Abstract
Automated medical report generation (MRG) faces a critical hurdle in seamlessly integrating detailed visual evidence with accurate clinical diagnoses. Current approaches often rely on static knowledge transfer, overlooking the complex interdependencies among pathological findings and their nuanced alignment with visual evidence, often yielding reports that are linguistically sound but clinically misaligned. To address these limitations, we propose a novel graph-driven medical report generation framework with adaptive knowledge distillation. Our architecture leverages a dual-phase optimization process. First, visual–semantic enhancement proceeds through the explicit correlation of image features with a structured knowledge network and their concurrent enrichment via cross-modal semantic fusion, ensuring that generated descriptions are grounded in anatomical and pathological context. Second, a knowledge distillation mechanism iteratively refines both global narrative flow and local descriptive precision, enhancing the consistency between images and text. Comprehensive experiments on the MIMIC-CXR and IU X-Ray datasets demonstrate the effectiveness of our approach, which achieves state-of-the-art performance in clinical efficacy metrics across both datasets.
1. Introduction
Automated medical report generation faces several critical challenges [1,2,3,4,5], particularly in producing accurate reports that demand a deep understanding of medical images and their underlying clinical significance. First, radiology images often exhibit low feature distinctiveness, limiting the reference information available for report generation models. Second, abnormal lesions may lack visually salient characteristics, making them difficult to detect even for experienced radiologists.
Beyond these inherent visual challenges, generating clinically accurate reports also requires a nuanced understanding of diagnostic semantics. A fundamental aspect of this process is disease classification, which is formulated as a supervised multi-label prediction task [6]. Here, the model must identify the presence or absence of multiple specific diseases based on radiological findings. Its performance is measured by precision, recall, and F1 score: a positive prediction indicates the model identifies a specific disease (e.g., pneumothorax) as present; precision measures the proportion of correct positive predictions, while recall measures the proportion of actual disease cases detected by the model [7]. The report generation task is a conditional language generation task, where the model must produce medical reports in free-text format [8]. Its linguistic quality and consistency with reference reports are evaluated using specialized metrics from the natural language generation (NLG) domain, such as BLEU, ROUGE, and METEOR [9,10,11]. In our report generation task, we must not only ensure the linguistic quality of the generated reports but also perform label extraction to guarantee their clinical accuracy. These two aspects correspond to the distinct tasks of disease classification and report generation. Consequently, an ideal medical report generation (MRG) system should possess the following capabilities: first, accurately identifying abnormalities in images, and then translating these findings into linguistically precise and clinically relevant text.
To enhance the performance of medical report generation (MRG), recent advancements in image captioning [12,13,14,15] have inspired various approaches. These include template retrieval frameworks [7,16], memory-driven networks [17,18], and knowledge-aware modules [19,20], all aimed at improving report accuracy. For instance, R2GenCMN [18] employs a shared memory mechanism to align visual and textual features, while GSKET [20] introduces a multi-attention framework to integrate visual features with external knowledge. Additionally, knowledge graphs have proven effective in boosting feature learning and diagnostic accuracy by embedding structured domain knowledge into models [21,22]. Multi-task learning [23,24,25] has further enhanced feature representation by jointly optimizing auxiliary tasks. Meanwhile, knowledge distillation is a model compression and optimization technique centered on transferring “knowledge” from a large, complex “teacher model” (or multiple models) to a smaller, more efficient “student model.” In traditional knowledge distillation, the teacher model’s output probabilities or intermediate feature representations guide the student model to learn the teacher’s knowledge, effectively improving the student model’s performance—sometimes even surpassing that of the teacher model. Despite these advances, a critical challenge persists: generating linguistically coherent reports that are also diagnostically precise remains an open problem.
However, existing methods often treat knowledge transfer as static, failing to adapt to varying clinical contexts or hierarchically structured medical knowledge. In radiological images, localized pathological changes are rarely independent; rather, they often reflect manifestations of a common underlying disease. These intrinsic relationships among pathological alterations are critical for radiologists’ diagnoses, yet they are frequently neglected by current AI-based methods. Furthermore, existing approaches often fail to fully capture the mapping between radiological images and textual reports, particularly in correlating localized pathologies with their corresponding lexical descriptions. This limitation hinders the generation of fine-grained, clinically accurate reports.
These shortcomings highlight the need for a framework that (1) explicitly models interdependencies among pathological findings and (2) ensures fine-grained alignment between visual features and diagnostic text. To address these challenges, we propose a graph-driven medical report generation framework that integrates knowledge graph mapping with adaptive knowledge distillation. Our approach enhances both the clinical accuracy and linguistic quality of generated reports while maintaining competitive clinical efficacy performance. Building upon an encoder–decoder architecture, we introduce a knowledge graph alongside image encoding to enable more accurate cross-modal alignment from medical multimodal data. This mapping allows the model to better correlate pathological regions with their corresponding textual descriptions, thereby improving the cueing effect of medical taxonomies for report generation. Furthermore, to prevent feature degradation and enhance supervision, we develop an information self-distillation strategy that iteratively optimizes global knowledge while incorporating intermediate-layer consistency losses to capture implicit disease relationships and temporal progression patterns.
The key contributions of our work can be summarized as follows:
(1) We leverage knowledge graph mapping to establish better cross-modal alignment between image features and pathological regions, enhancing the model’s ability to generate clinically accurate reports through taxonomy-aware feature representation.
(2) We propose a novel information self-distillation strategy that combines global knowledge transfer with local consistency constraints, improving the model’s sensitivity to both comprehensive diagnostic patterns and subtle pathological changes.
(3) Through extensive experiments on MIMIC-CXR [26] and IU X-Ray datasets [27], our framework demonstrates superior performance in generating clinically accurate and linguistically coherent reports, validating the effectiveness of both the knowledge graph integration and self-distillation strategy.
2. Related Work
2.1. Image Captioning
Conventional image captioning systems primarily employ encoder–decoder architectures trained on annotated image–text pairs to generate textual descriptions from visual inputs. The foundational work in this domain [28,29] established the paradigm of CNN-based visual feature extraction combined with RNN/LSTM-based sequential text generation. Subsequent advancements [12,13,30,31] integrated object detection frameworks to improve the representation of salient image regions. The field progressed further through the incorporation of attention mechanisms [13,32] and graph neural networks [33,34], which enhanced cross-modal alignment and reasoning capabilities.
Despite recent breakthroughs in large-scale vision–language pretraining models [15,35,36] that achieve state-of-the-art performance in general image captioning, these approaches face fundamental limitations in medical applications. While natural scene description typically emphasizes concise summarization, medical report generation requires comprehensive, clinically relevant interpretation of imaging findings. This critical distinction indicates that direct adaptation of conventional captioning methods may be suboptimal for radiological reporting.
2.2. Medical Report Generation
Medical report generation extends image description to the medical domain, requiring the generation of multiple descriptive sentences for radiological images. This task demands higher accuracy in report length and long-context descriptions. Research in medical report generation broadly falls into two main directions: one focuses on improving model architecture, while the other explores leveraging domain knowledge to guide report generation.
Regarding model architecture improvements, the primary focus lies in optimizing attention mechanisms or decoder structures. Early work adopted hierarchical approaches, utilizing layered LSTM networks to address the length requirements of medical reports, achieving effective progress in handling lengthy narratives. For instance, Liu et al. [37] proposed a hierarchical generation framework that first predicts topics relevant to the input and then generates sentences based on these topics. Xue et al. [38,39] adopted a structure incorporating both sentence-level and paragraph-level generative models, where the former guides the latter. Jing et al. [23] introduced a multi-task hierarchical model with collaborative attention, utilizing keyword prediction to assist long-paragraph generation. Subsequently, Transformers [40] were successfully applied: Hou et al. [41] developed a Transformer-based architecture utilizing attention to localize important image regions for generation, while Chen et al. [18] enhanced performance by combining Transformer-based models with a storage matrix for cross-modal feature alignment, employing memory-driven Transformers to record key generated information.
Regarding leveraging medical domain knowledge to guide report generation, some studies employ supplementary knowledge to assist report creation. Wang et al. [3] employed a shared cross-modal prototype matrix to capture cross-modal prototypes and embed cross-modal information. Yang et al. [20] integrated general (from pre-built knowledge graphs) and specific (from similar report retrieval) knowledge by mapping disease–organ relationships to align pathological and visual information. Wang et al. [42] proposed a medical concept generation network to consolidate semantic information. Liu et al. [22] distilled prior knowledge and posterior knowledge: the former mitigates textual data bias by leveraging prior medical knowledge and professional experience, while the latter reduces visual data bias by accounting for anomalous visual information. Furthermore, these studies exploited task-to-task correlations, using disease classification as a sub-task to guide the model toward more robust feature representations.
While existing approaches have made significant progress in medical report generation, several critical limitations remain unresolved. First, most current methods treat knowledge integration as a static process and disease classification as a parallel task with implicit expectations, failing to leverage diagnostic information for explicit report guidance. Second, the alignment between visual features and diagnostic text often lacks clinical reasoning depth, as exemplified by approaches like RGRG [43] that rely solely on regional visual features while neglecting diagnostic context. Third, existing frameworks typically maintain a separation between knowledge representation and generation optimization. Our work addresses these gaps through three key innovations: (1) a knowledge graph framework that explicitly models disease progression patterns while maintaining tight feature–text coupling, (2) an adaptive knowledge distillation mechanism that dynamically adjusts to clinical contexts, and (3) a novel architecture with diagnosis prompts generator that fundamentally differs from previous work by synergistically combining visual features with diagnostic cues. Unlike methods that treat classification as an auxiliary task, we explicitly incorporate diagnostic results as generative prompts. This approach establishes a more comprehensive knowledge representation paradigm that actively bridges medical knowledge with patient-specific findings through explicit clinical reasoning pathways.
3. Methods
In this section, we present the overall architecture of our graph-driven medical report generation framework, which integrates structured knowledge graphs with adaptive self-distillation to enhance both clinical accuracy and linguistic coherence. As illustrated in Figure 1, the model operates within an encoder–decoder paradigm. The input image is first processed through a Knowledge Integration and Fusion (KIF) module (Section 3.1), where visual features are enriched via structured knowledge mapping using a medical concept graph and further refined through cross-modal semantic fusion with retrieved clinical reports. The resulting representations are then utilized by a diagnosis prompts generator (DPG, Section 3.2) to produce diagnostic prompts that explicitly guide the subsequent decoding process. These prompts, along with the knowledge-augmented visual features, are fed into a decoder for initial report generation (Section 3.3). Finally, an information self-distillation (ISD) module (Section 3.4) applies a dynamic weight-shifting strategy between teacher and student networks to iteratively refine the model’s diagnostic and textual feature alignment, further enhancing the quality of the generated reports. A detailed pseudocode of the overall framework and specific implementation configurations are provided in Appendix A for reproducibility.
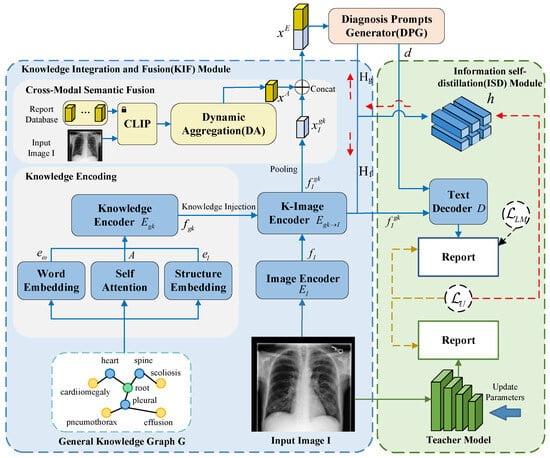
Figure 1.
Illustration of the proposed architecture. Input image I and general knowledge G undergo fusion through the KIF module to generate feature representations. These features are fed into the DPG to produce classification predictions, which are then combined with visual features and input into the text decoder D to generate reports. The solid blue line represents the data flow of the student model, the solid green line represents the data flow of the teacher model, and the dashed lines indicate the backward propagation path of gradients.
3.1. Knowledge Integration and Fusion (KIF)
As illustrated in Figure 1, the Knowledge Integration and Fusion (KIF) module is structured to systematically incorporate structured medical knowledge into visual feature learning through a multi-stage encoding and fusion pipeline. It comprises three integral components: knowledge encoding, which transforms discrete medical concepts into continuous vector representations that preserve semantic and anatomical relationships; image encoding, which extracts multi-scale spatial features from the input medical image using a deep convolutional architecture; and cross-modal semantic fusion, which aligns and integrates visual features with semantically relevant textual information retrieved from a clinical report database. Together, these components enable the model to construct a context-aware, diagnostically grounded feature representation that supports accurate and coherent medical report generation.
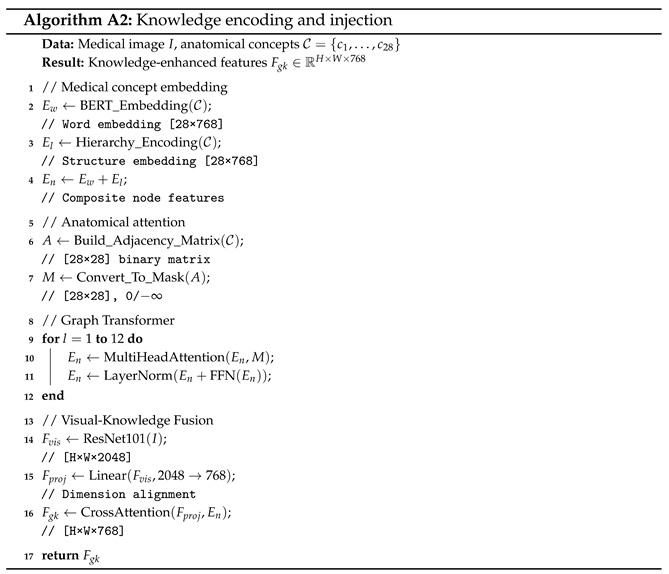
3.1.1. Knowledge Encoding
The knowledge encoding process transforms discrete medical concepts into continuous vector representations that preserve both semantic meanings and anatomical relationships. We consider a set of 28 medical concepts, denoted as , which includes anatomical structures (e.g., “heart”, “lung”) and pathological findings (e.g., “cardiomegaly”, “pneumonia”). To map these concepts into structured knowledge embeddings, we employ a modified BERT-base architecture incorporating domain-specific adaptations. As illustrated in Figure 1, the knowledge encoding component consists of four key elements: word embedding, structure embedding, self-attention, and a knowledge encoder.
Word Embedding. Each medical concept is tokenized and embedded through the BERT embedding layer:
where represents subword tokens, is the extended embedding matrix, and d = 768 denotes the hidden dimension of the model. The vocabulary was expanded from the original tokens to accommodate frequent medical terms from MIMIC-CXR reports.
Structure Embedding. Anatomical hierarchy is encoded through level-specific embeddings:
where maps concepts to predefined anatomical levels, and are learnable-type embeddings initialized orthogonally.
The composite node representation combines both information sources:
Self Attention. The core innovation lies in the anatomically constrained attention mechanism:
where represents the anatomical constraint mask that enforces
This constrained attention mechanism enables the model to focus specifically on medically relevant concept relationships during encoding.
Knowledge Encoder. Building upon this anatomically-aware attention, the knowledge encoder processes the enriched concept embeddings through 12 layers of Transformer architecture to produce the structured knowledge representation:
where the output captures the deep contextualized relationships among the 28 medical concepts, serving as a structured knowledge base to guide subsequent visual–semantic alignment and report generation.
3.1.2. Image Encoding
Assume a training dataset consisting of N samples, where each denotes an X-ray image and represents the corresponding medical report. Here, H, W, and C correspond to the height, width, and number of channels of the image, respectively. As shown in Figure 1, we detail the key components of our architecture for image encoding, which comprises an image encoder and a K-image encoder.
Image Encoder. Given an input chest X-ray image , visual features are extracted using a ResNet-101 backbone pre-trained on ImageNet. The encoder produces a spatial feature representation:
The output dimension is 2048, which is determined by the final global pooling layer of the ResNet-101 architecture.
K-Image Encoder. The K-image encoder employs a multi-head cross-attention design where visual features serve as queries and knowledge embeddings provide keys and values, and the encoder follows the standard Transformer cross-attention formulation with domain-specific adaptations:
where , , and serve as the query, the key, and the value, respectively, in the cross-attention calculation in . Enhanced by , the updated image feature can capture the global relations among different organs and diseases.
3.1.3. Cross-Modal Semantic Fusion
Single-modality diagnosis of medical images presents inherent limitations, reflecting how radiologists in clinical practice routinely integrate imaging data with supplementary medical records and diagnostic databases. To bridge this gap, our framework enhances visual features through semantically rich disease representations extracted from the training set’s medical report database.
As illustrated in Figure 2, our cross-modal semantic fusion framework employs a CLIP model pre-trained on the MIMIC dataset [44] to retrieve the k most clinically relevant report features for each input image I. These retrieved features undergo adaptive fusion via a dynamic aggregation (DA) module, which compresses them into a compact embedding representation . Crucially, this embedding is then concatenated with the knowledge-augmented visual features , which are pooled , forming a unified representation that simultaneously captures visual evidence and textual knowledge for the disease classification task.
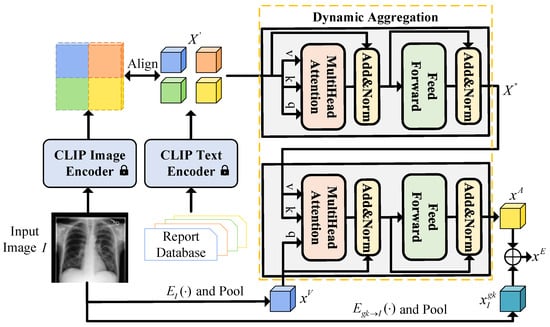
Figure 2.
Architecture of the cross-modal semantic fusion module. First, align image features with text features using CLIP to identify the most relevant report text feature . This feature is then input alongside the pooled feature from the encoder into the dynamic aggregation module and finally combines the pooled feature with the fused feature to obtain the enhanced feature .
This design intentionally mirrors radiologists’ diagnostic workflow while addressing the constraints of unimodal analysis through systematic cross-modal integration. The resulting framework not only improves classification accuracy but also provides more interpretable diagnostic support by maintaining the relationship between visual findings and their textual descriptions. Formally, the dynamic aggregation (DA) module comprises both self-attention and cross-attention mechanisms. Specifically, the self-attention layer first processes the set of retrieved top-k report features to capture inter-report relationships:
And then the cross-attention layer fuses report features with visual context, using pooled visual features of as query and as key/value:
The output retains the most clinically salient information. The final representation combines aggregated semantic features with knowledge-enhanced visual features :
where is the average pooled visual feature from , while is the average pooled from , and ⊕ represents the concatenation operation. To dynamically extract report features with respect to the visual feature , we implement DA as Transformer attention modules [40]. first goes through a self-attention layer, then the output is used as the key and value of a cross-attention layer while is the query.
We specifically set the DA module as a trainable component while keeping the CLIP model frozen during training. This design philosophy mimics the actual diagnostic process of radiologists, who often consult historical cases or medical databases for comparison when faced with uncertain medical images to enhance the reliability of their diagnoses.
3.2. Diagnosis Prompts Generator
Medical report generation (MRG) requires strict alignment between textual descriptions and diagnostic conclusions, as inconsistencies may directly impact clinical decision-making. Unlike generic image captioning, radiology reports must simultaneously achieve comprehensive findings summarization and precise clinical interpretation. Even minor diagnostic inaccuracies in generated reports risk propagating to downstream treatment workflows with potentially severe consequences.
To address this critical requirement, we use a diagnosis prompts generator (DPG) mechanism [45] that actively guides text generation using outputs d from the disease classification branch. The branch processes cross-modal enhanced visual features through L parallel classification heads (corresponding to 14 predefined chest diseases), each performing 4-class prediction: ’Blank’, ’Positive’, ’Negative’, and ’Uncertain’. Disease labels are automatically extracted from radiology reports using CheXbert [46], with optimization via standard cross-entropy loss .
During inference, classification results are converted into labeled prompts , with each prompt corresponding to a specific disease. To this end, four special tokens are introduced into the vocabulary: [BLA], [POS], [NEG], and [UNC], representing the four classification outcomes. This architectural innovation enables three key advantages: (1) explicit conditioning of report content on diagnostic evidence, (2) prevention of clinically contradictory statements, and (3) improved model interpretability through traceable prompt influence. By directly embedding diagnostic certainty levels into the generation process, the decoder produces reports with significantly higher clinical fidelity compared to implicit conditioning approaches.
3.3. Initial Report Generation
This module is responsible for producing an initial diagnostic report based on the graph-enhanced visual features and diagnostic prompts. While this draft report is already clinically relevant, it is subsequently refined by the self-distillation module described in Section 3.4 to further improve its accuracy and coherence. The representation , produced by the Knowledge Integration and Fusion (KIF) module, serves as input to the diagnosis prompts generator (DPG) to yield a sequence of diagnostic prompts . These prompts, together with the knowledge-enhanced visual features , guide a Transformer-based decoder D in auto-regressively generating the final medical report . Here, each denotes a token from the vocabulary , and T represents the total length of the report. The decoding process at each time step t is formally defined as
During inference, we employ a beam search strategy (beam size = 3) to approximate the optimal report sequence, effectively balancing generation quality and diversity. Finally, the language modeling loss () is indeed the core loss function used to measure the discrepancy between the generated report R by your model and the ground-truth report . It is a standard auto-regressive language modeling loss (also known as the cross-entropy loss). At each time step t, it computes the cross-entropy between the model’s predicted probability distribution over the vocabulary and the one-hot encoding of the actual target word (from the ground-truth report).
3.4. Report Optimization via Information Self-Distillation (ISD) Module
To prevent potential feature erosion, we employ a pre-trained model, with the same structure as our model, as the teacher network, taking the whole image as input to extract knowledge into the student model. Furthermore, our strategy differs from ordinary knowledge distillation where parameters are frozen. Instead, we shift the weights of teachers and students when students outperform the teacher network. We use KL scatter as distillation loss to align the probability distributions of the teacher and student models:
where and are the probability distributions of the teacher model and the student model for the word index c and the image I, respectively. N is the dimension of the word space.
In order to encode graphical, semantic, and visual information as feature representations, we link the visual features of the graphical embedding with the semantic features of the cue to obtain a hybrid feature representation , which is then sent to the text decoder D to generate a report. To further represent the middle layer output consistency, we stack column by column to shape it into such that the features of a batch of data can be represented as . We consider the Gram matrix as to reflect the sample correlation. Let ; can be rewritten as where is the column vector of corresponding to the training sample, and is the feature correlation representation. We reuse to represent for simplicity in , and is used to ensure the consistency between the intermediate layer output of the student model and the teacher model in each iteration, which is called the intermediate consistency loss :
Unlike traditional teacher–student model architectures, our distillation framework employs identical network architectures for both teacher and student models to fully capture the teacher model’s knowledge. The core mechanism involves leveraging the “high-quality knowledge” learned by the teacher model to guide the student model—sharing the same architecture—toward more efficiently capturing the associative patterns between medical images and report texts. This ultimately achieves iterative optimization of report generation performance. Furthermore, we employ KL divergence and intermediate-layer consistency loss to specifically guide the student model in learning the teacher model’s knowledge distribution across two core dimensions of the MRG task: ‘clinical phrase generation probability’ and ‘disease diagnosis probability’. The former aligns with the requirement for ‘textual clinical coherence’, while the latter addresses the need for ‘disease diagnosis accuracy’. This ensures the distillation objectives are precisely aligned with the core demands of the MRG task. The overall objective function of this framework is defined as
The total loss function for our whole framework is the combination of the loss from the initial report generation stage and the loss from the adaptive self-distillation stage:
This composite objective ensures the model not only learns to generate medically accurate and fluent text but also achieves iterative self-refinement through consistency constraints, enhancing both the clinical reliability and the textual quality of the final reports.
4. Experiments
Datasets. Our evaluation employs two benchmark datasets for medical report generation. For both datasets, each sample has one or more chest X-ray images taken from different angles. Following the method of Chen et al. [17], we excluded data without reports and selected samples with no more than two images. They use the same reporting information for X-ray images taken at different angles. The training/validation/test set was divided in a 7:1:2 ratio. Table 1 shows dataset statistics, including the number of images, reports, patients, and disease labels. We utilize the MIMIC-CXR dataset [26] (available at https://physionet.org/content/mimic-cxr/2.1.0/ (accessed on 9 October 2025)), the largest public dataset of chest X-rays and reports (first described by Johnson et al. [47] and sourced from PhysioNet [48]), preprocessed according to the protocol of Chen et al. [17]. This yields 270,790 samples for training, with 2130 and 3858 samples reserved for validation and testing, respectively. We additionally evaluate on the IU X-Ray dataset [27] (available at https://service.tib.eu/ldmservice/dataset/iu-x-ray-dataset (accessed on 9 October 2025)), another widely adopted public benchmark containing 3955 processed samples. However, our analysis revealed that the standard test split proposed by Chen et al. [17] contains insufficient positive samples for certain diseases to reliably assess diagnostic perception. Consequently, we adopt an alternative evaluation protocol where models trained exclusively on MIMIC-CXR [26] are tested across the entire IU X-Ray [27] collection, providing more statistically robust measurements of disease classification performance. All input chest X-ray images undergo a standardized transformation procedure to ensure consistency with pre-trained visual encoders. The transformation pipeline includes (1) resizing to 256 × 256 pixels while maintaining aspect ratio, (2) center cropping to 224 × 224 pixels for input standardization, (3) normalization using ImageNet precomputed statistics (mean = [0.485, 0.456, 0.406], std = [0.229, 0.224, 0.225]), and (4) conversion to PyTorch tensor format for GPU acceleration.This standardized processing ensures optimal compatibility with the ResNet-101 backbone pre-trained on ImageNet.

Table 1.
The statistics of the two benchmark datasets with respect to their training, validation, and test sets, including the numbers of images, reports, patients, and disease labels.
Evaluation Protocol. As illustrated in Figure 3. In our evaluation process, we first partition the dataset into an independent test set containing test images, corresponding true disease labels, and true report texts. Next, we input the test images into our medical report generation model to obtain the model-generated report texts. Finally, we process the report texts to extract disease labels, using both the generated report texts and extracted disease labels to compute the NLG metrics and CE metrics. CE serves as an evaluation metric for multi-label classification tasks, while NLG functions as an evaluation metric for conditional language generation tasks. Although they belong to distinct tasks, within our medical report generation model, both fundamentally aim to assess the quality of report generation.
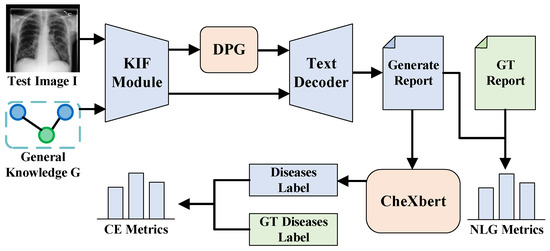
Figure 3.
Model evaluation flowchart: KIF denotes the Knowledge Integration and Fusion Module; DPG denotes the diagnosis prompts generator, which generates disease prompts. These prompts are input into the text decoder alongside features fused through KIF to generate reports. The resulting reports are then processed by CheXbert [46] to obtain the disease classification for diagnosis. Finally, metrics are calculated by comparing these outputs with ground truth (GT).
When using the CE metric, our goal is to evaluate the accuracy of disease diagnoses in reports generated by medical report generation models, which constitutes a text-to-label classification task. We follow the methodology outlined by Nicolson et al. [49], specifically assessing whether the model-generated reports accurately cover the chest disease information corresponding to the input images by matching categories with the ground-truth disease labels. We focus on 14 categories of chest diseases within the MIMIC-CXR and IU X-ray datasets, where ground-truth disease labels are provided. Model-predicted labels are obtained by inputting the generated report text into CheXbert [46] to yield disease labels matching the format of the ground-truth labels. When extracting labels using the CheXbert model, we employed CheXbert v1.0. After loading the pre-trained model, we input our generated radiology reports into the CheXbert model. The model automatically encodes the reports into input sequences of ≤512 tokens and outputs predictions for 14 label categories (POS/NEG/NEG/UNC) in inference mode. Finally, we perform report-level aggregation on the token-level labels (using majority voting). The NLG metric can be directly computed by comparing the generated reports with the ground-truth reports.
Evaluation Metrics
After obtaining the generated reports, we measure their quality by comparing them against authentic reports using NLG metrics on one hand. On the other hand, we extract disease labels from the generated reports via CheXbert [46] and calculate the CE metric against the authentic labels.
CE Metrics. The input is the set of 14 predicted chest disease labels extracted from the generated report using the CheXbert [46] labeler. These are compared against the 14 ground-truth disease labels from the original dataset to compute true positives (TPs), false positives (FPs), and false negatives (FNs) for each disease. When calculating the CE metrics, precision, recall, and F1 scores are computed separately for each of the 14 disease categories and then averaged using macro-averaging. This approach prevents common disease metrics from dominating the results, ensuring fairness in the evaluation of all diseases.
Precision represents the proportion of samples that are actually positive for a disease among those predicted positive by the model. True positives (TPs) are samples with a true disease label of positive and a model prediction label of positive, while false positives (FPs) are samples with a true disease label of negative but a model prediction label of positive.
Recall represents the proportion of samples correctly predicted as positive for a disease among all true positives for that disease. Here, FN denotes the number of samples with a true positive label for the disease but a false negative prediction by the model.
F1 is the harmonic mean of precision and recall, balancing the trade-off between them to comprehensively reflect the model’s disease classification performance.
NLG metrics. The input is the full sequence of words in the generated report . This sequence is compared against the sequence of words in the authentic, ground-truth physician report . We evaluate the textual quality and semantic consistency of generated radiology reports against authentic physician reports. The core measurement focuses on overlap in vocabulary, phrases, sentence structure, and semantic meaning. Generated content must align with authentic physician report structures, adhere to medical terminology standards, and produce logically coherent natural language text. We employ the BLEU-1 [9] metric to measure lexical accuracy at the word level.
This calculates the similarity by counting the number of individual words (1-grams) present in the generated report that also appear in the actual report, relative to the total number of 1-grams in the generated report. It incorporates a brevity penalty (BP) to prevent artificially high overlap rates from overly short reports.
The BLEU-4 [9] metric follows the same calculation logic as BLEU-1 but counts the number of consecutive 4-grams, reflecting the model’s logical coherence at the multi-word phrase level.
The METEOR [11] metric evaluates the ability to semantically match term variants and synonyms, addressing the limitation of BLEU metrics that focus solely on word forms.
The ROUGE-L [10] metric measures semantic structural consistency at the sentence level by calculating the proportion of the longest common subsequence (LCS) between generated and actual reports, reflecting the integrity of core semantic information.
where and .
We integrate CE and NLG metric results to analyze the comprehensive performance of model-generated reports in disease diagnosis accuracy and text generation quality, demonstrating that the model ensures diagnostic accuracy while maintaining clinical utility of the text.
Implementation Details. As shown in Table 2, all experiments were conducted using Python 3.8.13, with the model implemented based on PyTorch 2.4.1 (trained on an RTX 3090 GPU for approximately 24 h) and key dependent libraries specified with exact versions as follows: transformers 4.25.0 (for pre-trained language model loading and fine-tuning), torchvision 0.19.1 (for medical image preprocessing and data pipeline construction), opencv-python 4.10.0.84 (for image augmentation operations such as resizing and normalization), scipy 1.10.1 (for statistical calculations in evaluation metrics), pandas 2.0.3 (for organizing dataset metadata and label information), scikit-learn 1.3.2 (for computing classification metrics including precision, recall, and F1 score), and timm 0.6.12 (for loading pre-trained vision backbones); specifically, we used ImageNet pre-trained ResNet-101 [50] as the encoder and BERT base [51] as the decoder, set the coefficient k = 21 for cross-modal semantic fusion, and adopted AdamW [52] as the optimizer with a weight decay of 0.05 and an initial learning rate of 5 × 10−5. More details can be found in Appendix B and the code is available at https://github.com/ZIENxc/KIF-ISDforMRG/tree/v1.0 (accessed on 9 October 2025).

Table 2.
Environment. This table provides a centralized overview of the precise library versions used throughout the experiment.
5. Results
Qualitative results. As illustrated in Figure 4, we present two representative case studies that compare our model-generated reports with the ground truth (GT). The visualizations highlight semantic correspondences through gold-highlighted text (indicating alignment with GT) and blue-highlighted content (denoting clinically consistent model-generated findings). These examples underscore our model’s capability to accurately identify severe pathologies—such as pneumothorax and pleural effusion—which is reinforced by high CE scores. Furthermore, the model demonstrates an ability to capture nuanced observations (e.g., “low lung volumes”), contributing to its superior performance in BLEU-1 metrics and reflecting a fine-grained understanding of both visual and clinical features.
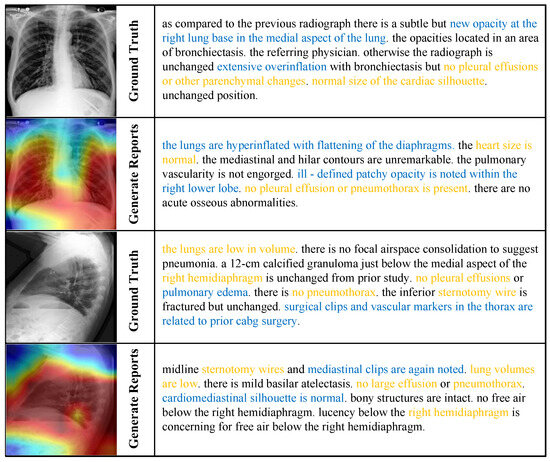
Figure 4.
Cross-modal attention visualization and report generation results on MIMIC-CXR [26]. Gold-highlighted text denotes semantic alignment with ground truth; blue-highlighted text indicates model-generated content with clinical consistency.
Quantitative results. As shown in Table 3, our model demonstrates superior performance compared to ten state-of-the-art methods across both MIMIC-CXR [26] and IU X-Ray datasets [27]. A complete list of all compared models and our study, along with details of their architectural configurations, key design innovations, and the operational workflows, is provided in Table A1 in Appendix C.

Table 3.
Performance comparison of different models on MIMIC [26] and IU X-Ray datasets [27]. * indicates the used image size is larger than 224. † indicates the performance evaluated by MIMIC model weights. # indicates the usage of a different data split. The best results are in bold.
On clinical efficacy (CE) metrics, since our CE metric is calculated by comparing extracted labels from generated reports against ground-truth labels, it differs from methods like image segmentation that achieve exceptionally high and accurate CE scores. This discrepancy arises because generated reports suffer from reduced label extraction performance due to word bias. Additionally, the metric represents a macro-average across 14 chest diseases, including relatively rare conditions. Consequently, the CE metric in the MRG task presents a challenging benchmark. The method proposed in this study achieved an F1 score of 0.480 in the MIMIC-CXR benchmark test, which outperforms existing technologies. There is a 10.7% absolute improvement over the recent method DCL [54] (F1 = 0.373) and a 3.3% absolute improvement over RGRG [43] (F1 = 0.447). It also has better performance than the recent method PromptMRG [45] (F1 = 0.476). This performance improvement fully demonstrates that the model we developed achieves a better balance between precision and recall in disease detection tasks, which is of great value in reducing misdiagnosis and missed diagnosis in clinical practice. Notably, on the smaller-scale IU X-Ray dataset, our model also demonstrated outstanding performance, except for the PromptMRG [45] method, which fully demonstrates the algorithm’s reliable generalization performance across datasets of different scales.
For natural language generation (NLG), the model achieved an excellent BLEU-1 score (assessing lexical accuracy) on the MIMIC-CXR dataset [26], confirming the model’s ability to accurately describe using specialized radiological terminology. More importantly, the model obtained the best results for all NLG metrics in the IU X-Ray test, further validating the adaptability of the method to different reporting styles. However, other NLG metrics on the MIMIC-CXR dataset [26] were not as good as on the IU X-Ray dataset [27]. Upon analysis, it is hypothesized that this is due to the model’s ability to generate high-frequency phrases when learning for longer reports (the average length of MIMIC-CXR reports is about twice as long as that of IU X-Ray reports, and the ability to generate high-frequency phrases still needs to be improved.
Unlike general image description generation, medical reports require precise integration of visual features with clinical knowledge—a capability that even state-of-the-art (SOTA) models struggle to fully master. In the MRG domain, extracting disease labels from generated reports and measuring diagnostic accuracy by calculating the accuracy of these extracted labels presents significant challenges. Even when generated reports closely mimic authentic reports in wording, a single misplaced word can lead to erroneous diagnostic conclusions. Table 3 compares our performance with other recent state-of-the-art models, demonstrating this benchmark’s difficulty. Furthermore, the goal of automated medical report generation is not to surpass human performance but to provide automated assistance, as reports generated by professional physicians remain irreplaceable. An F1 score of 0.48 represents a significant step forward in this field. These results do not indicate a model that merely produces “false positives,” but rather one that executes complex tasks with non-trivial accuracy.
Ablation Validation
To validate the effectiveness of each proposed module, we conducted ablation studies on the MIMIC-CXR test set (as shown in Table 4).

Table 4.
Ablation study of each module on different datasets. The best results are in bold.
With the addition of the ISD module, the F1 score improves to 0.480. This gain can be attributed to enhanced regional representation of image features through knowledge graph integration, which improves alignment with organ- and lesion-related terms. Corresponding improvements are also observed in precision (0.503) and recall (0.519).
When the KIF module is introduced, the F1 score further increases to 0.490, and BLEU-1 improves to 0.402. Although a slight decline in BLEU-4 is noted, these results indicate that adaptive knowledge distillation contributes positively to diagnostic accuracy and lexical-level text generation, despite a minor regression in longer phrasal matching.
Finally, when both ISD and KIF modules are incorporated, the F1 score remains stable at 0.480, while BLEU-1 rises to 0.408 and BLEU-4 to 0.114. Improvements are also observed in METEOR (0.158) and ROUGE (0.273). These findings demonstrate that our full method enhances the quality of generated reports while maintaining high diagnostic accuracy and improving linguistic coherence.
6. Conclusions
In this work, we propose a medical report generation (MRG) framework designed to enhance linguistic accuracy while maintaining high diagnostic performance. Our approach incorporates knowledge graph mapping to enrich image features and integrates diagnostic results from disease classification branches to provide semantic cues that guide the generation process. Furthermore, adaptive knowledge distillation is introduced to further improve linguistic quality. Extensive experiments on two benchmark datasets demonstrate the superiority of the proposed method, particularly in generating diagnostically accurate reports and narrowing the gap between current MRG models and real-world clinical requirements.
Although our experiments are conducted on chest X-ray data, the framework has the potential to be extended to other imaging modalities. Several challenges remain to be addressed: first, the training of disease classification branches relies on the availability of disease labels. Fortunately, most public MRG datasets include such annotations. In cases where labels are absent, unsupervised clustering methods could be applied to group reports into semantically meaningful categories. Additionally, we plan to incorporate richer and more fine-grained information to support the generation of longer and more detailed reports, which may further improve both linguistic precision and diagnostic accuracy.
Author Contributions
Conceptualization, X.H. and M.J.; methodology, J.C., X.H. and Y.L.; software, J.C.; validation, J.C. and Z.Z.; formal analysis, X.H.; investigation, J.C.; resources, X.H. and D.Q.; data curation, J.C. and Z.Z.; writing—original draft preparation, J.C.; writing—review and editing, X.H., M.J. and Y.L.; visualization, J.C. and X.H.; supervision, D.Q.; project administration, X.H.; funding acquisition, X.H. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by the Science Foundation of Zhejiang Sci-Tech University (ZSTU) (Grant No.22232337-Y) and the Key Research and Development Programs of Zhejiang Province (Grant No.2024SJCZX0026).
Informed Consent Statement
The source code developed for this study has been made publicly available on GitHub at https://github.com/ZIENxc/KIF-ISDforMRG/tree/v1.0 (accessed on 9 October 2025).
Data Availability Statement
This research utilizes exclusively publicly available datasets. The IU X-Ray dataset [27] was sourced from the TIB Leibniz Information Centre for Science and Technology (available at https://service.tib.eu/ldmservice/dataset/iu-x-ray-dataset (accessed on 9 October 2025)) under the CC-BY 4.0 license. The MIMIC-CXR dataset [26,47] was accessed from the PhysioNet repository [48] (available at https://physionet.org/content/mimic-cxr/2.1.0/ (accessed on 9 October 2025)) under a custom data use agreement, which permits research use. Both datasets are fully available for academic and research purposes, and the corresponding URLs are provided in the reference list.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations
The following abbreviations are used in this manuscript:
| MRG | Medical Report Generation |
| NLG | Natural Language Generation |
| CNNs | Convolutional Neural Networks |
| RNN | Recurrent Neural Network |
| LSTM | Long Short Term Memory |
| CE | Clinical Efficacy |
| KIF | Knowledge Integration and Fusion |
| DA | Dynamic Aggregation |
| ISD | Information Self-Distillation |
| DPG | Diagnosis Prompts Generator |
| BLA | Blank |
| POS | Positive |
| NEG | Negative |
| UNC | Uncertain |
| GT | Ground Truth |
| TP | True Positive |
| FP | False Positive |
| FN | False Negative |
| LCS | Longest Common Subsequence |
Appendix A. Algorithm Pseudocode
| Algorithm 1: Report generation |
Data: Medical image I, word embeddings W, CLIP memory M, generation parameters Result: Generated reports R, predicted classes C, class probabilities P
|
| Algorithm 3: ISD training process |
Data: Student model , teacher model (frozen), batch data , base class probabilities P Result: Total loss
|
Appendix B. Detailed Experimental Setup
(1) The KIF module integrates structured medical knowledge into visual features through a hierarchical attention mechanism. We utilize PyTorch (v2.4.1) for tensor operations and HuggingFace Transformers (v4.25) to implement the BERT-based knowledge encoder, which processes 28 medical concepts with domain-specific modifications including extended vocabulary and learnable positional embeddings. The visual encoder employs a ResNet-101 backbone from torchvision (v0.19.1) pre-trained on ImageNet, extracting 2048-dimensional spatial features that are projected to 768 dimensions using a linear layer. The cross-attention module computes 4-head attention between visual features (queries) and knowledge embeddings (keys/values) using PyTorch’s MultiheadAttention, followed by feature enhancement via a two-layer MLP (768→3072→768). For dynamic aggregation, we use OpenAI CLIP (ViT-B/32) to retrieve the top 21 relevant reports from the database, processing them through self-attention and visual-guided cross-attention. The final 1536-dimensional fused representation concatenates knowledge-enhanced visual features (768-d) with aggregated report embeddings (768-d).
(2) The ISD module optimizes feature consistency through teacher–student distillation implemented with PyTorch’s distributed training utilities. The teacher model weights are updated via exponential moving average using PyTorch’s inplace operations to maintain memory efficiency. Intermediate supervision compares 32-dimensional similarity matrices H, constructed by concatenating visual (16 × 16) and classification (16 × 16) similarity scores between samples in each batch, computed using PyTorch’s matrix multiplication (torch.matmul) and L2 normalization (torch.norm). The KL divergence loss is implemented with PyTorch’s KLDivLoss, aligning report generation distributions between teacher and student models. Training employs the AdamW optimizer from PyTorch with learning rate 5 × 10−5 and linear warm up over 10% of total steps.
(3) Prediction pipeline. The end-to-end pipeline first processes images through the KIF module to obtain knowledge-augmented visual features. We use torch.nn.Linear for all projection layers and torch.nn.LayerNorm for feature normalization. Generated reports are decoded autoregressively using the HuggingFace BertLMHeadModel with beam search (beam_size=3, max_length=100), while disease classification leverages both CheXbert [46] label extraction (implemented as a standalone PyTorch module) and direct prediction from pooled visual-knowledge features through a linear classifier (72-d output). Logit adjustment for class imbalance is implemented using NumPy (v1.23.4) to compute base probabilities. The system processes 224 × 224 chest X-rays at 16 samples/batch on an RTX 3090 GPU, with mixed-precision training enabled via PyTorch AMP (Automatic Mixed Precision). Key evaluation metrics including BLEU and ROUGE scores are computed using TorchMetrics (v0.6.12).
Appendix C. Comprehensive Comparison of Model Implementations

Table A1.
Comparison of medical report generation models.
Table A1.
Comparison of medical report generation models.
| Model | Innovation | Architecture | External Knowledge | Operational Workflow | Testing and Evaluation |
|---|---|---|---|---|---|
| Continued on next page | |||||
| R2GenCMN [18] | Memory-Driven Cross-Modal Attention: 1. Learnable Memory Matrix: A shared memory matrix stores frequently occurring clinical semantic units (e.g., “cardiomegaly,” “pulmonary opacity”) learned from the dataset. 2. Cross-Modal Interaction: At each decoding step, an attention mechanism dynamically retrieves relevant information from the memory matrix. This retrieved context is fused with the image features to guide the next word generation, ensuring terminological accuracy. | Encoder: CNN Decoder: LSTM | Implicit: Learned from the training data and stored in the memory matrix. | 1. Encode image into features using CNN. 2. Query Memory: Decoder queries the memory matrix via attention for relevant clinical context. 3. Fuse and Generate: Fuses memory context, image features, and previous hidden state to predict the next word. 4. Repeat until the report is complete. | Datasets: IU X-Ray [27], MIMIC-CXR [47]. Automated Metrics: Standard NLG scores (BLEU [9], ROUGE [10]). Clinical Metrics: CheXbert [46] F1 score. Ablation: Performance comparison against baselines (e.g., SAT, Transformer) and ablated model (w/o memory module) to prove its efficacy. |
| GSKET [20] | Gated Fusion of General and Specific Knowledge: 1. Dual Knowledge Sources: Incorporates “specific knowledge” (detected concepts from the image) and “general knowledge” (descriptions of concepts retrieved from an external medical KB like UMLS). 2. Gating Mechanism: A learned gating network dynamically computes weights for the two knowledge sources, adaptively controlling their contribution to generation for refined knowledge utilization. | Encoder: CNN Decoder: Transformer | Explicit: 1. Specific: Pre-trained visual concept detector. 2. General: Medical knowledge base (UMLS). | 1. Extract concepts from the image using a detector. 2. Retrieve information for concepts from UMLS. 3. Encode and Gate: Encode both knowledge types; the gating network fuses them. 4. Generate: Transformer decoder uses fused knowledge and image features to generate the report. | Datasets: IU X-Ray [27], MIMIC-CXR [47]. Metrics: Standard automated NLG metrics (BLEU [9], ROUGE [10], METEOR [11]) and clinical efficacy (CheXbert-based [46] recall, F1). Ablation: Tests variants (e.g., specific-only, general-only, no gate) to validate the necessity of both knowledge sources and the gating mechanism. |
| DCL [54] | Dual Contrastive Learning Strategy: 1. Image–Text Contrastive Learning: In a joint embedding space, pulls the features of matched image-report pairs closer and pushes unmatched pairs apart to enhance modality alignment. 2. Image–Image Contrastive Learning: Pulls “abnormal–abnormal” image pairs closer and pushes “abnormal–normal” pairs apart, forcing the model to focus on abnormal features and mitigating the dataset bias towards “normal” cases. | Encoder: CNN Decoder: LSTM | None. Learns better representations from the data itself via the contrastive strategy. | 1. Encode images and reports. 2. Construct Pairs: Build positive/negative pairs within a training batch. 3. Compute Losses: Calculate the dual contrastive losses and the standard generation loss. 4. Generate the report. | Datasets: IU X-Ray [27], MIMIC-CXR [47]. Metrics: Standard automated NLG metrics (BLEU [9], ROUGE [10], METEOR [11]) and clinical efficacy (CheXbert-based [46] recall, F1), with a strong focus on performance improvement on abnormal/rare disease cases compared to baselines. |
| KiUT [55] | Unified Transformer for Knowledge Injection: Abandons complex fusion designs. Instead, it unifies visual features (converted to tokens) and detected text concepts (keywords) by concatenating them into a single multimodal sequence. This sequence is processed by a standard Transformer, which uses its self-attention mechanism to automatically learn the intrinsic relationships and dependencies between visual and knowledge tokens. | Encoder: CNN + Concept Encoder Decoder: Transformer | Explicit: Pre-trained visual concept detector (provides keywords). | 1. Detect concepts from the image. 2. Tokenize: Map image features to visual tokens; embed concepts into knowledge tokens. 3. Concatenate: Form a single sequence of visual + knowledge tokens. 4. Process and Generate: The unified Transformer encodes the sequence and its decoder autoregressively generates the report. | Datasets: IU X-Ray [27], MIMIC-CXR [47]. Metrics: Standard automated NLG metrics (BLEU [9], ROUGE [10], METEOR [11]) and clinical efficacy (CheXbert-based [46] recall, F1). Comparison: Contrasted with models using complex cross-modal attention, demonstrating that its simple unified architecture can achieve superior or comparable performance. |
| METransformer [4] | Multi-Edge Sparse Attention: 1. Image as Graph: Treats image patches as nodes in a graph. 2. Sparse Attention: Does not compute attention between all nodes. Instead, it predefines connections (edges) between nodes (e.g., only between spatially adjacent or semantically similar patches). This drastically reduces computational complexity and enhances the model’s ability to model relationships between local key regions. | Encoder: ViT variant (Graph Attention Network) Decoder: Transformer | None. Focuses on internal image structure via sparse attention. | 1. Graph Construction: Image is split into patches; a sparse graph is built defining connections between relevant patches. 2. Sparse Encoding: Graph-based sparse attention is computed only between connected nodes. 3. Generation: The Transformer decoder generates the report from the encoded features. | Datasets: IU X-Ray [27], MIMIC-CXR [47]. Metrics: Standard automated NLG metrics (BLEU [9], ROUGE [10], METEOR [11]) and clinical efficacy (CheXbert-based [46] recall, F1). Comparison: Compared against standard ViT/Transformer to demonstrate superior computational efficiency and lower resource consumption while maintaining comparable generation quality. |
| RGRG [43] | Retrieval-Augmented Generation (RAG) Framework: 1. Two-Stage Paradigm: First, retrieves the most similar historical cases and their reports (reference templates) from a large database via image retrieval. Then, a generative model conditionally generates a new report based on the current image features and the retrieved report context. 2. Advantage: Generated reports maintain high professionalism (by referencing ground-truth reports) and specificity (by integrating current image features), avoiding generic or vague descriptions. | Encoder: CNN (for retrieval and generation) Decoder: Transformer | Explicit: The entire training set database used as a retrieval corpus. | 1. Retrieve: The query image is used to find K most similar images and their reports from the DB. 2. Encode: The query image and the retrieved report text are encoded. 3. Generate: The Transformer decoder is conditioned on both the query image features and the retrieved report encoding to produce the final report. | Datasets: IU X-Ray [27], MIMIC-CXR [47]. Metrics: Standard automated NLG metrics (BLEU [9], ROUGE [10], METEOR [11]) and clinical efficacy (CheXbert-based [46] recall, F1). Ablation: Analysis of retrieval quality impact (e.g., varying K, using different retrievers). Comparison to pure generative models to show advantage in generating clinically accurate terms. |
| PromptMRG [45] | Learnable Continuous Prompt Vectors: 1. Dynamic Prompt Generation: A dedicated module encodes detected medical concepts into a set of continuous prompt vectors (“soft prompts”), not natural language. 2. Prompt-Guided Generation: These learned vectors are prepended to the input of a standard Transformer decoder (the report generator). The decoder is specifically trained to attend to these prompts, which guide it to generate coherent and professional reports that include the specified medical findings. | Encoder: CNN (concept detector) Decoder: Transformer | Explicit: Pre-trained visual concept detector. | 1. Detect Concepts: A CNN-based concept detector analyzes the image to output keywords. 2. Generate Prompts: A prompt generator module maps these keywords to a set of continuous prompt vectors. 3. Generate Report: These prompt vectors are fed as a prefix to a Transformer decoder, which is trained to generate the full report conditioned on them. | Datasets: IU X-Ray [27], MIMIC-CXR [47]. Metrics: Standard automated NLG metrics (BLEU [9], ROUGE [10], METEOR [11]) and clinical efficacy (CheXbert-based [46] recall, F1). Human Evaluation: Radiologists rate generated reports for fluency, clinical accuracy, and usefulness. Ablation: Tests the impact of different prompt designs and the necessity of the prompt module. |
| Ours | 1. Knowledge Integration and Fusion: A knowledge graph framework that explicitly models disease progression patterns while maintaining tight feature–text coupling. 2. Information Self-Distillation: An adaptive knowledge distillation mechanism that dynamically adjusts to clinical contexts | Encoder: CNN(ResNet101) Decoder: Transformer | Explicit: General knowledge graph and report database | 1. Encode Knowledge and Vision: The knowledge encoder processes structured medical concepts into embeddings, while the image encoder extracts visual features from the input X-ray. 2. Inject Knowledge: The K-image encoder fuses these streams via cross-attention, producing knowledge-augmented visual features. 3. Retrieve and Aggregate: A CLIP-based retriever fetches relevant reports from a database, which are dynamically aggregated with visual context. 4. Generate Report: The aggregated features guide a Transformer decoder to produce the final diagnostic report auto-regressively. | Datasets: IU X-Ray [27], MIMIC-CXR [47]. Metrics: Standard automated NLG metrics (BLEU [9], ROUGE [10], METEOR [11]) and clinical efficacy (CheXbert-based [46] recall, F1). |
References
- Li, M.; Cai, W.; Verspoor, K.; Pan, S.; Liang, X.; Chang, X. Cross-modal clinical graph transformer for ophthalmic report generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 20656–20665. [Google Scholar]
- Li, Y.; Yang, B.; Cheng, X.; Zhu, Z.; Li, H.; Zou, Y. Unify, align and refine: Multi-level semantic alignment for radiology report generation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 1–6 October 2023; pp. 2863–2874. [Google Scholar]
- Wang, J.; Bhalerao, A.; He, Y. Cross-modal prototype driven network for radiology report generation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 563–579. [Google Scholar]
- Wang, Z.; Liu, L.; Wang, L.; Zhou, L. Metransformer: Radiology report generation by transformer with multiple learnable expert tokens. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Paris, France, 1–6 October 2023; pp. 11558–11567. [Google Scholar]
- Zhou, H.Y.; Chen, X.; Zhang, Y.; Luo, R.; Wang, L.; Yu, Y. Generalized radiograph representation learning via cross-supervision between images and free-text radiology reports. Nat. Mach. Intell. 2022, 4, 32–40. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar] [CrossRef]
- Demner-Fushman, D.; Kohli, M.; Rosenman, M.; Shooshan, S.; Rodriguez, L.; Antani, S.; Thoma, G.; McDonald, C. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 2016, 23, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Lebret, R.; Grangier, D.; Auli, M. Neural Text Generation from Structured Data with Application to the Biography Domain. arXiv 2016, arXiv:1603.07771. [Google Scholar] [CrossRef]
- Papineni, K.; Roukos, S.; Ward, T.; Zhu, W.J. Bleu: A method for automatic evaluation of machine translation. In Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics, Philadelphia, PA, USA, 7–12 July 2002; pp. 311–318. [Google Scholar]
- Lin, C.Y. Rouge: A package for automatic evaluation of summaries. In Proceedings of the Text summarization Branches Out, Barcelona, Spain, 25–26 July 2004; pp. 74–81. [Google Scholar]
- Denkowski, M.; Lavie, A. Meteor 1.3: Automatic metric for reliable optimization and evaluation of machine translation systems. In Proceedings of the Sixth Workshop on Statistical Machine Translation, Edinburgh, UK, 30 July 2011; pp. 85–91. [Google Scholar]
- Anderson, P.; He, X.; Buehler, C.; Teney, D.; Johnson, M.; Gould, S.; Zhang, L. Bottom-up and top-down attention for image captioning and visual question answering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6077–6086. [Google Scholar]
- Cornia, M.; Stefanini, M.; Baraldi, L.; Cucchiara, R. Meshed-memory transformer for image captioning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10578–10587. [Google Scholar]
- Hirota, Y.; Nakashima, Y.; Garcia, N. Quantifying societal bias amplification in image captioning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 13450–13459. [Google Scholar]
- Hu, X.; Gan, Z.; Wang, J.; Yang, Z.; Liu, Z.; Lu, Y.; Wang, L. Scaling up vision-language pre-training for image captioning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 17980–17989. [Google Scholar]
- Liu, F.; Ge, S.; Zou, Y.; Wu, X. Competence-based multimodal curriculum learning for medical report generation. arXiv 2022, arXiv:2206.14579. [Google Scholar]
- Chen, Z.; Song, Y.; Chang, T.H.; Wan, X. Generating radiology reports via memory-driven transformer. arXiv 2020, arXiv:2010.16056. [Google Scholar]
- Chen, Z.; Shen, Y.; Song, Y.; Wan, X. Cross-modal memory networks for radiology report generation. arXiv 2022, arXiv:2204.13258. [Google Scholar] [CrossRef]
- Li, M.; Liu, R.; Wang, F.; Chang, X.; Liang, X. Auxiliary signal-guided knowledge encoder-decoder for medical report generation. World Wide Web 2023, 26, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wu, X.; Ge, S.; Zhou, S.K.; Xiao, L. Knowledge matters: Chest radiology report generation with general and specific knowledge. Med. Image Anal. 2022, 80, 102510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Xu, Z.; Yu, Q.; Yuille, A.; Xu, D. When radiology report generation meets knowledge graph. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 12910–12917. [Google Scholar]
- Liu, F.; Wu, X.; Ge, S.; Fan, W.; Zou, Y. Exploring and distilling posterior and prior knowledge for radiology report generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13753–13762. [Google Scholar]
- Jing, B.; Xie, P.; Xing, E. On the automatic generation of medical imaging reports. arXiv 2017, arXiv:1711.08195. [Google Scholar]
- Wang, Z.; Han, H.; Wang, L.; Li, X.; Zhou, L. Automated radiographic report generation purely on transformer: A multicriteria supervised approach. IEEE Trans. Med. Imaging 2022, 41, 2803–2813. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Pei, M. Clinical-bert: Vision-language pre-training for radiograph diagnosis and reports generation. In Proceedings of the AAAI Conference on Artificial Intelligence, Online, 22 February–1 March 2022; Volume 36, pp. 2982–2990. [Google Scholar]
- Johnson, A.; Pollard, T.; Mark, R.; Berkowitz, S.; Horng, S. MIMIC-CXR Database (version 2.1.0). PhysioNet 2024, 13026, C2JT1Q. [Google Scholar] [CrossRef]
- Liu, W.; Xue, Y.; Lin, C.; Boumaraf, S. Dataset: IU X-Ray Dataset. 2025. Available online: https://service.tib.eu/ldmservice/dataset/iu-x-ray-dataset (accessed on 9 October 2025).
- Gu, J.; Wang, G.; Cai, J.; Chen, T. An empirical study of language cnn for image captioning. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 1222–1231. [Google Scholar]
- Vinyals, O.; Toshev, A.; Bengio, S.; Erhan, D. Show and tell: A neural image caption generator. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3156–3164. [Google Scholar]
- Huang, L.; Wang, W.; Chen, J.; Wei, X.Y. Attention on attention for image captioning. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, South Korea, 27 October–2 November 2019; pp. 4634–4643. [Google Scholar]
- Rennie, S.J.; Marcheret, E.; Mroueh, Y.; Ross, J.; Goel, V. Self-critical sequence training for image captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7008–7024. [Google Scholar]
- Zhang, X.; Sun, X.; Luo, Y.; Ji, J.; Zhou, Y.; Wu, Y.; Huang, F.; Ji, R. RSTNet: Captioning with adaptive attention on visual and non-visual words. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 15465–15474. [Google Scholar]
- Yang, X.; Tang, K.; Zhang, H.; Cai, J. Auto-encoding scene graphs for image captioning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 10685–10694. [Google Scholar]
- Yao, T.; Pan, Y.; Li, Y.; Mei, T. Exploring visual relationship for image captioning. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8 September 2018; pp. 684–699. [Google Scholar]
- Li, J.; Li, D.; Xiong, C.; Hoi, S. Blip: Bootstrapping language-image pre-training for unified vision-language understanding and generation. In Proceedings of the International Conference on Machine Learning, Baltimore, Maryland, USA, 17–23 July 2022; pp. 12888–12900. [Google Scholar]
- Yu, J.; Wang, Z.; Vasudevan, V.; Yeung, L.; Seyedhosseini, M.; Wu, Y. Coca: Contrastive captioners are image-text foundation models. arXiv 2022, arXiv:2205.01917. [Google Scholar]
- Liu, G.; Hsu, T.M.H.; McDermott, M.; Boag, W.; Weng, W.H.; Szolovits, P.; Ghassemi, M. Clinically accurate chest x-ray report generation. In Proceedings of the Machine Learning for Healthcare Conference, Ann Arbor, Michigan, USA, 9–10 August 2019; pp. 249–269. [Google Scholar]
- Xue, Y.; Xu, T.; Rodney Long, L.; Xue, Z.; Antani, S.; Thoma, G.R.; Huang, X. Multimodal recurrent model with attention for automated radiology report generation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 457–466. [Google Scholar]
- Xue, Y.; Huang, X. Improved disease classification in chest x-rays with transferred features from report generation. In Proceedings of the International Conference on Information Processing in Medical Imaging, Hong Kong, China, 2–7 June 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 125–138. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar] [CrossRef]
- Hou, B.; Kaissis, G.; Summers, R.M.; Kainz, B. Ratchet: Medical transformer for chest x-ray diagnosis and reporting. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 293–303. [Google Scholar]
- Wang, Z.; Tang, M.; Wang, L.; Li, X.; Zhou, L. A medical semantic-assisted transformer for radiographic report generation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 655–664. [Google Scholar]
- Tanida, T.; Müller, P.; Kaissis, G.; Rueckert, D. Interactive and explainable region-guided radiology report generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7433–7442. [Google Scholar]
- Endo, M.; Krishnan, R.; Krishna, V.; Ng, A.Y.; Rajpurkar, P. Retrieval-based chest x-ray report generation using a pre-trained contrastive language-image model. In Proceedings of the Machine Learning for Health, Online, 4 December 2021; pp. 209–219. [Google Scholar]
- Jin, H.; Che, H.; Lin, Y.; Chen, H. Promptmrg: Diagnosis-driven prompts for medical report generation. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver Convention Center, Vancouver, BC, Canada, 26–27 February 2024; Volume 38, pp. 2607–2615. [Google Scholar]
- Smit, A.; Jain, S.; Rajpurkar, P.; Pareek, A.; Ng, A.Y.; Lungren, M.P. CheXbert: Combining automatic labelers and expert annotations for accurate radiology report labeling using BERT. arXiv 2020, arXiv:2004.09167. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.y.; Mark, R.G.; Horng, S. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 2019, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, A.; Dowling, J.; Koopman, B. Improving chest X-ray report generation by leveraging warm starting. Artif. Intell. Med. 2023, 144, 102633. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, Minnesota, 2–7 June 2019; Volume 1 (Long and Short Papers). pp. 4171–4186. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Yang, S.; Wu, X.; Ge, S.; Zheng, Z.; Zhou, S.K.; Xiao, L. Radiology report generation with a learned knowledge base and multi-modal alignment. Med. Image Anal. 2023, 86, 102798. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lin, B.; Chen, Z.; Lin, H.; Liang, X.; Chang, X. Dynamic graph enhanced contrastive learning for chest x-ray report generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 3334–3343. [Google Scholar]
- Huang, Z.; Zhang, X.; Zhang, S. Kiut: Knowledge-injected u-transformer for radiology report generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 19809–19818. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).