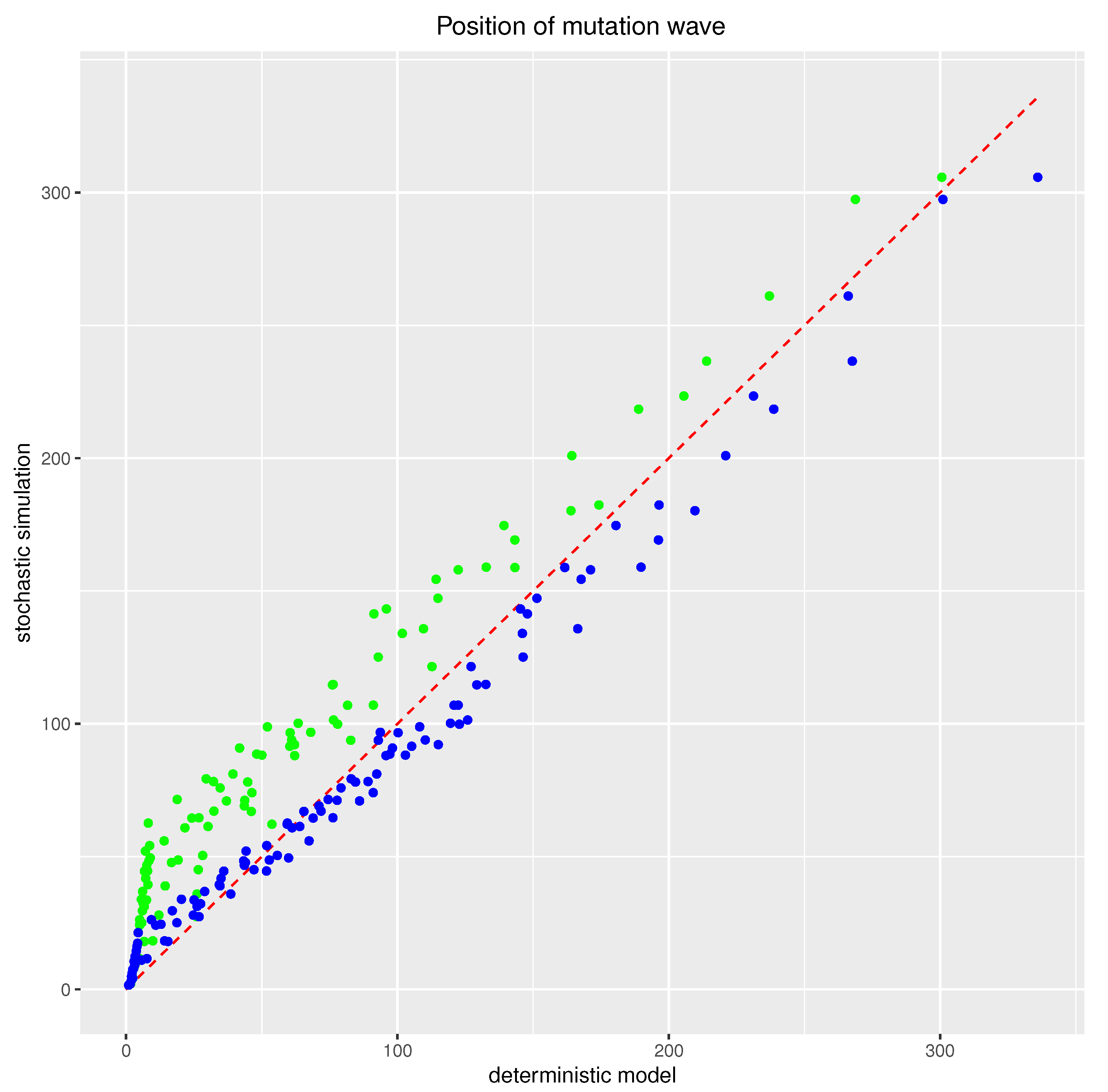
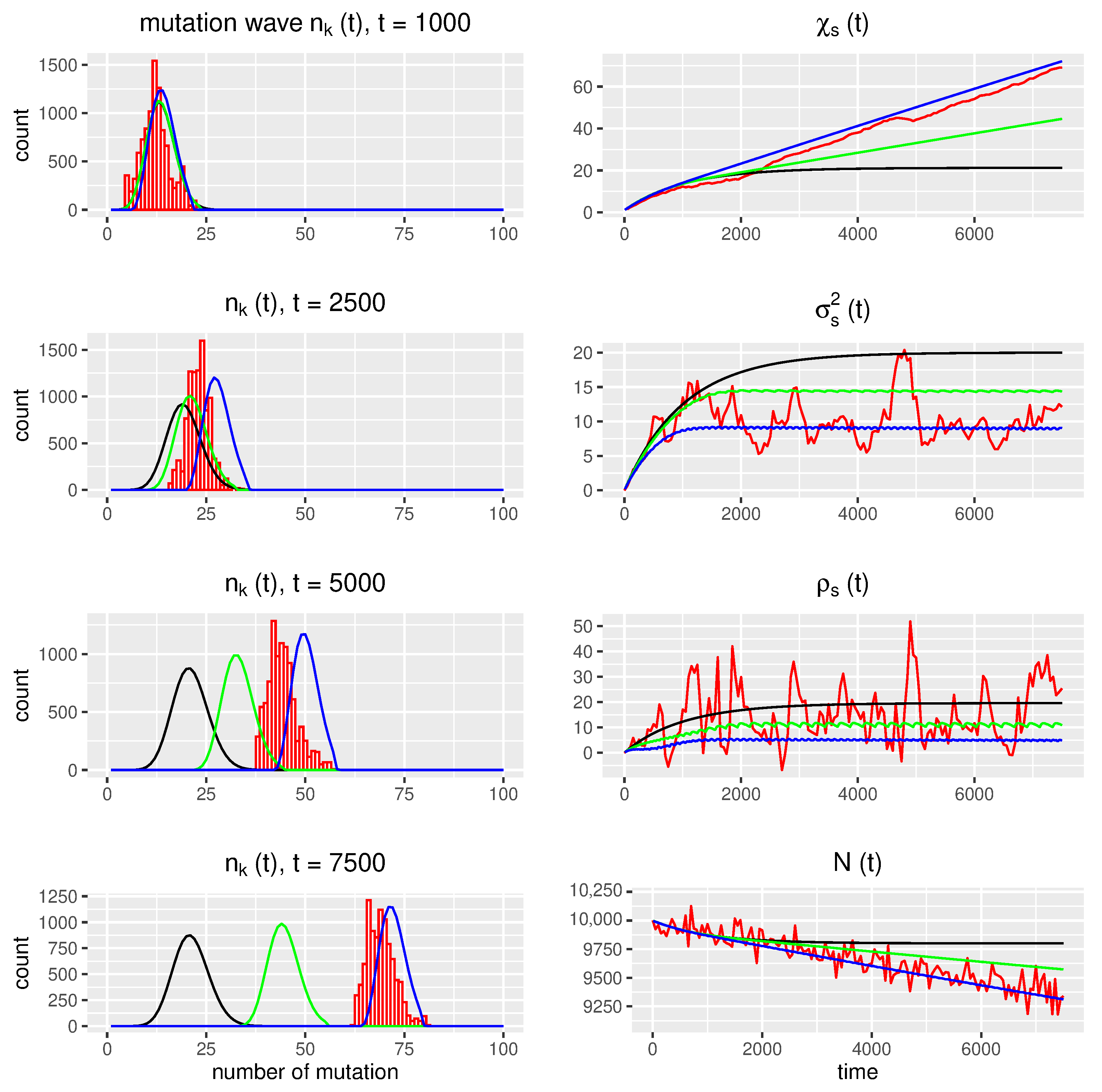
3.4.2. Numerical Computations for Comparative Analysis of the Positions of Mutation Waves at Various Time Points
4. Discussion
5. Conclusions
- In asexual evolution, where the population is under pressure from an environment of finite capacity, and random, weakly deleterious mutations take place, the deterministic balance equation model predicts the occurrence of a stationary mutation front, analogous to classical results [2].
- In the stochastic simulations corresponding to the above scenario, with a population of finite size, Muller’s ratchet effect of a slowly advancing mutation wave is seen.
- It is possible to introduce (heuristic) modifications to the deterministic model, allowing for better consistency between the deterministic model and stochastic simulations.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muller, H.J. Some genetic aspects of sex. Am. Nat. 1932, 66, 118–138. [Google Scholar] [CrossRef]
- Haigh, J. The accumulation of deleterious genes in a population—Muller’s ratchet. Theor. Popul. Biol. 1978, 14, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Bürger, R.; Butcher, D.; Gabriel, W. The mutational meltdown in asexual populations. J. Hered. 1993, 84, 339–344. [Google Scholar] [CrossRef]
- Gabriel, W.; Lynch, M.; Bürger, R. Muller’s ratchet and mutational meltdowns. Evolution 1993, 47, 1744–1757. [Google Scholar] [CrossRef]
- Sakamoto, T.; Innan, H. Muller’s ratchet of the Y chromosome with gene conversion. Genetics 2022, 220, iyab204. [Google Scholar] [CrossRef]
- Hill, G.E. Mitonuclear compensatory coevolution. Trends Genet. 2020, 36, 403–414. [Google Scholar] [CrossRef]
- Lodato, M.A.; Rodin, R.E.; Bohrson, C.L.; Coulter, M.E.; Barton, A.R.; Kwon, M.; Sherman, M.A.; Vitzthum, C.M.; Luquette, L.J.; Yandava, C.N.; et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 2018, 359, 555–559. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, X.; Lee, M.; Maslov, A.Y.; Wang, T.; Vijg, J. Single-cell whole-genome sequencing reveals the functional landscape of somatic mutations in B lymphocytes across the human lifespan. Proc. Natl. Acad. Sci. USA 2019, 116, 9014–9019. [Google Scholar] [CrossRef] [PubMed]
- Merleau, N.S.; Pénisson, S.; Gerrish, P.J.; Elena, S.F.; Smerlak, M. Why are viral genomes so fragile? The bottleneck hypothesis. PLoS Comput. Biol. 2021, 17, e1009128. [Google Scholar] [CrossRef] [PubMed]
- Brufsky, A.; Lotze, M.T. Ratcheting down the virulence of SARS-CoV-2 in the COVID-19 pandemic. J. Med. Virol. 2020, 92, 2379. [Google Scholar] [CrossRef]
- Dujon, A.M.; Gatenby, R.A.; Bramwell, G.; MacDonald, N.; Dohrmann, E.; Raven, N.; Schultz, A.; Hamede, R.; Gerard, A.L.; Giraudeau, M.; et al. Transmissible cancers in an evolutionary perspective. iScience 2020, 23, 101269. [Google Scholar] [CrossRef]
- McFarland, C.D.; Korolev, K.S.; Kryukov, G.V.; Sunyaev, S.R.; Mirny, L.A. Impact of deleterious passenger mutations on cancer progression. Proc. Natl. Acad. Sci. USA 2013, 110, 2910–2915. [Google Scholar] [CrossRef] [PubMed]
- McFarland, C.D.; Mirny, L.A.; Korolev, K.S. Tug-of-war between driver and passenger mutations in cancer and other adaptive processes. Proc. Natl. Acad. Sci. USA 2014, 111, 15138–15143. [Google Scholar] [CrossRef]
- Gordo, I.; Charlesworth, B. The degeneration of asexual haploid populations and the speed of Muller’s ratchet. Genetics 2000, 154, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Stephan, W.; Chao, L.; Smale, J.G. The advance of Muller’s ratchet in a haploid asexual population: Approximate solutions based on diffusion theory. Genet. Res. 1993, 61, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Rouzine, I.M. Mathematical Modeling of Evolution. In Mathematical Modeling of Evolution; De Gruyter: Berlin, Germany, 2020. [Google Scholar]
- Bräutigam, C.; Smerlak, M. Diffusion approximations in population genetics and the rate of Muller’s ratchet. J. Theor. Biol. 2022, 550, 111236. [Google Scholar] [CrossRef]
- Waxman, D.; Loewe, L. A stochastic model for a single click of Muller’s ratchet. J. Theor. Biol. 2010, 264, 1120–1132. [Google Scholar] [CrossRef]
- Lansch-Justen, L.; Cusseddu, D.; Schmitz, M.A.; Bank, C. The extinction time under mutational meltdown driven by high mutation rates. Ecol. Evol. 2022, 12, e9046. [Google Scholar] [CrossRef]
- Igelbrink, J.L.; Casanova, A.G.; Smadi, C.; Wakolbinger, A. Muller’s ratchet in a near-critical regime: Tournament versus fitness proportional selection. Theor. Popul. Biol. 2024, 158, 121–138. [Google Scholar] [CrossRef]
- Abrams, M. Infinite population models and random drift. Philos. Theory Pract. Biol. 2024, 16. [Google Scholar] [CrossRef]
- Strütt, S.; Excoffier, L.; Peischl, S. A generalized structured coalescent for purifying selection without recombination. Genetics 2025, 229, iyaf013. [Google Scholar] [CrossRef]
- Tsimring, L.; Levine, H.; Kessler, D. RNA virus evolution via a fitness-space model. Phys. Rev. Lett. 1996, 76, 4440. [Google Scholar] [CrossRef]
- Hallatschek, O. The noisy edge of traveling waves. Proc. Natl. Acad. Sci. USA 2010, 108, 1783–1787. [Google Scholar] [CrossRef]
- Brunet, E.; Derrida, B. Genealogies in simple models of evolution. J. Stat. Mech. Theory Exp. 2013, P01006. [Google Scholar] [CrossRef]
- Rouzine, I.M.; Wakeley, J.; Coffin, J.M. The solitary wave of asexual evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, C. Multiple Time Scale Dynamics; Springer: Cham, Switzerland, 2015; Volume 191. [Google Scholar]
- Neher, R.A. Genetic draft, selective interference, and population genetics of rapid adaptation. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 195–215. [Google Scholar] [CrossRef]
- Gillespie, D.T. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 1976, 22, 403–434. [Google Scholar] [CrossRef]
- Gillespie, D.T. Stochastic simulation of chemical kinetics. Annu. Rev. Phys. Chem. 2007, 58, 35–55. [Google Scholar] [CrossRef]
- Marchetti, L.; Priami, C.; Thanh, V.H. Simulation Algorithms for Computational Systems Biology; Springer: Cham, Switzerland, 2017; Volume 1. [Google Scholar]
- Filippov, A. Differential Equations with Discontinuous Righthand Sides; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Callier, F.M.; Desoer, C.A. Linear System Theory; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Gessler, D.D. The constraints of finite size in asexual populations and the rate of the ratchet. Genet. Res. 1995, 66, 241–253. [Google Scholar] [CrossRef]
- Polański, A.; Kania, M.; Gil, J.; Łabaj, W.; Lach, E.; Szcęsna, A. Modeling Propagation of Weakly Advantageous Mutations in Cancer Cells. Int. J. Appl. Math. Comput. Sci. 2025, 35, 479–492. [Google Scholar] [CrossRef]
- Perera, D.; Li, E.; Gordon, P.M.; van der Meer, F.; Lynch, T.; Gill, J.; Church, D.L.; de Koning, A.J.; Huber, C.D.; van Marle, G.; et al. Apollo: A comprehensive GPU-powered within-host simulator for viral evolution and infection dynamics across population, tissue, and cell. Nat. Commun. 2025, 16, 5783. [Google Scholar] [CrossRef] [PubMed]
- Zanini, F.; Puller, V.; Brodin, J.; Albert, J.; Neher, R.A. In vivo mutation rates and the landscape of fitness costs of HIV-1. Virus Evol. 2017, 3, vex003. [Google Scholar] [CrossRef] [PubMed]
- Chao, L. Fitness of RNA virus decreased by Muller’s ratchet. Nature 1990, 348, 454–455. [Google Scholar] [CrossRef] [PubMed]
- Boezen, D.; Ali, G.; Wang, M.; Wang, X.; van der Werf, W.; Vlak, J.M.; Zwart, M.P. Empirical estimates of the mutation rate for an alphabaculovirus. PLoS Genet. 2022, 18, e1009806. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łabaj, W.; Gil, J.; Kania, M.; Lach, E.; Szczęsna, A.; Polański, A. Deterministic Modeling of Muller’s Ratchet Effect in Populations Evolving in an Environment of Finite Capacity. Appl. Sci. 2025, 15, 11090. https://doi.org/10.3390/app152011090
Łabaj W, Gil J, Kania M, Lach E, Szczęsna A, Polański A. Deterministic Modeling of Muller’s Ratchet Effect in Populations Evolving in an Environment of Finite Capacity. Applied Sciences. 2025; 15(20):11090. https://doi.org/10.3390/app152011090
Chicago/Turabian StyleŁabaj, Wojciech, Jarosław Gil, Mateusz Kania, Ewa Lach, Agnieszka Szczęsna, and Andrzej Polański. 2025. "Deterministic Modeling of Muller’s Ratchet Effect in Populations Evolving in an Environment of Finite Capacity" Applied Sciences 15, no. 20: 11090. https://doi.org/10.3390/app152011090
APA StyleŁabaj, W., Gil, J., Kania, M., Lach, E., Szczęsna, A., & Polański, A. (2025). Deterministic Modeling of Muller’s Ratchet Effect in Populations Evolving in an Environment of Finite Capacity. Applied Sciences, 15(20), 11090. https://doi.org/10.3390/app152011090





