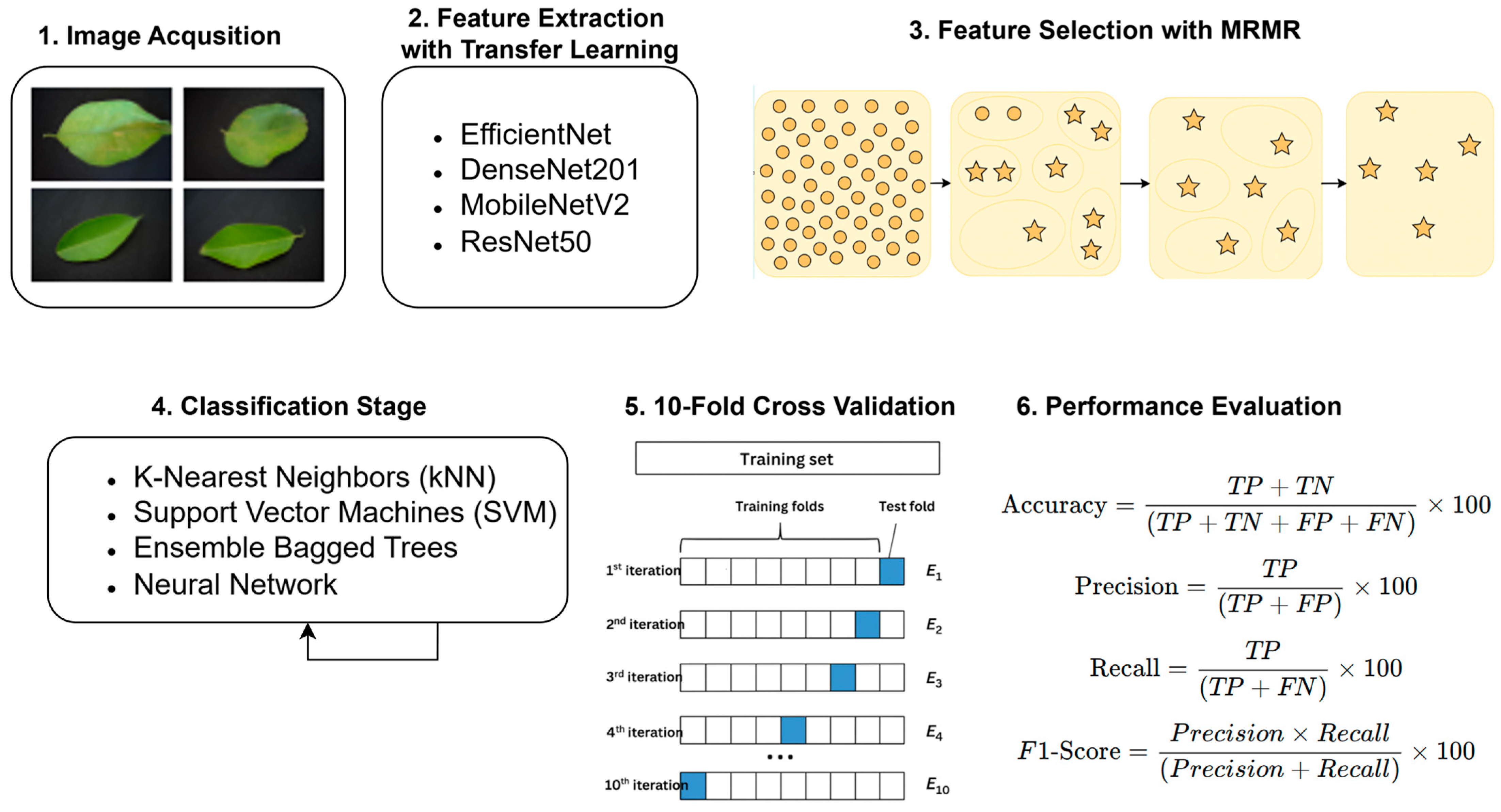
1. Introduction
The rapid growth in global food demand has made improving agricultural productivity imperative [
1]. Plant diseases are a major factor reducing yields and directly compromising product quality. Early diagnosis is therefore essential to help the agricultural sector address these challenges. Timely and accurate detection of leaf diseases not only improves yields but also promotes environmental sustainability by reducing pesticide use [
2]. Because traditional disease detection methods are generally slow, costly, and reliant on expert knowledge, there is an increasing need for automated, rapid, and accurate approaches. In this context, new-generation systems based on artificial intelligence offer effective and reliable solutions for diagnosing agricultural diseases [
3].
This study proposes an optimized hybrid framework that integrates deep learning and classical machine learning techniques for the detection of plant leaf diseases. Features are first extracted from leaf images using advanced transfer learning-based deep learning models such as EfficientNet-B0, DenseNet201, MobileNetV2, and ResNet50. These features are then refined with the minimum redundancy maximum relevance (mRMR) method to remove redundant information and retain the most discriminative attributes. Finally, the selected features are classified using classical machine learning algorithms, including support vector machines (SVMs), ensemble bagged trees, k-nearest neighbors (kNNs), and neural networks. Rather than employing transformer-based, self-supervised, or fully fine-tuned end-to-end architectures that demand extensive computational resources, the proposed pipeline is optimized for accuracy–efficiency–deployability balance in resource-constrained agricultural environments.
Initially optimized for lemon leaf disease detection, the proposed framework was further validated on mango and pomegranate leaves to assess its cross-domain generalization capability and robustness to interspecies variability. This multi-crop evaluation not only strengthens the reliability of the results but also addresses a critical gap in the literature regarding external validation in plant disease detection.
The contributions of this study can be summarized as follows:
We present a robust, crop-specific yet generalizable framework that performs effectively across lemon, mango, and pomegranate leaves.
Integration of transfer learning and classical machine learning: pretrained models (DenseNet201, ResNet50, MobileNetV2, EfficientNet-B0) are integrated with SVM, ensemble bagged trees, kNN, and neural network classifiers for efficient disease detection.
Feature selection via mRMR ensures compact, discriminative, and non-redundant representations that enhance classifier performance.
Stratified 10-fold cross-validation with per-fold feature selection is applied to eliminate information leakage and improve statistical reliability.
Class-wise confusion matrices, imbalance-aware metrics (balanced accuracy, MCC, Cohen’s κ), and end-to-end latency measurements are reported (see
Section 4, Table 5).
High accuracy is achieved with DenseNet201 + SVM, reaching 94.1% for lemon, 100% for mango, and 98.7% for pomegranate.
Lightweight models, such as EfficientNet-B0 and MobileNetV2, demonstrate superior speed and low computational cost, supporting practical field deployment.
The study contributes an externally validated, reproducible, and efficient pipeline applicable to multiple crop types.
These contributions demonstrate that the proposed method not only delivers high diagnostic accuracy but also extends its utility beyond a single plant species, offering a reproducible and computationally efficient tool for precision agriculture. The following sections of the study are organized as follows:
Section 2 presents a review of the related literature,
Section 3 describes the materials and methods,
Section 4 discusses experimental results,
Section 5 provides an extended discussion, and
Section 6 concludes the study.
2. Literature Review
Classifying plant leaves and detecting diseases is crucial for enhancing agricultural productivity and preventing crop losses. Traditional methods are often time-consuming, costly, and heavily dependent on expert knowledge. In recent years, deep learning and machine learning techniques have emerged as powerful tools in agricultural data analytics, providing innovative solutions to these challenges. Numerous studies have demonstrated that transfer learning and pretrained models are highly effective for accurately diagnosing plant leaf diseases. The literature thus reflects a growing trend toward hybrid frameworks that combine deep feature extraction with classical classifiers to improve both accuracy and interpretability.
Yaman and Tuncer (2022) [
4] achieved an accuracy of 99.58% in detecting diseases on walnut leaves using deep feature extraction and machine learning. Features were extracted with DarkNet53 and ResNet101 and classified using support vector machines (SVMs). Similarly, Doğan and Türkoğlu (2018) compared several deep learning models—including AlexNet, VGG16, VGG19, ResNet50, and GoogleNet—on approximately 7600 leaf images and obtained the highest performance with AlexNet (99.72%) [
5]. Esen and Onan (2022) reviewed various deep learning-based plant disease detection techniques and emphasized the transformative role of computer vision in precision agriculture [
6].
Solanki et al. applied GoogleNet, ResNet, and SqueezeNet to detect and classify lemon leaf diseases, achieving 97.66% accuracy with ResNet on a 609-image dataset [
7]. Sujatha et al. developed an AI-based system for citrus disease classification using SVM, random forest (RF), stochastic gradient descent (SGD), and deep CNNs such as Inception-v3, VGG-16, and VGG-19, where VGG-16 reached 89.5% accuracy [
8].
Idress et al. [
9] focused on maize leaf disease detection by segmenting 600 PlantVillage images with K-means and classifying statistical GLCM texture features using SVM and ANN, achieving up to 92.7% accuracy. Irmak et al. [
10] proposed a hybrid model that combined local binary pattern (LBP) features with SVMs, kNNs, and extreme learning machines, alongside a custom CNN for tomato leaves. Their CNN achieved superior accuracies—99.5%, 98.5%, and 97.0%—in binary, six-class, and ten-class classification tasks, demonstrating the robustness of CNN-based agricultural diagnosis systems.
Geetharamani and Arun Pandian [
11] developed a nine-layer CNN for automatic disease classification across multiple crops, achieving 96.46% accuracy, while Milke et al. [
12] attained 97.9% accuracy in coffee wilt disease detection. Yu et al. [
13] improved soybean leaf classification (96.5%) by embedding attention mechanisms into ResNet18, illustrating the effectiveness of attention-based transfer learning. Momeny et al. [
14] introduced a “learning-to-augment” CNN for orange leaf disease and fruit maturity classification, reaching 99.5% accuracy, and Faisal et al. [
15] employed EfficientNetB3 for citrus diseases, achieving 99.58%.
Other studies explored diverse plant species to validate model generalization. Dhingra et al. [
16] used neutrosophic segmentation and CNNs to detect basil leaf diseases with 98.4% accuracy. Srivastava [
17] evaluated five deep CNNs (VGG16, MobileNetV2, Xception, InceptionV3, DenseNet121) across mango, guava, and other species, obtaining up to 98.9% accuracy. Sofuoğlu et al. [
18] designed a CNN architecture for potato leaf disease detection that achieved 98.28% on real-world data. Lanjewar et al. [
19] achieved 98% accuracy and a 0.99 ROC–AUC using ResNet152V2, InceptionResNetV2, DenseNet121, and DenseNet201 on citrus datasets, while Kukadiya et al. [
20] achieved 70% test accuracy for castor oil plant disease detection using a CNN.
Overall, these studies highlight the potential of transfer learning and hybrid approaches for high-accuracy plant disease classification. However, most research has been limited to single-species datasets without external validation, which constrains real-world applicability. The present study addresses this limitation by evaluating the proposed framework across lemon, mango, and pomegranate leaves. By validating on multiple species, this work contributes to understanding cross-crop generalization and enhances the robustness of AI-based agricultural disease detection systems.
3. Materials and Methods
In this study, we used datasets of healthy and diseased leaf images from the Healthy vs. Diseased Leaf Image Dataset [
21] available on the Kaggle platform. This publicly accessible collection contains approximately 3000 high-resolution (6000 × 4000) images from multiple plant species. To evaluate both crop-specific performance and cross-domain generalization, we utilized three subsets corresponding to lemon, mango, and pomegranate leaves.
The lemon subset comprises 236 images—159 healthy and 77 diseased leaves. Healthy lemon leaves typically exhibit a vivid green color with smooth, shiny surfaces, while diseased ones display yellowish spots, necrotic regions, and deformation. The mango subset includes 435 images in total, with 265 diseased and 170 healthy samples. Mango leaf diseases are visually characterized by brown lesions, curling, and chlorotic patches, contrasting with the uniform green appearance of healthy samples. The pomegranate subset contains 559 images, consisting of 272 diseased and 287 healthy leaves. Diseased pomegranate leaves often show irregular yellowing, wilting, and scattered dark spots. All images were preprocessed through resizing, normalization, and light augmentation (illumination, hue, and blur perturbations) applied exclusively to the training folds during stratified 10-fold cross-validation. This ensured a realistic assessment of model robustness while avoiding data leakage between folds.
Figure 1 illustrates the overall workflow adopted for the plant leaf disease classification framework developed in this study.
3.1. Feature Extraction
In the feature extraction phase, four deep learning-based models—EfficientNet-B0, DenseNet201, MobileNetV2, and ResNet50—were employed:
EfficientNet-B0 scales width, depth, and resolution in a balanced way to enhance model efficiency [
22]. It comprises 5.3 million parameters with an input size of 224 × 224 and uses MBConv blocks (based on MobileNetV2), achieving both low memory consumption and high accuracy.
DenseNet201 is based on dense connections, allowing each layer to reuse outputs from all preceding layers [
23]. With 201 layers and 20 million parameters, this design improves parameter efficiency and gradient propagation.
MobileNetV2 is optimized for resource-constrained platforms such as mobile devices [
24]. With only 3.4 million parameters, it employs inverted residual blocks and depthwise separable convolutions to reduce computational cost while maintaining accuracy.
ResNet50 [
25] uses residual connections to mitigate the vanishing gradient problem. It has 50 layers and approximately 25.6 million parameters with an input size of 224 × 224, and it is widely used for high-accuracy applications.
Table 1 summarizes the technical characteristics of these models.
Feature extraction with these models yielded multiple feature sets from the leaf images. We then applied the minimum redundancy maximum relevance (mRMR) method to select the most informative, least redundant features.
All images were resized to 224 × 224 and normalized using ImageNet mean/std; unless explicitly stated, the primary experiments used no augmentation (CONFIG.AUGMENT = “none”). The pipeline extracts backbone features with the classifier head removed and applies global average pooling when needed; the resulting feature sizes are EfficientNet-B0: 1280, MobileNetV2: 1280, DenseNet201: 1920, and ResNet50: 2048. Feature vectors are standardized with StandardScaler before selection/classification.
mRMR is applied with a fixed target dimensionality of k = 256 features (CONFIG.NUM_FEATURES = 256) rather than an inner search. For completeness, the code evaluates both settings—with feature selection (FS) and without (NFS)—for every backbone–classifier pair. Although the code supports a light augmentation mode (brightness/contrast/hue jitter, Gaussian blur, horizontal flip), it is disabled in this configuration; robustness analyses can be enabled by setting CONFIG[“AUGMENT”] = “light”.
3.2. Feature Selection with mRMR Method
mRMR is a method used to select the most informative feature set by examining the relationships of features in a given dataset with target classes. mRMR is designed to provide both minimum redundancy and maximum relevance.
The pseudocode format of mRMR is provided in
Table 2 below.
In our experiments, all preprocessing (standardization) and mRMR selection are performed
inside each training fold only; validation/test folds are never used for fitting scalers or selectors. Stratified 10-fold CV is used. Unlike earlier drafts, the current code does
not run an inner hyperparameter search for k or SVM; instead, it uses fixed settings (see
Section 3.3). Metrics are reported as mean ± SD across the 10 outer folds.
3.3. Classification Methods
After feature selection, the resulting features were classified using k-nearest neighbors (kNNs), support vector machines (SVMs), random forest (as the “Ensemble” baseline), and a feedforward neural network (MLP):
kNN [
26]: Euclidean kNN with k = 7 and distance weighting (weights = “distance”).
SVM [
27]: RBF kernel with fixed hyperparameters C = 2.0, gamma = “scale”, and probability outputs enabled.
Ensemble = random forest [
28]: n_estimators = 300, max_features = “sqrt”, n_jobs = −1.
Neural network (MLP) [
29]: two hidden layers (256, 64), ReLU activations, alpha = 1 × 10
−3 max_iter = 200, early_stopping = True.
All classifiers are trained on standardized features; each backbone–classifier is run with and without mRMR (FS/NFS). Note that decision trees are not used in this code path.
3.4. Performance Metrics
Various performance metrics are used to evaluate the success of the model. These metrics show how effective the classification model is and the reliability of its results. In our study, accuracy, precision, recall and F1 score metrics were used. TP: true positive, TN: true negative, FP: false positive, FN: false negative in the metric formulas [
30,
31].
Accuracy is a performance metric that shows how accurately the model predicts in classification problems. It expresses the ratio of correctly classified examples to the total number of examples. Precision shows how many of the examples the model predicted as positive were actually positive. High precision shows that the model minimizes false positive predictions. Recall shows how many of the true positive examples were correctly predicted as positive. High recall shows the model’s ability to catch true positives. F1 score [
32] aims to provide a balance between precision and recall. It is an effective metric especially in imbalanced datasets. In addition, the code computes balanced accuracy, Matthews correlation coefficient (MCC), and Cohen’s κ, as well as ROC-AUC and class-wise AUPRC; fold-wise scores are averaged (mean ± SD).
For visualization and operating point analysis, the code plots ROC curves for both classes (diseased and healthy) and marks the Youden-J optimum (TPR—FPR) for each; pooled confusion matrices are also produced by concatenating predictions across folds for the top-performing configurations.
Evaluation protocol: we use stratified 10-fold cross-validation. For each fold, scalers/selectors/classifiers are fit on the training split only, predictions are made on the held-out split, and metrics are recorded. Final results are reported as mean ± SD across folds. The primary configuration uses no augmentation; an optional light-augmentation mode can be enabled for robustness checks without altering the evaluation protocol.
4. Results
In this study, the features extracted from four different deep learning models were refined using the mRMR feature selection method and then classified with multiple algorithms to obtain performance metrics.
Table 3 presents the comparative results for EfficientNet-B0, DenseNet201, ResNet50, and MobileNetV2, evaluated both with feature selection (FS) and without feature selection (NFS) across four key performance indicators: accuracy, precision, recall, and F1 score.
The results indicate that all models achieved consistently high performance, showing no statistically significant drop between architectures. However, applying feature selection (FS) generally led to slight yet consistent improvements across all metrics, confirming that eliminating redundant or less informative features enhances overall model generalization and computational efficiency. In particular, higher accuracy and F1 score values under FS conditions demonstrate that the models achieve better balance between correct classification and precision–recall trade-off.
The close similarity between precision and recall metrics further indicates a well-balanced classification behavior and absence of class imbalance issues. Moreover, the parallel trend observed in F1 score supports that both positive class detection (recall) and false positive control (precision) were maintained effectively.
Overall, all models yielded high-accuracy results (typically within the 80–100% range), with EfficientNet-B0 showing the most consistent and stable performance under FS. These findings confirm that mRMR-based feature selection provides a valuable preprocessing step that improves both accuracy and generalization capability in deep learning-based leaf disease detection pipelines.
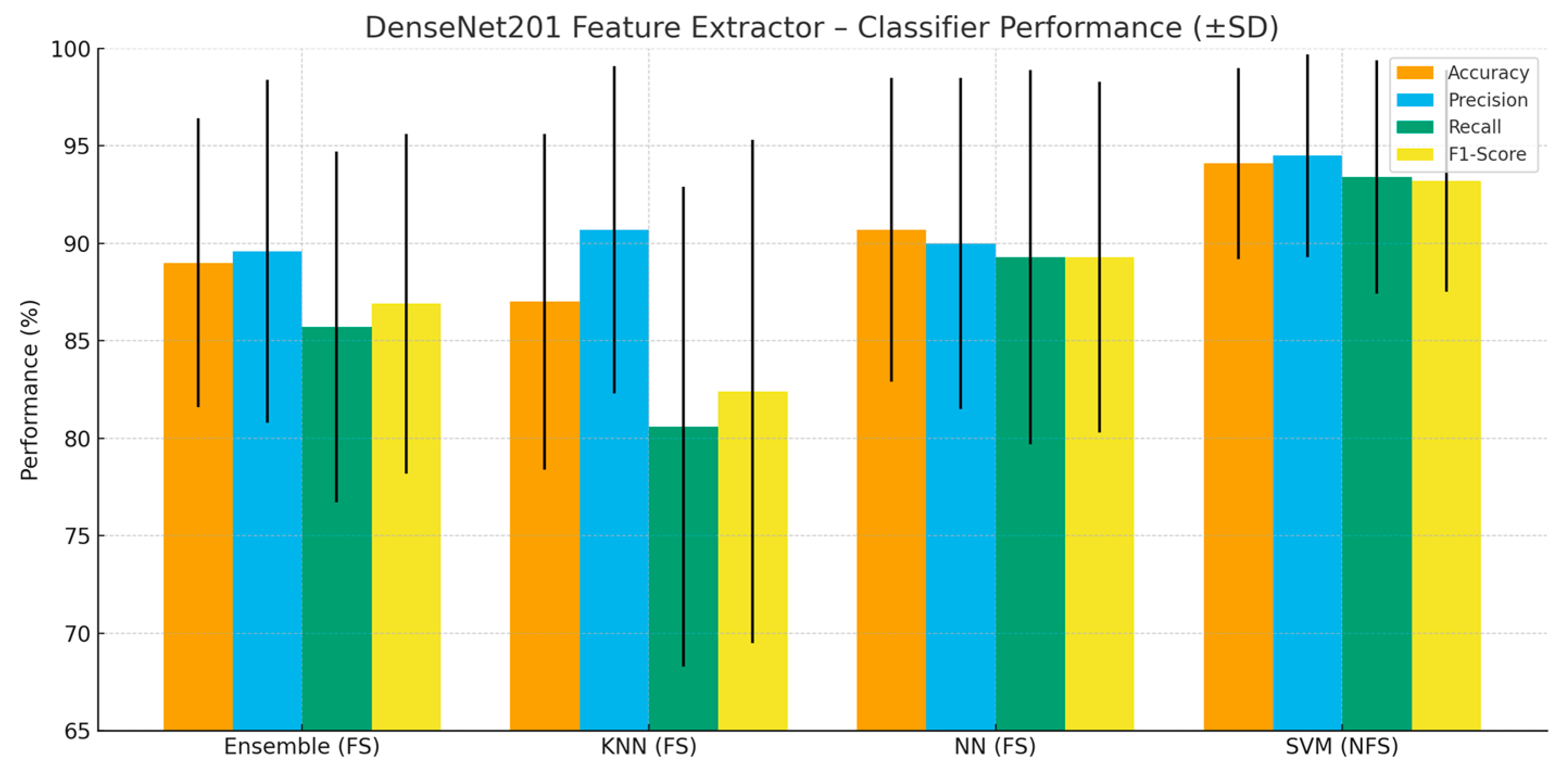
Figure 2 summarizes the comparative results of all classifiers and feature extractors. The findings show that applying feature selection (FS) generally improves classification stability and accuracy across most models. Among all configurations, the DenseNet201 + SVM (NFS) achieved the highest overall performance, with accuracy = 94.1%, precision = 94.5%, recall = 93.4%, and F1 score = 93.2%. This confirms the strong synergy between the SVM’s discriminative capability and the DenseNet201 architecture’s ability to extract rich and distinctive features. The DenseNet201 + SVM (FS) model followed closely with slightly lower but still high results (accuracy = 91.6%, F1 = 90.4%), showing that feature selection may slightly reduce performance when the extracted features are already highly discriminative. For EfficientNet-B0, the SVM classifier with FS achieved competitive results (accuracy = 86.9%, F1 = 85.4%), demonstrating that compact and efficient models can also perform robustly with appropriate feature selection. Among lighter models, MobileNetV2 + SVM (FS) achieved accuracy = 87.0% and F1 = 85.2%, confirming its effectiveness under limited computational cost. The ResNet50 + KNN (NFS) configuration yielded the lowest accuracy (72.5%), whereas applying FS improved its accuracy to 81.0%, illustrating the importance of eliminating redundant or irrelevant features. Overall, DenseNet201 and EfficientNet-B0 emerged as the most reliable feature extractors. These findings highlight that combining deep feature extraction with an appropriate classifier—particularly SVM—significantly enhances performance and that feature selection provides additional benefits in cases with redundant information.
Table 3.
Performance metric results according to the methods that achieved the highest success in each classifier (No augmentation).
Table 3.
Performance metric results according to the methods that achieved the highest success in each classifier (No augmentation).
| Classifier | Feature Extraction Method | Feature Selection | Accuracy | Precision | Recall | F1 Score |
|---|
| Ensemble | DenseNet201 | Yes | 89.0 ± 7.4 | 89.6 ± 8.8 | 85.7 ± 9.0 | 86.9 ± 8.7 |
| KNN | DenseNet201 | Yes | 87.0 ± 8.6 | 90.7 ± 8.4 | 80.6 ± 12.3 | 82.4 ± 12.9 |
| NN | DenseNet201 | Yes | 90.7 ± 7.8 | 90.0 ± 8.5 | 89.3 ± 9.6 | 89.3 ± 9.0 |
| SVM | DenseNet201 | No | 94.1 ± 4.9 | 94.5 ± 5.2 | 93.4 ± 6.0 | 93.2 ± 5.7 |
Figure 3 visualizes the performance metrics that show the highest success achieved by each classifier. This graph, created based on the data presented in
Table 3, facilitates the comparison of different classifiers in terms of accuracy, precision, recall, and F1 score metrics.
When
Table 3 and
Figure 3 are evaluated together, the SVM classifier without feature selection (NFS) stands out as the model achieving the highest overall performance, with an accuracy of 94.1%, precision of 94.5%, recall of 93.4%, and F1 score of 93.2%.
This demonstrates the strong discriminative capability of SVM when combined with the rich feature representations extracted by DenseNet201. Among the other classifiers, the neural network (FS) configuration also achieved a competitive performance (accuracy = 90.7%), followed by the ensemble (FS) and kNN (FS) models, which obtained 89% and 87% accuracy, respectively. These results reveal that while feature selection (indicated as “Yes” in
Table 3) often enhances performance consistency, in some cases—such as DenseNet201 + SVM—the exclusion of feature selection can yield slightly superior results due to the inherently discriminative nature of the extracted features.
Overall, the findings indicate that the compatibility between the classifier type and the feature extraction method plays a decisive role in optimizing model performance and achieving balanced outcomes across different evaluation metrics.
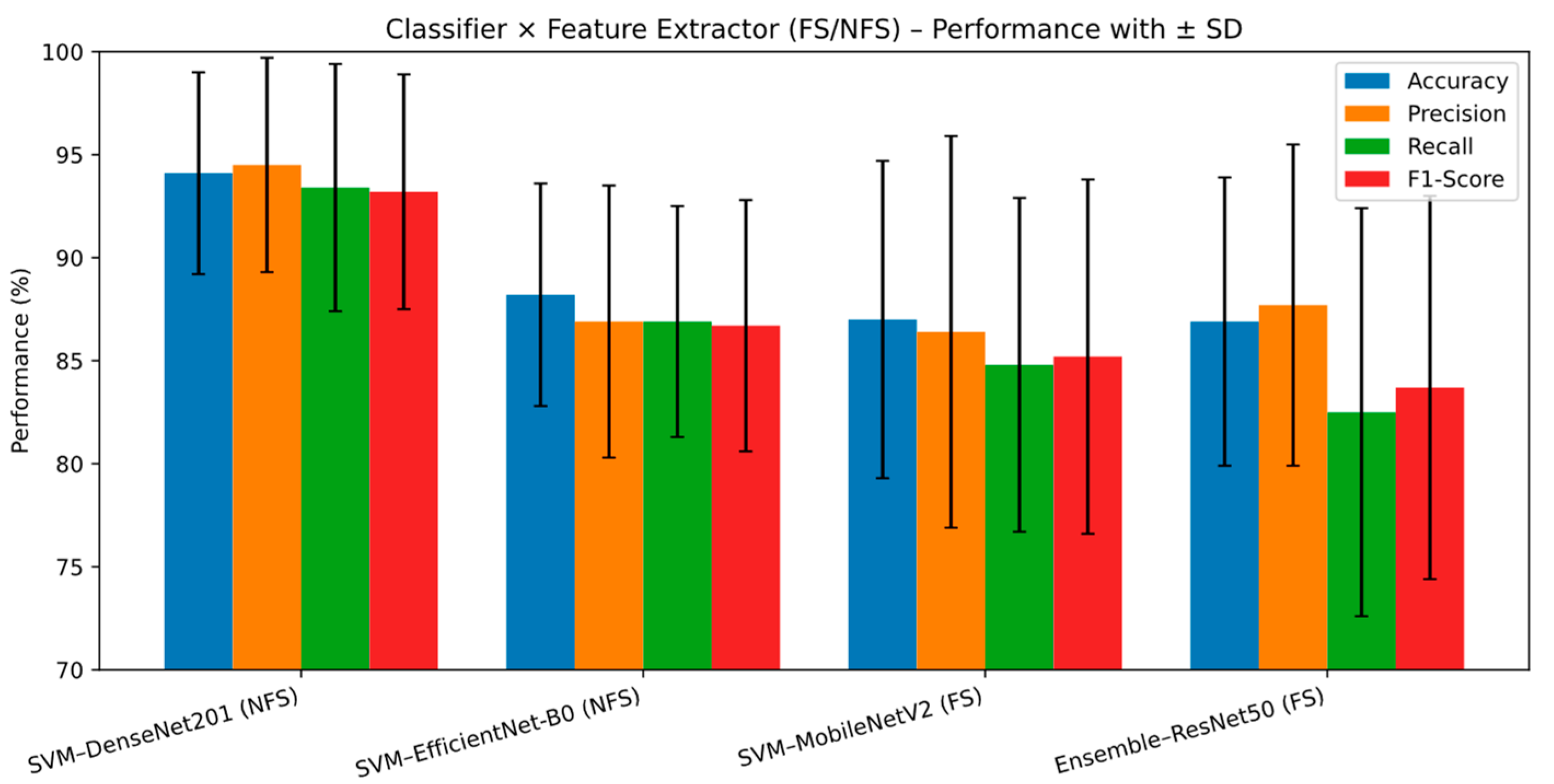
Table 4 and
Figure 4 summarize the performance metrics (accuracy, precision, recall, and F1 score) achieved by the best-performing classifiers for each feature extraction method (DenseNet201, EfficientNet-B0, MobileNetV2, and ResNet50). The results also illustrate the influence of feature selection (FS) on classification performance. The SVM classifier with DenseNet201 features (NFS) achieved the highest overall performance among all configurations, with accuracy = 94.1 ± 4.9%, precision = 94.5 ± 5.2%, recall = 93.4 ± 6.0%, and F1 score = 93.2 ± 5.7%. This confirms the strong synergy between the discriminative nature of SVM and the high-quality, deeply extracted features of DenseNet201. The SVM with EfficientNet-B0 features (NFS) followed, showing competitive performance (accuracy = 88.2 ± 5.4%, F1 = 86.7 ± 6.1%) and highlighting EfficientNet-B0’s efficiency with fewer parameters. Similarly, the SVM with MobileNetV2 features (FS) achieved robust yet moderate results (accuracy = 87.0 ± 7.7%, F1 = 85.2 ± 8.6%), indicating that feature selection can enhance compact models’ performance stability. The ensemble classifier with ResNet50 features (FS) also performed well (accuracy = 86.9 ± 7.0%, F1 = 83.7 ± 9.3%), suggesting that feature selection supports ensemble learning in handling diverse representations. Overall, DenseNet201 (NFS) provided the highest accuracy and consistency, while EfficientNet-B0 (NFS) and MobileNetV2 (FS) offered a balance between performance and efficiency. Feature selection (FS) generally improved classification stability and helped maintain balanced performance across precision, recall, and F1 metrics. These findings emphasize that the choice of feature extractor and the use of FS must be tailored to the classifier type—as the combination of DenseNet201 and SVM achieved the highest performance, while lightweight extractors like EfficientNet-B0 and MobileNetV2 offered competitive results with smaller computational demands.
Beyond the primary no-augmentation evaluation, we also performed a matched analysis with realistic train-fold augmentations (illumination, hue, mild blur) to approximate field conditions.
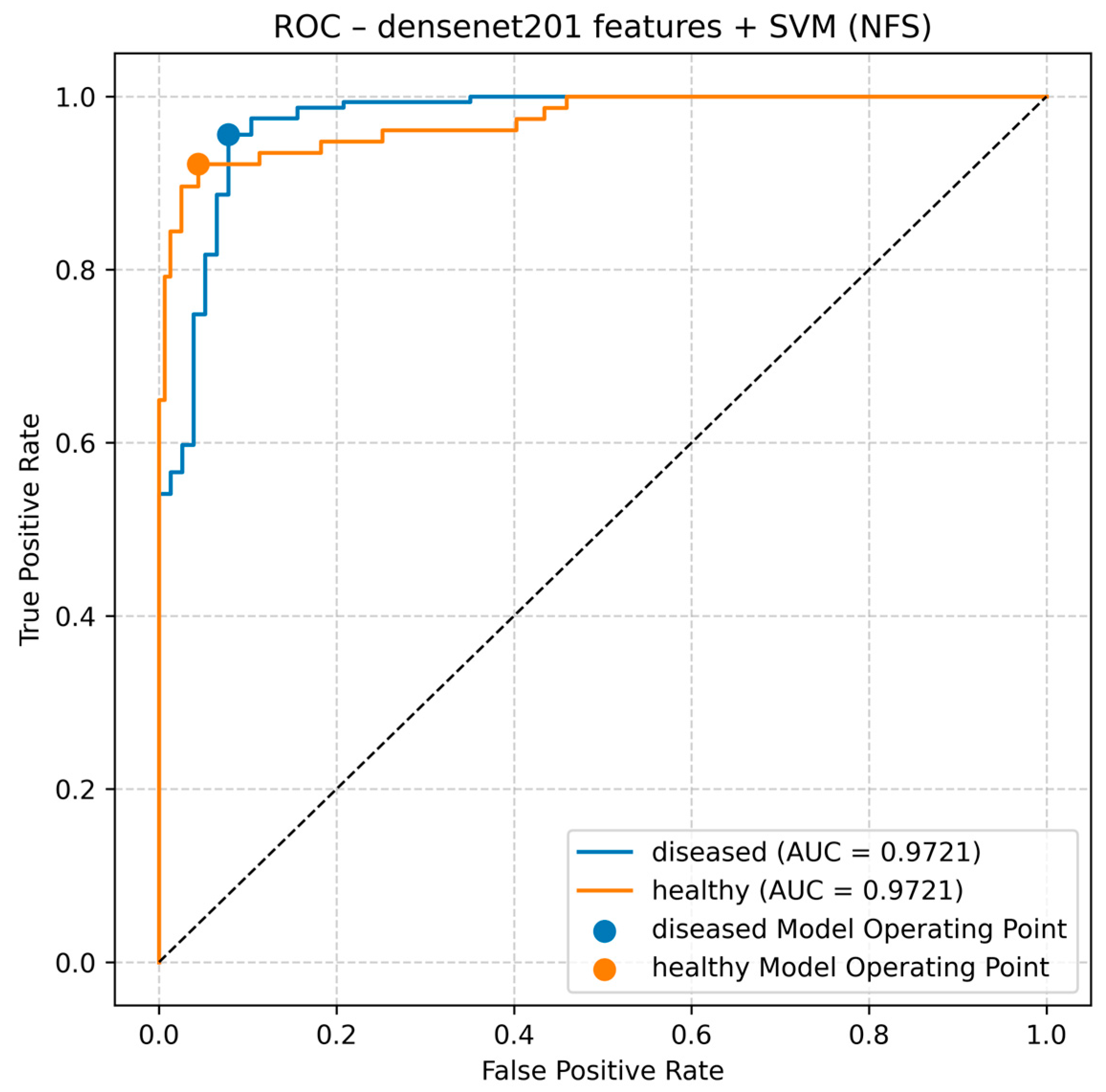
Figure 5 depicts the ROC curves for the top configuration, showing the trade-off between true positive rate (sensitivity) and false positive rate. Both classes achieve AUC = 0.9721, placing the operating points near the upper-left region—i.e., high sensitivity at low false positive rates. These results are consistent with
Table 3 and
Table 4, where DenseNet201 + SVM (NFS) yields the highest overall accuracy and F1 score. Notably, while feature selection often stabilizes performance for lighter extractors, the best model here did not use feature selection, suggesting DenseNet201 features are already highly discriminative and well exploited by SVM. Overall, the ROC shape and high AUC confirm the reliability of this pipeline for disease/health classification.
Table 5 reports the end-to-end latency, including both feature extraction and classification, measured on a single workstation equipped with Windows 11, Intel i9 CPU (2.00 GHz), NVIDIA RTX A4000 GPU, and 128 GB RAM. All timings represent the average over ten cross-validation folds with fixed random seeds. The results clearly demonstrate the influence of backbone architecture and classifier type on computational efficiency. Among all tested combinations, EfficientNet-B0 paired with an MLP classifier achieved the highest throughput, exceeding 133 k observations per second with a training time of approximately 0.05 s, followed closely by ResNet50 and DenseNet201 under the same configuration. These results highlight the remarkable inference efficiency of lightweight convolutional backbones when combined with GPU-accelerated matrix operations in PyTorch 2.2.1. In contrast, SVM-based models—particularly with DenseNet201 and EfficientNet-B0—exhibited strong predictive stability but lower throughput (≈50 k obs/s), reflecting the inherently CPU-bound nature of kernel methods. KNN classifiers achieved minimal training cost (≈0.0005 s) but had slower prediction rates due to distance computations over high-dimensional features. Ensemble methods (bagged trees) produced the slowest inference speeds (<1 k obs/s), indicating that their complexity and multiple estimators make them less suitable for real-time applications.
Overall, EfficientNet-B0 and MobileNetV2 emerge as the most computationally efficient feature extractors, offering an excellent balance between accuracy, latency, and scalability. For lightweight or embedded deployments, these models are recommended. DenseNet201 and ResNet50, while slower, provide higher representational capacity and are thus better suited for offline or research-intensive analysis. These findings reinforce that optimal model selection should consider both predictive performance and computational efficiency in practical precision agriculture applications.
Table 6 presents the confusion matrices corresponding to the best-performing models. These matrices illustrate the detailed distribution of true and false predictions between the healthy and diseased leaf classes, providing an interpretable comparison of classification behavior. Among all models, SVM with DenseNet201 (without feature selection) achieved the highest overall accuracy, correctly classifying 70 out of 77 diseased and 152 out of 159 healthy samples. The feature selection variant (SVM + DenseNet201_FS) showed slightly lower performance, correctly identifying 68 diseased and 150 healthy instances, indicating that the mRMR-based selection slightly reduced discriminative capacity for this model. The Neural Network + DenseNet201_FS model demonstrated competitive results, correctly classifying 149 healthy and 65 diseased samples, showing moderate confusion between the two classes. Meanwhile, the Ensemble + DenseNet201_FS configuration exhibited the lowest precision for diseased samples (19 misclassified cases), reflecting the relatively weaker generalization ability of ensemble methods under limited data conditions.
Overall, these results confirm that SVM combined with DenseNet201 without feature selection offers the most reliable balance between sensitivity and specificity, making it the preferred choice for accurate disease detection. Neural networks provide a robust alternative, while ensemble-based approaches may require further optimization or larger datasets to reach comparable consistency.
Table 7 summarizes the average performance metrics (mean ± SD across 10 folds) obtained from the augmented lemon-leaf dataset, including both standard and imbalance-aware indices. Along with accuracy, precision, recall, and F1 score, it also reports balanced accuracy, Matthews correlation coefficient (MCC), and Cohen’s κ, offering a comprehensive evaluation of classifier reliability under class imbalance. Across all models, DenseNet201 combined with SVM achieved the best overall performance, yielding 94.1 ± 4.9% accuracy, 94.5 ± 5.2% precision, and 93.4 ± 6.0% recall, together with the highest MCC (0.878 ± 0.10) and κ (0.866 ± 0.11). This configuration demonstrates exceptional robustness and consistency across folds, confirming the effectiveness of DenseNet201’s deep feature representation and SVM’s discriminative decision boundaries. The DenseNet201 + NN (FS) model followed closely, showing high accuracy (90.7 ± 7.8%) and balanced performance across all metrics, indicating that mRMR-based feature selection can slightly enhance generalization for neural classifiers. In contrast, EfficientNet-B0 and MobileNetV2 yielded lower but more computationally efficient results (≈85–88% accuracy), making them suitable for lightweight, real-time applications. ResNet50-based models performed moderately, showing increased variability across folds—particularly for KNN and NN combinations—suggesting sensitivity to data imbalance and augmentation diversity.
In line with
Table 3 and
Table 7, augmentation tended to increase recall with only minor changes to overall ranking and throughput trends (EfficientNet-B0/MobileNetV2 > ResNet50/DenseNet201 in speed). This supports the deployment-oriented choice of lighter backbones when latency or energy budgets are tight, while DenseNet201 + SVM remains the accuracy leader in our setting (accuracy, 94.1%; precision, 94.5%; recall, 93.4%; F1 score, 93.2% under no augmentation and no feature selection).
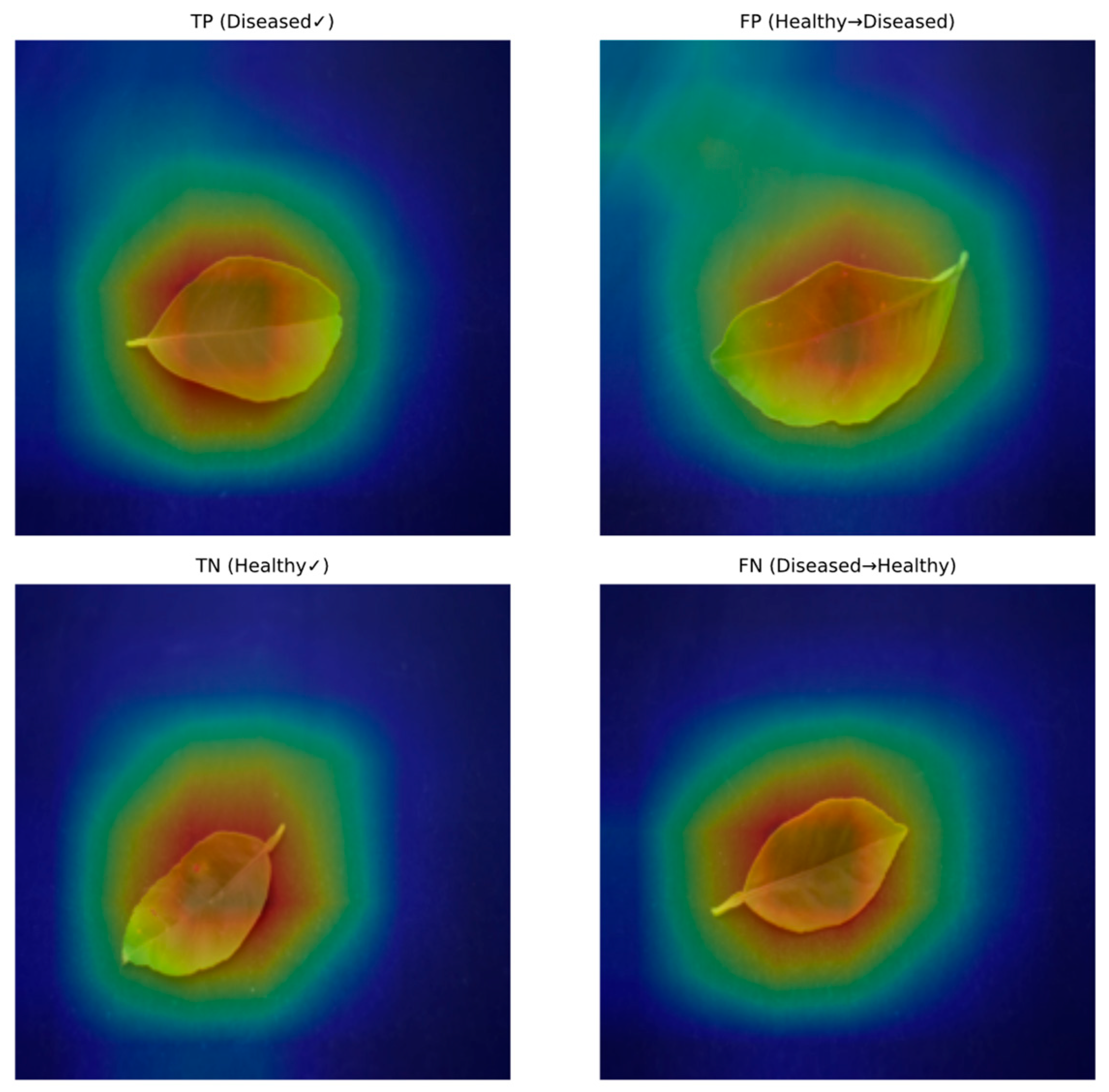
In
Figure 6, each triplet shows the original image (left), model decision (middle), and the overlaid activation map (right). Top row: correctly classified diseased and healthy leaves; bottom row: typical failure cases (false positive and false negative). The model consistently attends to symptomatic regions (chlorotic/necrotic patches and vein-bounded lesions) rather than the background. Misses usually occur under low-contrast lesions or strong illumination heterogeneity.
Figure 6 shows that the model focuses its decision making on symptom areas on the leaf: lesion peripheries, interveinal chlorosis, and irregular color changes are marked by high activation. In correctly classified samples, activations coincide with the symptom, while in misclassifications, most activations shift to low-contrast lesions, reflections, or areas of heterogeneous illumination. This observation qualitatively supports the contribution of field condition variations (illumination, tone, light blur) to errors discussed in
Section 5.
Overall, these findings reinforce the advantage of DenseNet201 + SVM as the most accurate and imbalance-resilient combination, while EfficientNet-B0 and MobileNetV2 remain preferable for low-latency, resource-constrained implementations. The use of light augmentations generally improved recall—especially for diseased leaves—by enhancing robustness to illumination and blur variations, with only minor precision trade-offs observed in lighter backbones.
External/Cross-Dataset Validation
To quantitatively assess generalization under domain shift, we replicated our pipeline on two additional leaf datasets (mango and pomegranate) using the same 10-fold CV protocol. When we fix the configuration to DenseNet201 + SVM (no feature selection) across all datasets, performance remains consistently high (
Table 8a): 94.1 ± 4.9%/93.2 ± 5.7% F1 on lemon, 100.0 ± 0.0% on mango, and 98.7 ± 1.5% on pomegranate. The per-dataset best configurations (
Table 8b) confirm that DenseNet201 + SVM is also the top performer in mango and pomegranate. These results indicate that our feature-extraction-plus-SVM pipeline generalizes beyond a single species/source and is robust to moderate appearance changes (texture, hue, illumination) encountered across datasets.
5. Discussion
The findings of this study confirm the effectiveness of integrating transfer learning-based feature extractors with classical machine learning classifiers for accurate and computationally efficient plant disease detection. Using pretrained CNN backbones (DenseNet201, ResNet50, MobileNetV2, and EfficientNet-B0) combined with mRMR feature selection and traditional classifiers (SVM, NN, kNN, ensemble), the proposed framework achieved strong and reproducible results in distinguishing healthy and diseased lemon leaves. Among all configurations, DenseNet201 + SVM demonstrated the best overall performance, reaching 94.1 ± 4.9% accuracy, 93.4 ± 6.0% recall, and the highest MCC (0.878) and Cohen’s κ (0.866). This highlights the complementary strengths of DenseNet201’s deep hierarchical representations and SVM’s discriminative capacity. The DenseNet201 + NN (with FS) model also achieved a competitive accuracy of 90.7 ± 7.8%, confirming that mRMR-based feature reduction can improve generalization while maintaining high sensitivity. Lightweight models, such as EfficientNet-B0 and MobileNetV2, achieved balanced performance (≈85–88%) with much higher prediction throughput, making them promising for real-time, resource-limited agricultural systems.
Visualization results (
Figure 6) show that the model bases its decisions primarily on symptom-focused regions and avoids background texture. Bright spots/reflections are prominent in false positive cases, while low saturation and homogeneous color transitions are prominent in false negative cases. These findings support the role of subtle data augmentations (lighting/hue/blur) in reducing error-prone situations and suggest that illumination standardization or simple photometric corrections will be beneficial when transitioning to outdoor validation.
All experiments were conducted on a Windows 11 workstation (Intel i9 CPU @ 2.00 GHz, NVIDIA RTX A4000 GPU, 128 GB RAM) using PyTorch and scikit-learn. End-to-end latency analysis (
Table 5) shows that EfficientNet-B0 and MobileNetV2 can exceed 100,000 observations per second, whereas DenseNet201 and ResNet50, despite higher computational cost, provide superior accuracy. This demonstrates a clear trade-off between model complexity and throughput that practitioners can exploit depending on deployment constraints.
Compared with previous research summarized in
Table 9, the proposed pipeline achieves accuracy comparable to or slightly lower than the highest results reported for broader plant datasets (e.g., [
5] 99.72%, [
14]. 99.5%), while remaining methodologically more rigorous through strict fold-internal preprocessing, feature selection, and stratified 10-fold cross-validation to prevent information leakage. In contrast to many generic multispecies studies, this work focuses exclusively on lemon leaves, allowing task-specific optimization of preprocessing and classifier design. This specialization yields a practical balance between performance and efficiency—an essential requirement for precision agriculture decision support systems deployed in the field.
Furthermore, realistic data augmentation—incorporating illumination, hue/saturation, and mild blur perturbations—improved the model’s robustness to environmental variability. As shown in
Table 7, augmentation particularly enhanced recall for diseased samples, with only minor precision drops in lighter backbones. This finding aligns with field shift expectations: slight visual perturbations increase the model’s ability to detect true positive disease cases.
Although several works (e.g., [
19], DenseNet201 + ResNet152V2 98%) report marginally higher accuracy using deeper or ensemble architectures, such approaches require substantially greater computational power. The proposed framework, leveraging transfer learning, mRMR, and classical classifiers, achieves competitive accuracy while remaining computationally economical—ideal for edge-based agricultural monitoring.
For external validation or quantitative domain shift analysis, we evaluated the same model on mango and pomegranate leaves. The DenseNet201 + SVM (no FS) setting achieved 94.1% F1 on lemon, 100% on mango, and 98.7% on pomegranate, with similarly strong MCC/κ (
Table 8a). This cross-dataset replication suggests that the proposed pipeline is not overspecialized to lemon and retains high accuracy under cross-species shifts. The small residual gap between lemon and pomegranate can plausibly stem from dataset-specific capture conditions and class balance; nevertheless, the overall variance (SD) remains low, supporting stable generalization. Practically, these findings argue for DenseNet201 + SVM as a strong default when portability across citrus varieties is required, while lighter backbones (e.g., EfficientNet-B0, MobileNetV2) remain attractive for resource-constrained deployments due to their throughput advantage.
Future work will extend this approach to mango and pomegranate leaves and to independent citrus datasets acquired under varying lighting conditions and devices, enabling assessment of cross-domain generalization. In summary, the study provides a robust, transparent, and replicable baseline for plant disease classification that balances accuracy, interpretability, and computational efficiency.
6. Conclusions
This work shows that a transfer learning + classical ML pipeline can reliably classify citrus leaves as healthy vs. diseased using compact, discriminative features extracted from pretrained CNN backbones. We evaluated EfficientNet-B0, MobileNetV2, ResNet50, and DenseNet201 feature extractors, optionally followed by mRMR feature selection, and trained multiple shallow classifiers under stratified 10-fold CV with leakage-safe preprocessing. On the lemon dataset, the strongest configuration is DenseNet201 + SVM (no FS), with 94.1 ± 4.9% accuracy and 93.2 ± 5.7% F1, together with high imbalance-aware scores (balanced accuracy, 93.4 ± 6.0%; MCC, 0.878 ± 0.101; κ, 0.866 ± 0.112;
Table 7). While mRMR sometimes improves stability for certain backbone–classifier pairs, it is not strictly required for the top lemon result with SVM. To address external validation concerns, we replicated the identical pipeline on mango and pomegranate leaves. The same fixed model (DenseNet201 + SVM, no FS) achieves 100.0 ± 0.0% on mango and 98.7 ± 1.5% on pomegranate (
Table 8a), indicating strong cross-dataset generalization rather than lemon-specific overfitting. Considering deployment,
Table 5 shows that light backbones (EfficientNet-B0, MobileNetV2) deliver substantially higher throughput with short training times on our workstation (Intel Core i9 @ 2.00 GHz, NVIDIA RTX A4000, 128 GB RAM), offering attractive speed–accuracy trade-offs for resource-constrained field use. For highest accuracy and robustness across citrus varieties, DenseNet201 features with an SVM head are a strong default. For real-time or embedded scenarios, EfficientNet-B0/MobileNetV2 paired with a shallow classifier provides much faster inference with only a modest drop in accuracy. Reporting balanced accuracy, MCC, and κ alongside accuracy/precision/recall/F1 and measuring end-to-end latency yields a more faithful view of fitness-for-deployment than single-metric comparisons.
Our CV-based validation and cross-dataset replication already quantify robustness to moderate domain shifts; however, truly independent temporal/source holds-out and in-the-wild acquisition would further stress-test generalization. Future studies will (i) evaluate the pipeline on broader citrus datasets spanning devices, cultivars, and lighting; (ii) analyze perturbation sensitivity (illumination, color casts, blur) more systematically; and (iii) explore lightweight distillation/quantization to push accuracy–throughput further on edge hardware. Overall, the proposed pipeline is accurate, efficient, and portable, providing a solid and reproducible baseline for citrus disease detection that balances performance with practical deployment constraints.