Towards Trustful Machine Learning for Antimicrobial Therapy Using an Explainable Artificial Intelligence Dashboard
Abstract
1. Introduction
2. Background
3. Materials and Methods
3.1. Use Case and Explainability Requirements
3.2. XAI Methods
3.3. Dashboard Visualization
3.4. Architecture, Software, and Dashboard Development
3.5. Participant Recruitment and Dashboard Evaluation Form
4. Results
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nazar, M.; Alam, M.M.; Yafi, E.; Su’ud, M.M. A Systematic Review of Human–Computer Interaction and Explainable Artificial Intelligence in Healthcare With Artificial Intelligence Techniques. IEEE Access 2021, 9, 153316–153348. [Google Scholar] [CrossRef]
- 2023. Available online: https://ezra.com/en-gb (accessed on 15 November 2023).
- Zhang, J.; Whebell, S.; Gallifant, J.; Budhdeo, S.; Mattie, H.; Lertvittayakumjorn, P.; Lopez, M.P.A.; Tiangco, B.J.; Gichoya, J.W.; Ashrafian, H.; et al. An interactive dashboard to track themes, development maturity, and global equity in clinical artificial intelligence research. medRxiv 2021. [Google Scholar] [CrossRef]
- Lång, K.; Josefsson, V.; Larsson, A.M.; Larsson, S.; Högberg, C.; Sartor, H.; Hofvind, S.; Andersson, I.; Rosso, A. Artificial intelligence-supported screen reading versus standard double reading in the Mammography Screening with Artificial Intelligence trial (MASAI): A clinical safety analysis of a randomised, controlled, non-inferiority, single-blinded, screening accuracy study. Lancet Oncol. 2023, 24, 936–944. [Google Scholar] [CrossRef]
- Yusof, M.M.; Kuljis, J.; Papazafeiropoulou, A.; Stergioulas, L.K. An evaluation framework for Health Information Systems: Human, organization and technology-fit factors (HOT-fit). Int. J. Med. Inform. 2008, 77, 386–398. [Google Scholar] [CrossRef] [PubMed]
- De Corte, T.; Van Maele, L.; Dietvorst, J.; Verhaeghe, J.; Vandendriessche, A.; De Neve, N.; Vanderhaeghen, S.; Dumoulin, A.; Temmerman, W.; Dewulf, B.; et al. Exploring Preconditions for the Implementation of Artificial Intelligence-based Clinical Decision Support Systems in the Intensive Care Unit—A Multicentric Mixed Methods Study. J. Crit. Care, 2025; under review. [Google Scholar]
- Cutillo, C.M.; Sharma, K.R.; Foschini, L.; Kundu, S.; Mackintosh, M.; Mandl, K.D. Machine intelligence in healthcare—perspectives on trustworthiness, explainability, usability, and transparency. npj Digit. Med. 2020, 3, 47. [Google Scholar] [CrossRef] [PubMed]
- Tonekaboni, S.; Joshi, S.; McCradden, M.D.; Goldenberg, A. What Clinicians Want: Contextualizing Explainable Machine Learning for Clinical End Use. arXiv 2019, arXiv:1905.05134. [Google Scholar] [CrossRef]
- Pierce, R.L.; Van Biesen, W.; Van Cauwenberge, D.; Decruyenaere, J.; Sterckx, S. Explainability in medicine in an era of AI-based clinical decision support systems. Front. Genet. 2022, 13, 903600. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems 30; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; pp. 4765–4774. [Google Scholar]
- A., S.; R., S. A systematic review of Explainable Artificial Intelligence models and applications: Recent developments and future trends. Decis. Anal. J. 2023, 7, 100230. [Google Scholar] [CrossRef]
- Wismüller, A.; Stockmaster, L. A prospective randomized clinical trial for measuring radiology study reporting time on Artificial Intelligence-based detection of intracranial hemorrhage in emergent care head CT. In Proceedings of the Medical Imaging 2020: Biomedical Applications in Molecular, Structural, and Functional Imaging, Houston, TX, USA, 15–20 February 2020; SPIE: Bellingham, WA, USA, 2020; Volume 11317, pp. 144–150. [Google Scholar] [CrossRef]
- Zhang, W.-f.; Li, D.-h.; Wei, Q.-j.; Ding, D.-y.; Meng, L.-h.; Wang, Y.-l.; Zhao, X.-y.; Chen, Y.-x. The Validation of Deep Learning-Based Grading Model for Diabetic Retinopathy. Front. Med. 2022, 9, 839088. [Google Scholar] [CrossRef]
- Ni, Y.; Bermudez, M.; Kennebeck, S.; Liddy-Hicks, S.; Dexheimer, J. A Real-Time Automated Patient Screening System for Clinical Trials Eligibility in an Emergency Department: Design and Evaluation. JMIR Med. Inform. 2019, 7, e14185. [Google Scholar] [CrossRef]
- Norrie, C. Explainable AI Techniques for Sepsis Diagnosis: Evaluating LIME and SHAP Through a User Study. Master’s Thesis, University of Skovde, Skovde, Sweden, 2021. [Google Scholar]
- Verhaeghe, J.; Dhaese, S.A.M.; De Corte, T.; Vander Mijnsbrugge, D.; Aardema, H.; Zijlstra, J.G.; Verstraete, A.G.; Stove, V.; Colin, P.; Ongenae, F.; et al. Development and evaluation of uncertainty quantifying machine learning models to predict piperacillin plasma concentrations in critically ill patients. BMC Med. Inform. Decis. Mak. 2022, 22, 224. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gandomi, A.H.; Chen, F.; Holzinger, A. Evaluating the Quality of Machine Learning Explanations: A Survey on Methods and Metrics. Electronics 2021, 10, 593. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Anchors: High-Precision Model-Agnostic Explanations. Proc. AAAI Conf. Artif. Intell. 2018, 32, 1527–1535. [Google Scholar] [CrossRef]
- Ustun, B.; Rudin, C. Supersparse linear integer models for optimized medical scoring systems. Mach. Learn. 2016, 102, 349–391. [Google Scholar] [CrossRef]
- Sharchilev, B.; Ustinovsky, Y.; Serdyukov, P.; de Rijke, M. Finding Influential Training Samples for Gradient Boosted Decision Trees. arXiv 2018, arXiv:1802.06640. [Google Scholar] [CrossRef]
- Baniecki, H.; Kretowicz, W.; Piatyszek, P.; Wisniewski, J.; Biecek, P. dalex: Responsible Machine Learning with Interactive Explainability and Fairness in Python. J. Mach. Learn. Res. 2021, 22, 1–7. [Google Scholar]
- Mothilal, R.K.; Sharma, A.; Tan, C. Explaining machine learning classifiers through diverse counterfactual explanations. In Proceedings of the 2020 Conference on Fairness, Accountability, and Transparency, Barcelona, Spain, 27–30 January 2020; pp. 607–617. [Google Scholar]
- Wickramanayake, B.; Ouyang, C.; Xu, Y.; Moreira, C. Generating multi-level explanations for process outcome predictions. Eng. Appl. Artif. Intell. 2023, 125, 106678. [Google Scholar] [CrossRef]
- Galanti, R.; de Leoni, M.; Monaro, M.; Navarin, N.; Marazzi, A.; Di Stasi, B.; Maldera, S. An explainable decision support system for predictive process analytics. Eng. Appl. Artif. Intell. 2023, 120, 105904. [Google Scholar] [CrossRef]
- Knowledge Centre Data & Society HC-XAI Evaluation Inventory. 2023. Available online: https://data-en-maatschappij.ai/en/publications/xai-decision-tool-2021 (accessed on 27 November 2023).
- De Bus, L.; Gadeyne, B.; Steen, J.; Boelens, J.; Claeys, G.; Benoit, D.; De Waele, J.; Decruyenaere, J.; Depuydt, P. A complete and multifaceted overview of antibiotic use and infection diagnosis in the intensive care unit: Results from a prospective four-year registration. Crit. Care 2018, 22, 241. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.L.; Sakr, Y.; Singer, M.; Martin-Loeches, I.; Machado, F.R.; Marshall, J.C.; Finfer, S.; Pelosi, P.; Brazzi, L.; Aditianingsih, D.; et al. Prevalence and Outcomes of Infection Among Patients in Intensive Care Units in 2017. JAMA 2020, 323, 1478–1487. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Ten Health Issues WHO Will Tackle This Year. 2023. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 17 October 2023).
- Roberts, J.A.; De Waele, J.J.; Dimopoulos, G.; Koulenti, D.; Martin, C.; Montravers, P.; Rello, J.; Rhodes, A.; Starr, T.; Wallis, S.C.; et al. DALI: Defining Antibiotic Levels in Intensive care unit patients: A multi-centre point of prevalence study to determine whether contemporary antibiotic dosing for critically ill patients is therapeutic. BMC Infect. Dis. 2012, 12, 152. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Aziz, M.H.; Alffenaar, J.W.C.; Bassetti, M.; Bracht, H.; Dimopoulos, G.; Marriott, D.; Neely, M.N.; Paiva, J.A.; Pea, F.; Sjovall, F.; et al. Antimicrobial therapeutic drug monitoring in critically ill adult patients: A Position Paper. Intensive Care Med. 2020, 46, 1127–1153. [Google Scholar] [CrossRef]
- Pai Mangalore, R.; Ashok, A.; Lee, S.J.; Romero, L.; Peel, T.N.; Udy, A.A.; Peleg, A.Y. Beta-Lactam Antibiotic Therapeutic Drug Monitoring in Critically Ill Patients: A Systematic Review and Meta-Analysis. Clin. Infect. Dis. 2022, 75, 1848–1860. [Google Scholar] [CrossRef]
- Verhaeghe, J.; De Corte, T.; De Waele, J.J.; Ongenae, F.; Van Hoecke, S. Designing a Pharmacokinetic Machine Learning Model for Optimizing Beta-Lactam Antimicrobial Dosing in Critically Ill Patients. In Proceedings of the ICMHI ’24: 2024 8th International Conference on Medical and Health Informatics, New York, NY, USA, 17–19 May 2024; pp. 311–317. [Google Scholar] [CrossRef]
- Grinberg, M. Flask Web Development: Developing Web Applications with Python; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2018. [Google Scholar]
- Meta. React: A JavaScript Library for Building User Interfaces. 2023. Available online: https://legacy.reactjs.org/ (accessed on 16 February 2023).
- Kaur, H.; Nori, H.; Jenkins, S.; Caruana, R.; Wallach, H.; Wortman Vaughan, J. Interpreting Interpretability: Understanding Data Scientists’ Use of Interpretability Tools for Machine Learning. In Proceedings of the CHI ’20: 2020 CHI Conference on Human Factors in Computing Systems, New York, NY, USA, 25–30 April 2020; pp. 1–14. [Google Scholar] [CrossRef]
- Gkatzia, D.; Lemon, O.; Rieser, V. Natural Language Generation enhances human decision-making with uncertain information. arXiv 2016, arXiv:1606.03254. [Google Scholar] [CrossRef]
- Sokol, K.; Flach, P. One Explanation Does Not Fit All. KI-Künstliche Intell. 2020, 34, 235–250. [Google Scholar] [CrossRef]
- Werner, C. Explainable AI through Rule-based Interactive Conversation. In Proceedings of the EDBT/ICDT Workshops, Copenhagen, Denmark, 30 March–2 April 2020. [Google Scholar]
- Chromik, M. Making SHAP Rap: Bridging Local and Global Insights Through Interaction and Narratives. In Human-Computer Interaction—INTERACT 2021, Proceedings of the 18th IFIP TC 13 International Conference, Bari, Italy, 30 August–3 September 2021; Ardito, C., Lanzilotti, R., Malizia, A., Petrie, H., Piccinno, A., Desolda, G., Inkpen, K., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; pp. 641–651. [Google Scholar] [CrossRef]
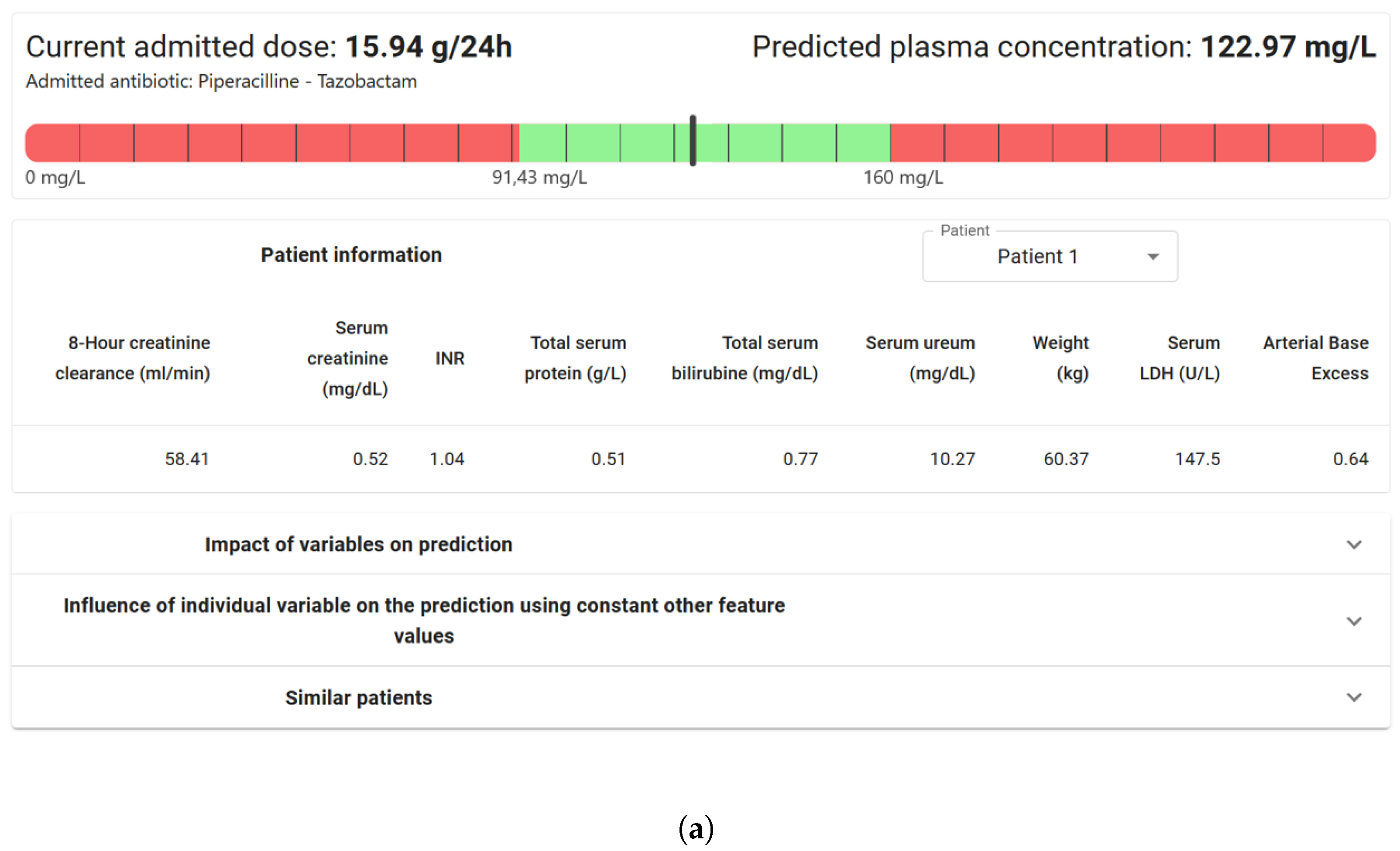

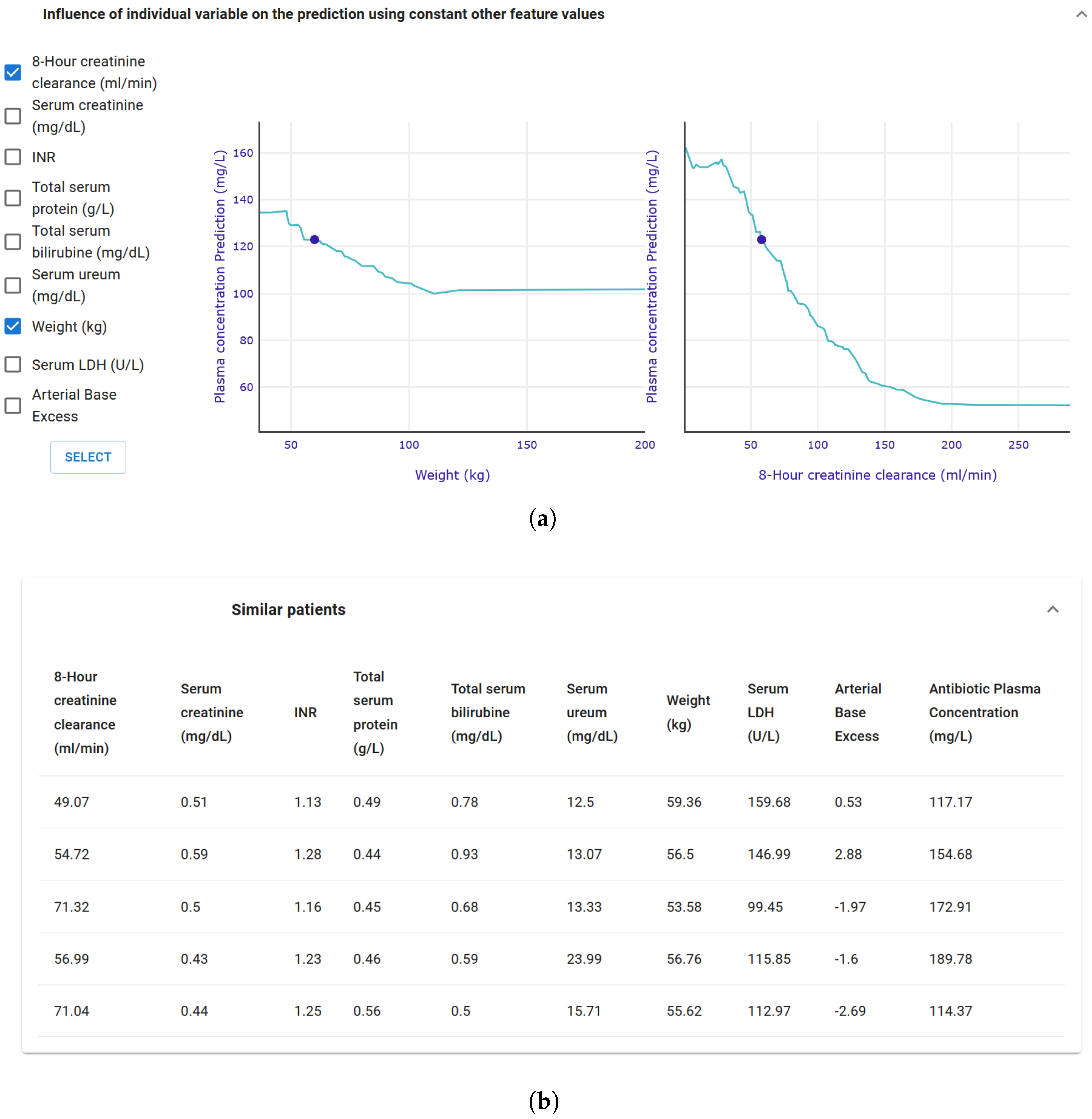

| Measured Antimicrobial Concentration (mg/L) | Range Measured Concentration | Predicted Antimicrobial Concentration (mg/L) | Range Prediction Concentration |
|---|---|---|---|
| 125.1 | Therapeutic | 122.97 | Therapeutic |
| 90.41 | Lower bound of | 92.03 | Lower bound of |
| therapeutic range | therapeutic range | ||
| 118.95 | Therapeutic | 216.4 | Supratherapeutic |
| 108.8 | Therapeutic | 67.6 | Subtherapeutic |
| 226.4 | Supratherapeutic | 218.7 | Supratherapeutic |
| Component | Score/10 | |
|---|---|---|
| Predicted concentration | 8.14 | 1.57 |
| Last given dose | 8.14 | 1.57 |
| Toxic–therapeutic margin | 7.57 | 1.51 |
| CP Profile | 7.00 | 1.83 |
| Patient information | 6.29 | 1.70 |
| SHAP | 5.43 | 3.50 |
| Leaf influence | 5.43 | 1.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Corte, T.; Verhaeghe, J.; Ongenae, F.; De Waele, J.J.; Van Hoecke, S. Towards Trustful Machine Learning for Antimicrobial Therapy Using an Explainable Artificial Intelligence Dashboard. Appl. Sci. 2025, 15, 10933. https://doi.org/10.3390/app152010933
De Corte T, Verhaeghe J, Ongenae F, De Waele JJ, Van Hoecke S. Towards Trustful Machine Learning for Antimicrobial Therapy Using an Explainable Artificial Intelligence Dashboard. Applied Sciences. 2025; 15(20):10933. https://doi.org/10.3390/app152010933
Chicago/Turabian StyleDe Corte, Thomas, Jarne Verhaeghe, Femke Ongenae, Jan J. De Waele, and Sofie Van Hoecke. 2025. "Towards Trustful Machine Learning for Antimicrobial Therapy Using an Explainable Artificial Intelligence Dashboard" Applied Sciences 15, no. 20: 10933. https://doi.org/10.3390/app152010933
APA StyleDe Corte, T., Verhaeghe, J., Ongenae, F., De Waele, J. J., & Van Hoecke, S. (2025). Towards Trustful Machine Learning for Antimicrobial Therapy Using an Explainable Artificial Intelligence Dashboard. Applied Sciences, 15(20), 10933. https://doi.org/10.3390/app152010933








