Abstract
Image quality plays a critical role in medical image analysis, significantly impacting diagnostic outcomes. Sharp and detailed images are essential for accurate diagnoses, but acquiring high-resolution medical images often demands sophisticated and costly equipment. To address this challenge, this study proposes a convolutional neural network (CNN)-based super-resolution architecture, utilizing a melanoma dataset to enhance image resolution through deep learning techniques. The proposed model incorporates a convolutional self-attention block that combines channel and spatial attention to emphasize important image features. Channel attention uses global average pooling and fully connected layers to enhance high-frequency features within channels. Meanwhile, spatial attention applies a single-channel convolution to emphasize high-frequency features in the spatial domain. By integrating various attention blocks, feature extraction is optimized and further expanded through subpixel convolution to produce high-quality super-resolution images. The model uses L1 loss to generate realistic and smooth outputs, outperforming existing deep learning methods in capturing contours and textures. Evaluations with the ISIC 2020 dataset—containing 33126 training and 10982 test images for skin lesion analysis—showed a 1–2% improvement in peak signal-to-noise ratio (PSNR) compared to very deep super-resolution (VDSR) and enhanced deep super-resolution (EDSR) architectures.
1. Introduction
Deep learning has emerged as a critical tool in image recognition and medical applications, including melanoma diagnosis. Melanoma is a highly lethal skin cancer, making early detection essential for improving patient outcomes [1]. However, accurate diagnosis remains challenging due to subtle lesion appearances and diverse clinical presentations. According to a pre-COVID-19 analysis report, as of 2015, 3.1 million people had the active disease, resulting in approximately 60,000 deaths. Therefore, dermatologists have developed extensive data and algorithms for diagnosing melanoma, primarily based on deep learning. Meanwhile, artificial intelligence is currently making innovative advancements through its complementary relationship with big data. The demand for data processing and understanding increases substantially as the volume of data increases. Consequently, artificial intelligence can provide better insights and predictive capabilities using big data. Big data and artificial intelligence can synergize and result in unpredictable changes and innovations. Among these, super-resolution imaging using artificial intelligence is investigated. High-resolution images contain more detail than low-resolution ones. However, acquiring high-resolution images is expensive. Therefore, super-resolution techniques have been incorporated to reconstruct high-resolution images from low-resolution ones. These approaches have shown potential for addressing challenges in medical imaging, such as low spatial resolution and diagnostic limitations, but their application remains complex and demanding [2,3,4,5]. However, super-resolution techniques are challenging [6,7]. Generating high-resolution images is computationally complex, requires significant computational resources and time, and may not be suitable for real-time applications. However, in medical imaging, fine details such as small anatomical structures can convey essential information for diagnosis. For instance, applying artificial intelligence in the diagnosis, treatment, and prognosis assessment of pancreatic cancer has been discussed. Artificial intelligence has been used to aid in the imaging-based diagnosis of pancreatic cancer by automatically identifying and classifying pancreatic tumors based on imaging data from computed tomography and magnetic resonance imaging [8]. It can also predict drug treatment responses and propose personalized treatment plans. Models and algorithms that predict the prognosis of pancreatic cancer have been developed by integrating clinical information, genetic data, and histological tissue characteristics [9]. Therefore, artificial intelligence introduced through super-resolution techniques can impact diagnosis substantially [10].
Therefore, deep learning and artificial intelligence technologies considerably assist in diagnosis and analysis across various medical disciplines through appropriate data training and parallel computing. This study aimed to apply super-resolution techniques to improve the accuracy and efficiency of melanoma diagnosis. The subsequent section discusses the proposed architecture.
1.1. Deep Learning-Based Methods
High-resolution image generation is essential in computer vision, with the aim of converting low-resolution images into high-quality, high-resolution images. Previous research has attempted to enhance image resolution using traditional interpolation and upscaling techniques. However, these methods often do not adequately recover detailed image information or amplify high-frequency noise. In contrast, deep learning can effectively extract and restore high-frequency and fine-grained information from images through neural network training based on large-scale datasets [11]. Various approaches—such as machine learning, prediction-, reconstruction-, and boundary-based methods—have been explored. In addition, hybrid techniques that combine deep learning with models and signal processing have also emerged. These studies aimed to improve the accuracy, color fidelity, and quality of images generated. Overcoming the limitations of the existing methods for high-resolution image generation provides a novel approach to generating high-quality images with realistic and sharp details. High-resolution images are applicable in various fields, including high-quality video production, medical image analysis, and security, emphasizing their importance. Among these approaches, very deep super-resolution (VDSR) [12], enhanced deep super-resolution (EDSR) [13], and bicubic interpolation have garnered attention for generating high-resolution images. VDSR employs a deep neural network architecture to generate high-resolution images by learning the residuals between the low- and high-resolution images. This allows the extraction of high-frequency details from low-resolution input images to generate high-resolution images. However, using a deep network in VDSR increases the risk of overfitting and often requires voluminous training data. Additionally, the training and inference processes of VDSR can be computationally intensive and time-consuming. Similarly to VDSR, EDSR is a deep learning-based method for generating high-resolution images that uses a deep and robust neural network. However, EDSR further improves the structure of the residual blocks and employs a larger receptive field to generate more accurate and natural high-resolution images. This enables the extraction of more high-frequency details. However, the increased complexity of the EDSR structure can lead to many parameters, high resource requirements, and challenges such as gradient fading. Bicubic interpolation is a traditional method that is used to generate high-resolution images [14]. It is used to enlarge low-resolution input images into high-resolution images by interpolating high-resolution values using neighboring pixel values. However, bicubic interpolation may not accurately reproduce high-frequency details during image generation, resulting in overly smooth images. Moreover, as an interpolation technique used in upsampling, it may not accurately model the complexity and diversity of real images. These deep learning-based methods can be challenging for diagnosing and analyzing melanoma. Therefore, this study proposes a channel–spatial block-based super-resolution (CS-SISR) method, an artificial intelligence technique for melanoma diagnosis and high-resolution image generation. CS-SISR is a deep learning-based approach that converts low-resolution images into high-resolution images. CS-SISR addresses the limitations of high-frequency regions and feature extraction using a convolutional self-attention (CS) block that combines channel and spatial attention. The core components and their roles in the CS-SISR method are summarized in Table 1 below.

Table 1.
Key components of CS-SISR.
Channel attention learns the importance of each channel in the input feature map to emphasize high-frequency features. Spatial attention learns the importance of each spatial position in the input feature map to enhance the spatial information of the high-frequency features. The feature maps extracted through the CSBlock were used to generate high-resolution images using subpixel convolution and residual blocks. Subpixel convolution is used for upsampling low-resolution input images to high-resolution output images, and the residual block helps generate natural high-resolution images by calculating the residual between the high-resolution and low-resolution images. CS-SISR is trained by minimizing the L1 loss using 48 × 48-sized low- and high-resolution patches. This method emphasizes the high-frequency features and utilizes them to generate natural high-resolution images. CS-SISR is a novel deep learning-based method that can generate more accurate and natural high-resolution images than existing methods by emphasizing high-frequency features and utilizing diverse feature information.
This can enhance the diagnostic accuracy and time efficiency of medical images, as shown in Figure 1, which presents a melanoma image (ISIC 2020 Challenge dataset).
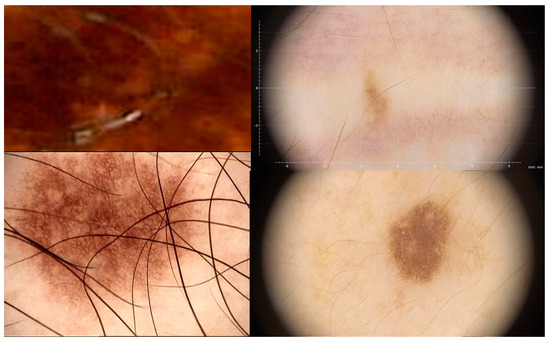
Figure 1.
ISIC 2020 Challenge dataset.
1.2. Data Availability
The experiments in this study were conducted using the ISIC 2020 Challenge dataset, provided by the International Skin Imaging Collaboration (ISIC). The dataset includes de-identified dermoscopic images, which were used in accordance with the guidelines provided by the ISIC.
The dataset is publicly available and can be accessed by researchers for further experimentation through the following repositories:
- ISIC Archive;
- ISIC 2020 Challenge.
1.3. Ethical Considerations
The ISIC 2020 dataset consists of de-identified medical images, and the dataset providers ensured that all necessary ethical approvals and patient consents were obtained prior to data distribution. The ISIC organization guarantees that all required ethical guidelines are followed, in compliance with international medical standards. Researchers utilizing this dataset adhere to the ethical guidelines outlined by the ISIC and the relevant medical authorities. This dataset is publicly available for further experimentation, ensuring reproducibility and transparency in research.
2. Related Work
Before delving into specific attention mechanisms, it is essential to examine core deep learning architectures that tackle the challenges posed by increasingly deeper networks. The following subsection introduces ResNet, a widely adopted neural network design known for its residual connections. This architecture effectively mitigates issues such as vanishing gradients, enabling the construction of more robust and high-performing networks for image recognition and other computer vision tasks.
2.1. ResNet
ResNet is a neural network architecture that is widely employed in deep learning for image classification and computer vision tasks [15]. Residual learning is a methodology that addresses the increasing difficulty in learning as the network becomes deeper. This involves adding an input value to the output value of the network. This approach enables the effective training of deep networks while ensuring network stability. The ResNet architecture exhibits the following characteristics:
- Expansion to deep networks: ResNet introduces residual connections to enable the stacking of networks to deeper levels, thereby alleviating the gradient vanishing that occurs in deep networks. Residual connections construct a network by learning the residuals between the input and output and adding them to the output. This allows for a better performance as the network depth increases.
- Performance: With its ability to construct deep networks, ResNet can learn and extract more complex and abstract image features, leading to high classification accuracy.
- Efficient learning: Residual connections preserve information from the previous layers, ensuring stable gradients and a smooth learning process.
- Reusable block structure: The block structure is composed of stacked basic blocks, constituting a reusable form that facilitates model construction.
- Neural network architecture: Generally, ResNet is built by stacking basic blocks consisting of convolution layers and rectified linear units (ReLUs) as activation functions. These basic blocks are connected by residual connections, enabling the neural network to learn the residuals between the inputs and outputs. Convolution layers with various kernel sizes and strides are employed for each layer, enabling feature extraction at different scales.
As the number of layers in a neural network increases, higher-level features are expected to be extracted. However, when the neural network becomes excessively deep, gradient vanishing occurs, leading to training difficulties. Additionally, several disadvantages have been observed (Figure 2):
- Computational and memory requirements: Deep networks have high computational and memory requirements during training and inference. Training with larger models requires significant resources and increases memory usage.
- Model Size: Deep structures result in relatively large models. Therefore, models with extensive structures may encounter storage space and transmission speed constraints in certain scenarios.
- Overfitting Issue: Constructing models with deep structures requires more parameters. Consequently, overfitting might arise in cases with limited training data.
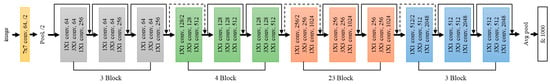
Figure 2.
ResNet architecture.
Thus, caution is required regarding computational resources and model size. Therefore, proper data management and the prevention of overfitting must be considered. Hence, a residual block that employs skip connections is proposed to solve these problems.
In Figure 3, (a) represents the conventional neural network, and (b) represents the residual block structure. The primary difference lies in the configuration of residual connections. In a conventional neural network, the input is transformed into features, and multiple layers are traversed to compute the final output. Each layer learns a transformation function between the input and output and maps the input into the desired form. However, as the network deepens, traditional neural networks must progressively learn the transformations between the input and output, which can lead to vanishing or exploding gradients.
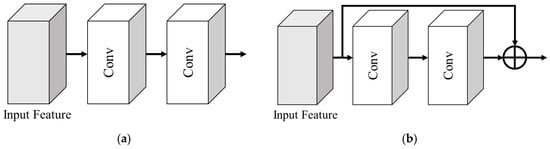
Figure 3.
Network model: (a) existing CNN; (b) CNN-based ResNet residual block.
In contrast, residual blocks explicitly learn the residuals between the input and output. After passing through the input, a residual block learns the difference between the input and output through a residual connection and adds this difference to the output. Consequently, the neural network only needs to learn the difference between the input and output, ensuring stable gradients and facilitating smoother training, even as the network deepens.
Equation (1) represents the formulation of a conventional neural network structure. This equation extracts features from input through the convolution operation H(). Next, the ReLU function introduces nonlinearity by converting negative values to 0 and keeping positive values unchanged. Finally, another H function is applied to extract the features and compute the final output.
Equation (2) represents the formulation of the ResNet. This equation extracts features from input using the convolution operation—H(). The ReLU function then introduces nonlinearity to calculate the activation values. Subsequently, the previous computation result is added to input . This is referred to as a residual connection, allowing for the explicit learning of the residuals between the input and output. This residual connection alleviates the vanishing gradient problem as the network deepens and enables more accurate predictions. Thus, ResNet enhances the network performance by explicitly learning the difference between the input and output through residual connections. The residual connection stabilizes the gradients and facilitates smoother training. In addition, it improves the representation capability of the network by learning the residuals between the input and output. Thus, ResNet can construct deeper networks than traditional neural network structures and exhibits high accuracy and performance in various vision tasks such as image segmentation, object detection, and classification [16,17,18,19].
2.2. Attention Mechanism
The attention mechanism can be applied to machine translation, question answering, and natural language processing tasks [20]. This technique allows the model to focus more on specific parts and assign weights, allowing it to concentrate more on the relevant information within the input sequence. It has been used in the sequence-to-sequence (seq2seq) model to apply features to previous inputs differentially. It improves the traditional encoder–decoder structure and regulates the relevance between the input and output. This effectively mitigates the loss and gradient vanishing issues that occur during the compression of fixed-size vectors.
The mechanism consists of the following stages:
- Encoder stageThis involves embedding each element (e.g., word or token) of the input sequence to represent its meaning in a vector space. These embeddings contribute to extracting and representing input features. Encoder structures are typically implemented using recurrent neural networks (RNNs) or modified functions (e.g., transformers). The output of the encoder serves as a context vector and is used in the subsequent attention stage.
- Attention stageHerein, a weighted average is computed based on the output from the encoder and the previously generated state from the decoder. This process focuses on the relevant parts of the input sequence. The query, key, and value concepts are commonly used. The query is generated based on the previous state or output of the decoder, whereas the key and value are generated from the output of the encoder. Attention weights are used to calculate the relevance between the query and key, with the attention-weighted sum summarizing the information of specific parts of the input.
- Decoder stageThe decoder utilizes the computed weighted average from the attention stage and combines it with the previous state to generate the subsequent output. This process is iteratively repeated with the decoder, using the previous output as the input to complete the sequence. Similarly to the encoder, the decoder is implemented using RNNs or structures such as transformers. The output of the decoder serves as the final prediction, and can be used in various ways, depending on the objective of the model.
This attention mechanism dynamically allocates weights to each element in the input sequence, thereby assigning relevance. It can efficiently recognize relevant information by focusing on specific parts of the input sequence and adjusting the relevance between the input and output. Recent attention mechanisms include seq2seq and mechanisms such as Reformer [21], LinFormer [22], and Swin Transformer [23], which utilize transformer structures [24]. These attention mechanisms play a role in efficient attention operations that reduce memory usage and optimize computational costs. They also demonstrate superior performance in natural language processing tasks using self-attention instead of RNNs. Moreover, attention in computer vision is applied in object detection and semantic segmentation. In such tasks, the objects of interest are often distributed in specific regions rather than uniformly across the entire image. Therefore, attention is employed to focus on crucial object regions and extract features by disregarding unnecessary regions. This approach emphasizes spatial and semantic information and improves network performance by leveraging the meaning and positional characteristics of objects. In addition, this method even demonstrated good performance with noisy inputs. In the convolutional neural network (CNN), attention mechanisms such as the convolutional block attention module (CBAM) are used [25]. CBAM provides an effective attention mechanism for computer vision tasks, such as image classification and object detection. As shown in Figure 4, it combines a CNN for extracting image features with an attention mechanism to focus on important object regions and allocate attention to spatial and channel dimensions. CBAM consists of two main components: channel and spatial attention. Figure 5 illustrates the channel attention module, which helps focus on important channel features by modeling the relationships between channels in the input feature map. Spatial attention models the relationships between spatial positions in the input feature map to concentrate on important parts of the objects. CBAM models the interaction between the channel and spatial positions in the input feature map through these two attention mechanisms, enabling stronger attention and meaning towards specific objects.
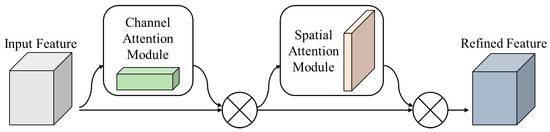
Figure 4.
Convolutional block attention module.
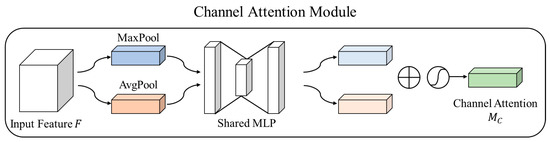
Figure 5.
Channel attention module of the convolutional block attention module.
Channel attention generates an attention map by leveraging the relationships between channels in the features. First, channel attention calculates the average value for each channel in the input feature map. Spatial compression was performed using max pooling and avg pooling to efficiently compute the operations and transform them into a one-dimensional vector to achieve this. A multi-layer perceptron (MLP) learns the important weights and calculates the relevance of channel features among different feature maps using the channel-wise average values. The MLP also models interactions between channels. The obtained weights are then transformed into an appropriate form using an activation function. Typically, the sum of two outputs and a Sigmoid function are used to generate an attention map within the range of 0 to 1.
Equation (3) represents the channel attention module of CBAM. Here, σ denotes the Sigmoid activation function, MLP refers to the multi-layer perceptron, avg pooling represents average pooling, max pooling denotes maximum pooling, and F represents the input feature map.
As shown in Figure 6, spatial attention was used to model the spatial information across channels. This is typically achieved by multiplying the computed weights through convolutional layers and activation functions with the input feature map. The most common approach is to generate a one-channel feature map using a 1 × 1 convolution, which is then incorporated into the input feature map. Unlike channel attention, spatial attention calculates the relevance between pixels. Subsequently, a Sigmoid function is used to generate an attention map within the range of 0 to 1.
Equation (4) represents the spatial attention module of CBAM. Here, denotes a convolution.
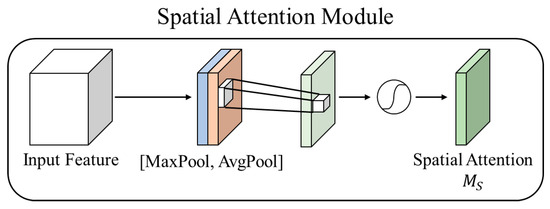
Figure 6.
Spatial attention module.
2.3. Super-Resolution Approaches
Recent advancements in medical image super-resolution have introduced innovative techniques leveraging attention mechanisms and wavelet-based architectures. For instance, multimodal-boost, a multimodal medical image super-resolution using a multi-attention network with wavelet transform, integrates multi-attention mechanisms with wavelet transforms to effectively enhance high-frequency details across diverse modalities [26]. Similarly, multimodal multi-head convolutional attention with various kernel sizes for medical image super-resolution utilizes convolutional attention mechanisms with adaptive kernel sizes to improve spatial and frequency domain learning [27].
Furthermore, wavelet-based methods, such as a super-resolution network for medical imaging via transformation analysis of wavelet multi-resolution and analysis of medical images super-resolution via a wavelet pyramid recursive neural network constrained by wavelet energy entropy, have achieved state-of-the-art performance by employing multi-resolution wavelet analysis and recursive structures [28,29]. These approaches demonstrate the growing emphasis on domain-specific optimization in medical imaging.
In comparison, the proposed CS-SISR model builds upon these advancements by incorporating lightweight channel and spatial attention mechanisms. This design ensures computational efficiency while maintaining high diagnostic relevance, particularly for melanoma image enhancement.
3. Proposed Method
Recent advancements in super-resolution methods, such as RCAN [30] and SwinIR [31], have demonstrated exceptional performance on standard datasets, like DIV2K and Set14. These models utilize advanced attention mechanisms and transformer-based architectures to achieve state-of-the-art results. However, such approaches often involve complex architectures and high computational costs, which may limit their applicability in real-time scenarios, particularly in specialized domains such as medical imaging. To address these limitations, we propose the CS-SISR model, which integrates lightweight channel and spatial attention mechanisms optimized for melanoma diagnosis. This model is specifically designed to restore diagnostically relevant features, such as lesion boundaries and textures, while maintaining computational efficiency.
The CS-SISR model employs the convolutional self-attention block (CSBlock) to emphasize high-frequency features. High-frequency features, such as edges and textures, significantly impact the visual quality of an image and are critical for accurate diagnosis in medical imaging. Self-attention, a method that emphasizes some values and suppresses others, effectively handles the high-frequency and low-frequency regions that influence image quality. Therefore, we introduce the CSBlock structure, which combines channel and spatial attention to emphasize high-frequency features effectively. The CSBlock structure employs a hierarchical feature extraction process that integrates both shallow feature extraction and deep feature extraction to enhance the reconstruction quality of images. The architecture of the proposed method, as shown in Figure 7, illustrates the flow of feature extraction and upsampling.
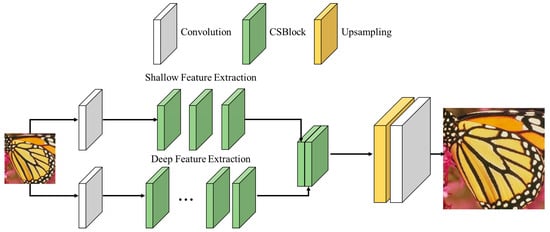
Figure 7.
Architecture of the proposed super-resolution method.
- Shallow feature extraction focuses on capturing low-level features such as edges, textures, and basic patterns in the initial layers of the network. These features are extracted using convolution operations, which process the input data to highlight essential visual elements. To ensure that the shallow features contribute effectively to the final reconstruction, skip connections are employed. These connections directly link the output of the shallow layers to the deeper layers, preserving critical base-level information necessary for accurate image reconstruction.
- Deep feature extraction involves learning complex and abstract features in the later stages of the network. This process focuses on capturing high-level patterns and relationships that are essential for reconstructing detailed and high-quality images. To enhance feature representation, skip connections and concatenation are utilized. Skip connections ensure that shallow, low-level features are retained and seamlessly integrated with deeper, high-dimensional features. By combining these features, the network effectively preserves low-level details while learning high-level abstractions, resulting in an improved and comprehensive feature representation.
The CSBlock structure combines shallow and deep features through skip connection and concatenation.
- Skip connection directly connects the output of a previous layer to a current layer within the network. This design helps mitigate the problem of gradient vanishing, which often occurs in deep networks, by ensuring that gradients can flow uninterrupted through the layers during backpropagation. By maintaining this direct connection, the network is able to learn deeper and more complex features effectively, contributing to an enhanced image reconstruction quality.
- Concatenation combines feature maps from different depths of the network, enabling the extraction of a wider range of features. This approach allows the model to integrate both low-level and high-level features, creating a richer and more diverse feature representation. By leveraging features at varying depths, concatenation enhances the network’s ability to capture fine details and complex patterns, improving the overall performance of the super-resolution method.
Subsequently, the features are expanded through upsampling to achieve super-resolution. Upsampling increases the size of low-resolution images to restore high-resolution features. By employing subpixel convolution, the proposed method avoids checkerboard artifacts, ensuring smooth outputs and preserving high-frequency details. The proposed super-resolution method can generate detailed and sharp images with emphasized high-frequency features. This hierarchical feature extraction process ensures that both shallow and deep features contribute to the final output, enhancing the model’s ability to preserve fine details and reconstruct high-quality images.
3.1. Attention Block
The high-frequency region is a crucial part of an image, as it significantly influences the visual quality, including contour lines and texture information. Therefore, restoring high-frequency components during the super-resolution process is vital. Therefore, we introduced an attention mechanism that uses CSBlock to emphasize high-frequency features. As shown in Figure 8, connecting the CSBlocks in a series, we strengthened the emphasis on high-frequency features. Skip connection structures in series connections help mitigate gradient vanishing, which can occur in deeper networks. With this architecture, we can effectively emphasize high-frequency features and train without encountering gradient-vanishing.
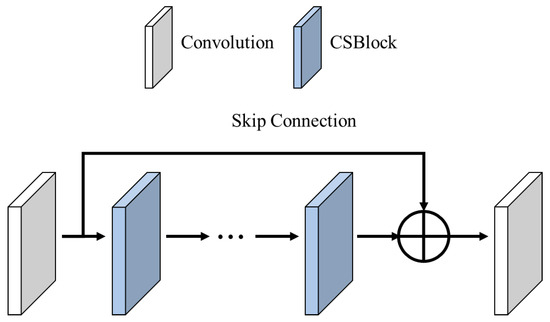
Figure 8.
Attention block structure.
Equation (5) presents the formula for the attention block, which emphasizes super-resolution. denotes an attention block, while denotes a convolutional operation. indicates the nth CSBlock, and is the input feature map.
In this study, we used ten CSBlocks constructed with skip connection structures and convolutional operations to prevent gradient vanishing and aid network training.
3.2. CSBlock
The CSBlock is effective for emphasizing high-frequency features in image analysis and processing tasks, and can achieve high accuracy and performance. Conventional convolution operations extract features by measuring the pattern similarity between the center and surrounding pixel values. However, this method does not effectively extract the patterns of pixels that exceed the kernel size, which is particularly striking for high-frequency features, and obstructs accurate image analysis. The CSBlock is designed to address this issue by combining channel and spatial attention with an input feature map to emphasize high-frequency features. Channel attention emphasizes necessary features by calculating the correlation and importance between channels of the input feature map. However, spatial attention emphasizes spatial features by calculating the correlation and importance among pixels. Thus, a CSBlock can effectively extract high-frequency features and achieve accurate image analysis results using this approach.
Figure 9 illustrates a CSBlock, which combines channel and spatial attention.
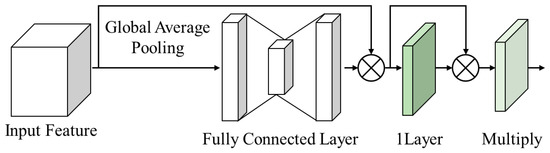
Figure 9.
Channel spatial block.
This structure progresses through the following steps:
Step 1: The input feature map is reduced using global average pooling (GAP), and the representative feature values of each channel are obtained by averaging the input feature map.
Step 2: The reduced feature map is used with fully connected layers to calculate the correlation and importance between channels, allowing us to grasp the importance of each channel and conduct channel attention by multiplying it by the input feature map.
Step 3: A feature map is generated by calculating the correlation between pixels using the convolution of one channel. This process extracted features by considering the spatial patterns of the input feature map.
Step 4: Spatial attention is performed by multiplying the feature map generated by calculating the correlation between pixels with the feature map on which channel attention is performed.
This method emphasizes the spatial features of the input feature map. Through this combined structure of channel and spatial attention, the CSBlock emphasizes high-frequency features and enables efficient image analysis and processing with fewer parameters and operations. This structure helps us clearly understand the core functions of the CSBlock and apply them to the analysis.
Figure 10 presents the channel attention structure of the proposed method.
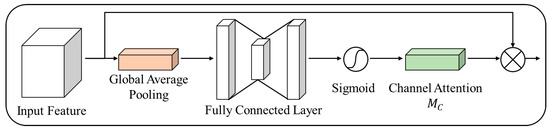
Figure 10.
Channel attention structure.
In contrast to the conventional CBAM, we removed the max pooling component and used the average pooling. This architectural adjustment reduced the emphasis on high-frequency features while achieving efficient channel attention with lower computational requirements. The previous CBAM accentuated high- and low-frequency features by incorporating max pooling and average pooling. However, this approach often resulted in an excessive emphasis on high-frequency features, which hindered the accurate extraction of the desired characteristics. Hence, in the proposed structure, we eliminated the max pooling component to mitigate the emphasis on high-frequency features, relying solely on the average pooling. Consequently, this modification helped enhance computational efficiency while effectively conducting channel attention. This facilitated the accentuation of relevant features and improved the ability to discern patterns within the input images. Such structural modifications contribute to better comprehension and yield superior image analysis results.
Equation (6) represents the GAP process, which is used to perform channel attention. H and W denote the height and width of the feature map, respectively, whereas HGP (*) represents the step of obtaining a representative value for each channel in the feature map by computing the average of all pixel values within that channel. This enables the extraction of representative feature values for each channel. GAP helped to obtain representative feature maps containing channel-specific information without requiring numerous computations or parameters. GAP reduced the number of parameters and computational complexity by calculating the average instead of processing every pixel value in the feature map. Consequently, GAP is leveraged to generate representative feature maps that consider interchannel relationships, thereby facilitating efficient feature extraction.
Equation (7) presents the calculation of the interchannel correlation and importance based on the GAP result, , obtained from Equation (6). Here, f and δ denote the Sigmoid and ReLU activation functions, respectively, while and represent the weights of the fully connected layers with output feature map channel sizes of d and d/4, respectively. We computed the interchannel correlation and its importance using two fully connected layers. The results were transformed into attention values ranging from 0 to 1 using a Sigmoid activation function. The attention values obtained indicate the importance of each channel in the feature map. Furthermore, by employing a fully connected layer that reduces the number of channels in the middle, we effectively reduced the computational complexity and number of parameters. This simultaneously enhanced the computational efficiency while accurately inferring the interchannel correlation and importance. This design enables the efficient computation of channel attention.
Equation (8) generates an emphasis feature map through multiplication operation of the input feature map on the channel attention feature map obtained from Equation (7). Here, x represents the input feature map, and s represents the channel attention feature map obtained from Equation (7). This process adjusts each channel of the input feature map by emphasizing high-frequency features and suppressing irrelevant ones. The channel attention feature map output values of 0–1, through a Sigmoid activation function, indicate the level of importance for each channel. The necessary channels are preserved by multiplying them with the input feature map. However, irrelevant channels are suppressed with values closer to 0. This enables feature refinement by emphasizing specific regions of the input feature map based on interchannel importance while inhibiting less important areas. The channel attention map considers interchannel importance, allowing for the accentuation of relevant parts and suppression of less significant areas, thereby enabling effective feature extraction. This provides a clear understanding and can be applied to the input feature maps for more efficient feature extraction.
Figure 11 illustrates the spatial attention structure of the proposed method.
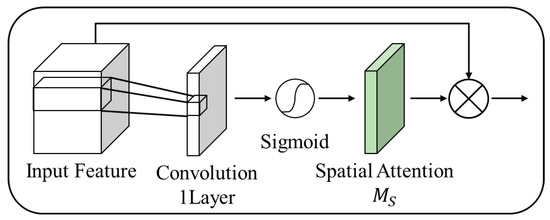
Figure 11.
Spatial attention structure.
In contrast to the conventional CBAM, this structure does not undergo a pooling process, but instead forms a convolution output as a single-channel representation. This minimizes the loss of spatial information and emphasizes spatial features. In the conventional CBAM, the pooling process reduced spatial information in some instances. However, in the proposed method, we omitted the pooling process and used convolution to reduce information loss while emphasizing the spatial features.
To provide a comprehensive view of the convolutional self-attention block (CSBlock), Figure 9, Figure 10 and Figure 11 have been consolidated into an integrated representation, as shown in Figure 12. This unified diagram illustrates the sequential processes of channel attention and spatial attention within the CSBlock, emphasizing how these mechanisms complement each other to enhance high-frequency features for efficient image analysis and processing.
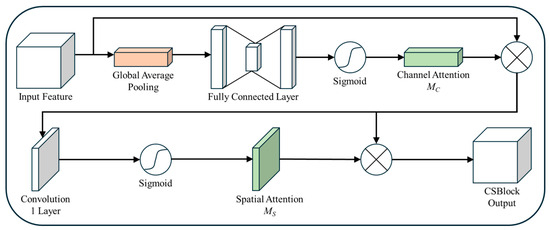
Figure 12.
Integrated architecture of the CSBlock with channel and spatial attention.
This structural modification facilitates a more accurate extraction of spatial features and provides comprehensible explanations. This strengthened the proposed spatial attention structure and enabled efficient feature extraction.
Equation (9) is the multiplication between the input feature map and the spatial attention feature map to generate the latter. Here, represents a convolution operation that results in a single-channel output, and represents the input feature map. is used to calculate the interpixel correlation within the input feature map. The previously performed channel attention ensures that the emphasized feature map with enhanced high-frequency components is preserved, while suppressing low-frequency or unnecessary features. From this emphasized feature map, Equation (10) calculates the interpixel correlation through spatial attention on regions with significant differences in neighboring pixel values. Equation (10) emphasizes the critical parts of the input feature map and extracts features by considering the correlation between pixels. This further enhances the emphasis on the spatial features, and the spatial attention structure enables effective feature extraction.
Equation (10) represents the equation for the CSBlock, which incorporates channel and spatial attention. Here, SA represents the spatial attention process from Equation (9), and CA denotes the channel attention obtained from Equation (8). Additionally, a residual scale is used to ensure stable learning. The CSBlock is the result of sequentially applying channel and spatial attention to the input feature map, denoted as . The CSBlock refines the essential parts and spatial features of the input feature map through the SA and CA processes, and the resulting feature map is multiplied by the residual scale. This design addresses the instability and performance degradation that can occur in deep networks. The gradient values were constrained using a residual scale, allowing for stable learning. Equation (10) is the key formula for the CSBlock, which combines channel and spatial attention. It captures the important features of the input feature map and implements strategies for stable learning to address performance degradation issues effectively.
3.3. Upsampling
The conventional approach to image super-resolution typically involves deconvolution to expand the feature map. However, this method often introduces checkerboard artifacts, which appear as grid-like patterns in reconstructed images. These artifacts are primarily caused by an uneven overlap of convolutional kernels during the deconvolution process. Such artifacts can significantly degrade image quality, particularly in tasks where high precision is required, such as medical imaging. The impact of checkerboard artifacts on super-resolution tasks has been extensively discussed in the literature [32].
To address this issue, the proposed CS-SISR model utilizes a subpixel convolution technique. Subpixel convolution rearranges the feature map to expand its resolution without introducing artifacts, effectively eliminating checkerboard patterns. This approach minimizes redundant operations and provides an efficient method for feature map expansion while maintaining high image quality. Super-resolution involves restoring a high-resolution image from its low-resolution counterpart. Traditional methods rely on convolution filters or multiple convolution operations to extract features. However, these methods often face challenges, such as increased network complexity and a high number of parameters. Subpixel convolution resolves these issues by rearranging multiple feature maps to generate an expanded feature map, which increases the redundancy of low-resolution patches. This process ensures efficient feature extraction and expansion while preserving image quality. In the proposed CS-SISR model, subpixel convolution is used to expand feature maps and generate RGB super-resolution images. A combination of residual blocks and three-channel convolutions is employed to enhance high-frequency details, further improving the quality of the reconstructed images.
Figure 13 presents images of the expanded feature map at each step of the feature map expansion process.
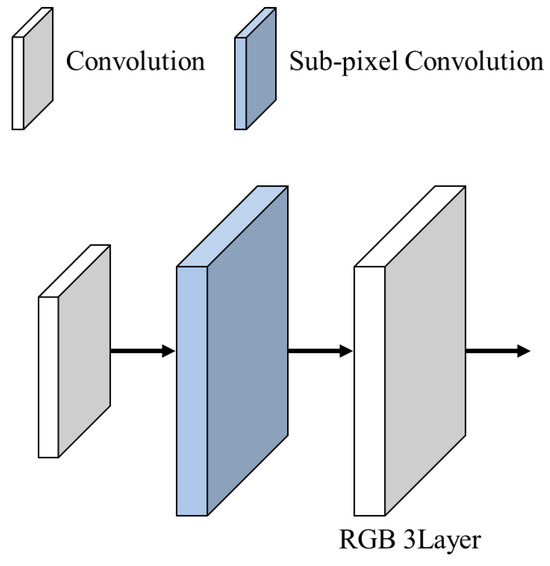
Figure 13.
Upsampling.
Equation (11) represents the formula for the feature map expansion. denotes the expanded feature map, UP represents the subpixel convolution, represents the feature map with a depth of 3, and represents the feature map with a depth of 10. represents the convolution, and represents the low-resolution image. Using , and are obtained by performing convolution operations using as the input. The subpixel convolution generates the expanded feature map, , using the low-resolution image, , to expand the feature map. The use of subpixel convolution varies according to the scale factor to minimize the parameters, loss during expansion, and computational complexity. For 2× and 3× expansion, a subpixel convolution is used to expand the feature map. For a 4× expansion, a subpixel convolution is applied twice to expand the feature map. Adjusting the subpixel convolution according to different scale factors allows for the efficient execution of the expansion. This method optimizes the parameter usage, reduces the computational complexity, and maintains the quality of the expanded feature map.
4. Experimental Results
4.1. Training Configurations and Evaluation Metrics
The L1 loss function used in this study is defined as follows:
Here, L1(x, f(x′)) represents the pixel-wise absolute difference between the ground truth image (x) and the reconstructed super-resolved image f(x′). This loff function is computed as:
where N is the total number of pixels in the image, xi represents the pixel value in the ground truth image, and f(x′)i is the corresponding pixel value in the reconstructed image. The L1 loss minimizes the absolute error, preserving texture details and mitigating over-smoothing effects often seen with L2 loss.
The Adam optimizer [33] was used for training, with a learning rate of 0.001, β1 of 0.9, and β2 of 0.999. The input data were divided into 48 × 48 sized patches, and the resulting images were trained according to the ratio. In this paper, we used the ISIC (2020) dataset to evaluate the proposed method, comparing it with bicubic interpolation, VDSR, and EDSR. For the training data configuration, we downsampled the images to one-half, one-third, and one-quarter of their original sizes. The ISIC consists of high-resolution images with resolutions of 1000 × 1000 or higher. We used the original images and images downscaled using bicubic interpolation as training data. To provide further clarity on the experimental environment and ensure reproducibility, the hardware and software configurations used in this study are summarized in Table 2.

Table 2.
Hardware and software configuration.
The training parameters, including patch size, upscaling factors, and initialization settings, are detailed in Table 3. These configurations were optimized to balance model performance and computational efficiency.

Table 3.
Training parameters.
The training time varied depending on the dataset size. For the ISIC 2020 dataset, training required approximately 3–4 days to complete on the specified hardware. For smaller datasets such as DIV2K, training was significantly faster, taking less than a day. Models for each upscaling factor (2×, 3×, and 4×) were trained separately to optimize performance for each scale.
The peak signal-to-noise ratio (PSNR) measures the quality loss of an image by comparing the noise ratio to the maximum image value. A higher PSNR value indicates less loss compared with the original image [34]. Similarly, the structural similarity index (SSIM) evaluates human perceptual quality by assessing the structural integrity of the image.
Here, refers to the maximum value of the image, and (mean squared error) represents the squared difference between the original image and the reconstructed image.
The structural similarity index measure (SSIM) is a metric that evaluates human perception. The human vision specializes in extracting structural information, which impacts image quality considerably, from an image. SSIM utilizes luminance, contrast, and structure to measure similarity; a higher SSIM value indicates less loss.
Here, represents luminance, represents contrast, and represents structural.
Figure 14 presents the super-resolution results for 3371618 images from the ISIC 2020 challenge dataset. The images within the red boxes are the downscaled versions of the original images, which were then 2× upscaled using the super-resolution algorithm. The images within the blue boxes represent the 3× upscaled versions, and the images within the green boxes represent the 4× upscaled versions. Bicubic interpolation restores the values of the surrounding 16 pixels using a 2D equation, resulting in a smoother image but lacking high-frequency information, leading to blurry results in regions with similar patterns, such as dark lesions. VDSR, which uses 20 convolution layers and residual learning, improves the performance of edge preservation compared to bicubic interpolation. EDSR, with its deeper architecture comprising 69 convolution layers, produces sharper results than VDSR. The proposed CSBlock and attention-block methods handled high-frequency information better than existing networks. However, the numerical values were similar to those of EDSR. Nonetheless, the EDSR structure, which mainly consists of convolution layers, lacks emphasis on specific high-frequency features. In contrast, our proposed method optimizes the network, allowing for improved performance in specific high-frequency regions and faster learning.
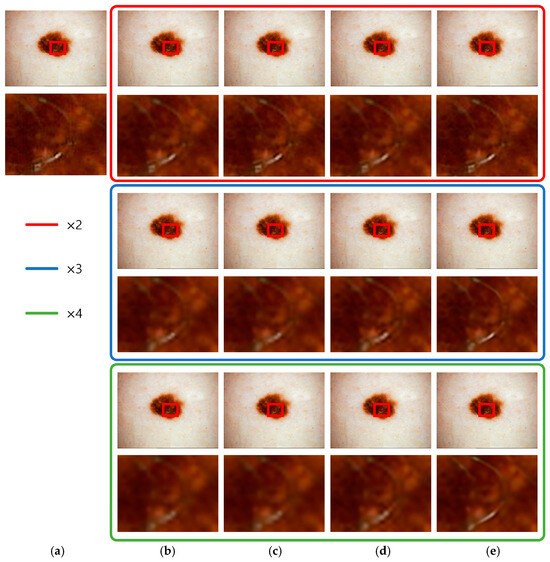
Figure 14.
ISIC_3371618 result: (a) original image, (b) bicubic interpolation image, (c) VDSR, (d) EDSR, and (e) the proposed method.
Table 4 displays the PSNR and SSIM results for the ISIC dataset of 3371618 images. Owing to the complex and similarly colored patterns, our proposed method demonstrates improved results with higher values and more high-frequency components than the existing algorithms.

Table 4.
ISIC_3371618 PSNR/SSIM for scale factors ×2, ×3, and ×4.
Figure 15 presents the super-resolution results for 4382016 images from the ISIC 2020 challenge dataset. In cases with straight patterns, such as fur, bicubic interpolation results in staircase artifacts and blurry edges. VDSR produces improved results compared to bicubic interpolation but still exhibits blurriness. EDSR enhances the sharpness of the fur area but introduces staircase artifacts and unnatural regions. The proposed method demonstrates improved results in addressing these issues.
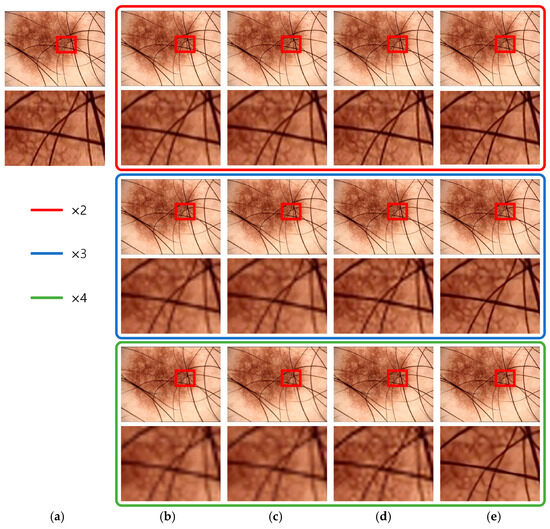
Figure 15.
ISIC_4382016 results: (a) original image, (b) bicubic interpolation image, (c) VDSR, (d) EDSR, and (e) the proposed method.
Table 5 displays the PSNR and SSIM results for ISIC_4382016. The results are improved compared to existing methods in addressing straight patterns, like fur and skin textures.

Table 5.
ISIC_4382016 PSNR/SSIM for scale factors ×2, ×3, and ×4.
Table 6 presents the PSNR and SSIM results for the ISIC dataset. Owing to the characteristics of the ISIC dataset, which consists mainly of skin and low-frequency regions, it often exhibits irregularities with high-frequency patterns limited to certain areas. The proposed method exhibits superior performance compared to the existing EDSR and VDSR methods by utilizing high-frequency components and diverse features.

Table 6.
Average PSNR/SSIM for scale factors ×2, ×3, and ×4 (the boldened values indicate the best performance).
4.2. Convergence Analysis
To provide an insight into the training behavior of the proposed CS-SISR model, we present convergence graphs illustrating the training loss and validation PSNR across epochs. These graphs demonstrate the stability and efficiency of the training process, as well as the consistent performance improvement over the course of training.
- Training Loss: As shown in Figure 16, the training loss steadily decreases, reflecting the model’s effective optimization. The use of the L1 loss function contributes to the preservation of fine details while mitigating over-smoothing.
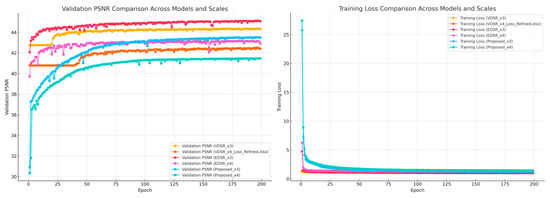 Figure 16. Training loss and validation PSNR comparison across models and scales.
Figure 16. Training loss and validation PSNR comparison across models and scales. - Validation PSNR: Figure 16 shows the validation PSNR values improving consistently across epochs, highlighting the model’s ability to generalize effectively to unseen data. This result further validates the robustness of the proposed approach.
By comparing these metrics with other baseline models, it is evident that the CS-SISR model achieves superior convergence properties, balancing computational efficiency and high performance.
4.3. Diagnostic Accuracy Assessment
This study proposes a CNN-based super-resolution technique incorporating an attention mechanism to enhance the resolution and quality of medical images. However, evaluating the impact of super-resolution techniques on diagnostic accuracy in real clinical settings is essential. In this section, we discuss diagnostic accuracy based on previous research findings.
4.4. Diagnostic Accuracy in Related Studies
Deep learning-based super-resolution techniques have been shown to be effective in diagnosing diseases such as melanoma, lung cancer, and breast cancer. For example, the study by Fayaz Ali Dharejo et al. (multimodal-boost) demonstrated that integrating multi-attention mechanisms with wavelet transform significantly improved image quality across various medical imaging modalities. This enhancement contributed to the restoration of fine details, such as lesion texture and boundary features, thereby increasing diagnostic accuracy in experimental settings [26]. Similarly, Mariana-Iuliana Georgescu et al. employed a multi-head convolutional attention mechanism to improve the super-resolution performance of CT and MRI images. By utilizing multiple kernel sizes, they effectively captured spatial and channel-wise attention, leading to significant improvements in PSNR and SSIM metrics compared to conventional methods. Their study emphasized how restored high-frequency textures in lesion areas could directly enhance diagnostic accuracy [27].
4.5. Relevance to This Study
The proposed CS-SISR (convolutional self-attention block-based single image super-resolution) method aligns with similar principles, emphasizing high-frequency information to enhance medical image quality. Experimental results using the ISIC 2020 dataset demonstrated a 1–2% improvement in PSNR and SSIM metrics compared to conventional super-resolution methods such as VDSR and EDSR. This indicates that the proposed method effectively restores fine details in melanoma images, validating its efficacy for clinical applications.
4.6. Clinical Applicability
The findings of this study further highlight the potential of super-resolution techniques to improve diagnostic accuracy. For instance, the proposed CS-SISR method enhances boundary clarity and texture restoration, playing a critical role in melanoma diagnosis. Preliminary evaluations by medical experts suggest that images generated using the proposed method provide more defined lesion boundaries than existing approaches, reducing diagnostic ambiguity.
However, the impact of super-resolution techniques on diagnostic accuracy can vary depending on the diversity of datasets and clinical contexts. Therefore, additional validation is required to ensure that the proposed method maintains its effectiveness across different medical imaging modalities and patient populations. Future research should focus on verifying the generalizability of the proposed model in diverse clinical scenarios.
5. Discussion
In this study, we proposed a novel method for enhancing the resolution and quality of medical images using a convolutional self-attention block (CSBlock). The experimental results demonstrated that the proposed method significantly improves PSNR and SSIM metrics compared to conventional approaches, such as VDSR and EDSR. This indicates that our model effectively preserves fine details, such as texture and contours, which are critical for medical image analysis. The structural innovations introduced in the proposed method play a key role in achieving these improvements:
- Dynamic Attention Mechanisms: The CSBlock effectively combines channel and spatial attention to prioritize diagnostically relevant features. This allows the model to emphasize high-frequency details, such as lesion boundaries, while suppressing irrelevant noise.
- Subpixel Convolution for Upsampling: Unlike deconvolution-based methods, the use of subpixel convolution reduces artifacts and improves the clarity of restored images, particularly in high-frequency regions.
- Stable Training with L1 Loss: The use of L1 loss mitigates over-smoothing effects, resulting in sharper and more realistic outputs compared to VDSR and EDSR.
From a clinical perspective, enhanced image resolution can significantly improve the detection of subtle lesions and structural abnormalities. For example, sharper lesion boundaries and improved texture clarity assist dermatologists in distinguishing malignant melanomas from benign lesions. Preliminary qualitative feedback from medical experts has highlighted the diagnostic value of these improvements, though further blinded evaluations are required to validate these findings quantitatively.
Moreover, we performed a comparative analysis of PSNR and SSIM values across multiple subsets of the ISIC 2020 dataset to assess the statistical significance of the observed improvements. Repeated measurements revealed consistent gains (within a confidence interval of 95%), suggesting that the proposed approach reliably outperforms baseline methods. However, further statistical tests on larger and more diverse datasets are needed to confirm the robustness of our model. Future work could incorporate advanced statistical methods (e.g., ANOVA or mixed-effects modeling) to better account for inter-sample variability and further validate the clinical relevance of our findings.
6. Conclusions and Future Directions
This paper presented a novel deep learning-based super-resolution method that leverages a convolutional self-attention block (CSBlock) to emphasize high-frequency features in medical images. Experimental results showed significant improvements in PSNR and SSIM metrics, with an increase of approximately 1–2% on the ISIC 2020 dataset compared to existing methods such as VDSR and EDSR. The structural innovations in the proposed method, including dynamic attention mechanisms and subpixel convolution layers, effectively preserve fine details like texture and contours. These findings underscore the potential of the proposed method to enhance diagnostic accuracy in melanoma detection and other medical imaging applications.
Despite these promising results, several limitations must be addressed. First, the reliance on a single dataset (ISIC 2020) may restrict the generalizability of our findings to other medical imaging modalities and diverse patient populations. Second, the computational complexity of the model could hinder its real-time clinical deployment, especially in resource-constrained environments.
To overcome these limitations and to further advance the field, we suggest the following directions:
- Dataset Diversity: Incorporating additional datasets, such as PH2 and HAM10000, which include diverse lesion types and higher variability in image resolution, to validate the model’s generalizability across diverse patient populations.
- Model Simplification and Scalability: Developing lightweight architectures, such as MobileNet-based designs, to reduce the computational overhead while maintaining high performance. This approach aims to enable deployment on portable diagnostic devices and in telemedicine applications.
- Expanding Evaluation Metrics: Including diagnostic metrics, such as sensitivity, specificity, and AUC, along with qualitative assessments by medical experts. These evaluations will provide a more comprehensive understanding of the model’s clinical relevance.
By implementing these improvements, the proposed super-resolution technique can evolve to provide even greater accuracy and efficiency in melanoma detection and beyond. Ultimately, these advancements will contribute to improved patient outcomes and more effective diagnostic workflows in medical imaging.
Author Contributions
Conceptualization, S.Y.C.; methodology, S.Y.C.; software, D.Y.L.; validation, S.Y.C., J.Y.K. and D.Y.L.; formal analysis, J.Y.K.; investigation, S.Y.C.; resources, S.Y.C.; data curation, D.Y.L.; writing—original draft preparation, S.Y.C.; writing—review and editing, S.Y.C., J.Y.K. and D.Y.L.; visualization, J.Y.K. and D.Y.L.; supervision, S.Y.C.; project administration, S.Y.C.; funding acquisition, Kwangwoon University. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Kwangwoon University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are publicly available and can be accessed through the ISIC Archive and the ISIC 2020 Challenge repositories. Further details regarding the dataset are provided in Section 1.2 of the article.
Acknowledgments
The present Research has been conducted by the Research Grant of Kwangwoon University in 2022.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, N.K.; Raza, K. Progress in deep learning-based dental and maxillofacial image analysis: A systematic review. Expert Syst. Appl. 2022, 199, 116968. [Google Scholar] [CrossRef]
- Zhao, M.; Naderian, A.; Sanei, S. Generative Adversarial Networks for Medical Image Super-resolution. In Proceedings of the 2021 International Conference on e-Health and Bioengineering (EHB), Iasi, Romania, 18–19 November 2021; pp. 1–4. [Google Scholar]
- Park, J.; Hwang, D.; Kim, K.Y.; Kang, S.K.; Kim, Y.K.; Lee, J.S. Computed tomography super-resolution using deep convolutional neural network. Phys. Med. Biol. 2018, 63, 145011. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sixou, B.; Peyrin, F. A review of the deep learning methods for medical images super resolution problems. IRBM 2021, 42, 120–133. [Google Scholar] [CrossRef]
- Rajeshwari, P.; Shyamala, K. Pixel attention based deep neural network for chest CT image super resolution. In Advanced Network Technologies and Intelligent Computing, Proceedings of the Second International Conference, ANTIC 2022, Varanasi, India, 22–24 December 2022; Proceedings, Part II; Springer Nature: Cham, Switzerland, 2023; pp. 393–407. [Google Scholar]
- Yang, W.; Zhang, X.; Tian, Y.; Wang, W.; Xue, J.H.; Liao, Q. Deep learning for single image super-resolution: A brief review. IEEE Trans. Multimed. 2019, 21, 3106–3121. [Google Scholar] [CrossRef]
- Jo, Y.; Oh, S.W.; Vajda, P.; Kim, S.J. Tackling the ill-posedness of super-resolution through adaptive target generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2021, Nashville, TN, USA, 20–25 June 2021; pp. 16236–16245. [Google Scholar]
- Liu, W.; Zhang, B.; Liu, T.; Jiang, J.; Liu, Y. Artificial Intelligence in Pancreatic Image Analysis: A Review. Sensors 2024, 24, 4749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liang, G.; Pan, S.; Zheng, L. A fast medical image super resolution method based on deep learning network. IEEE Access 2018, 7, 12319–12327. [Google Scholar] [CrossRef]
- Bai, Y.; Zhuang, H. On the comparison of bilinear, cubic spline, and fuzzy interpolation techniques for robotic position measurements. IEEE Trans. Instrum. Meas. 2005, 54, 2281–2288. [Google Scholar] [CrossRef]
- Dong, C.; Loy, C.C.; Tang, X. Accelerating the super-resolution convolutional neural network. In Computer Vision—ECCV 2016, Proceeding of the 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; Proceedings, Part II; Springer: Cham, Switzerland, 2016; pp. 391–407. [Google Scholar]
- Kim, J.; Lee, J.K.; Lee, K.M. Accurate image super-resolution using very deep convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 1646–1654. [Google Scholar]
- Lim, B.; Son, S.; Kim, H.; Nah, S.; Mu Lee, K. Enhanced deep residual networks for single image super-resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 136–144. [Google Scholar]
- Keys, R. Cubic convolution interpolation for digital image processing. IEEE Trans. Acoust. Speech Signal Process. 1981, 29, 1153–1160. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV) 2018, Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision 2017, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2018, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. In Proceedings of the Advances in Neural Information Processing Systems 27, Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- Kitaev, N.; Kaiser, Ł.; Levskaya, A. Reformer: The efficient transformer. arXiv 2020, arXiv:2001.04451. [Google Scholar]
- Wang, S.; Li, B.Z.; Khabsa, M.; Fang, H.; Ma, H. Linformer: Self-attention with linear complexity. arXiv 2020, arXiv:2006.04768. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision 2021, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems 30, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Dharejo, F.A.; Zawish, M.; Deeba, F.; Zhou, Y.; Dev, K.; Khowaja, S.A.; Qureshi, N.M.F. Multimodal-boost: Multimodal medical image super-resolution using multi-attention network with wavelet transform. IEEE ACM Trans. Comput. Biol. Bioinform. 2022, 20, 2420–2433. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, M.I.; Ionescu, R.T.; Miron, A.I.; Savencu, O.; Ristea, N.C.; Verga, N.; Khan, F.S. Multimodal multi-head convolutional attention with various kernel sizes for medical image super-resolution. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision 2023, Waikoloa, HI, USA, 2–7 January 2023; pp. 2195–2205. [Google Scholar]
- Yu, Y.; She, K.; Liu, J.; Cai, X.; Shi, K.; Kwon, O.M. A super-resolution network for medical imaging via transformation analysis of wavelet multi-resolution. Neural Netw. 2023, 166, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; She, K.; Shi, K.; Cai, X.; Kwon, O.M.; Soh, Y. Analysis of medical images super-resolution via a wavelet pyramid recursive neural network constrained by wavelet energy entropy. Neural Netw. 2024, 178, 106460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, K.; Li, K.; Wang, L.; Zhong, B.; Fu, Y. Image super-resolution using very deep residual channel attention networks. In Proceedings of the European Conference on Computer Vision (ECCV) 2018, Munich, Germany, 8–14 September 2018; pp. 286–301. [Google Scholar]
- Liang, J.; Cao, J.; Sun, G.; Zhang, K.; Van Gool, L.; Timofte, R. Swinir: Image restoration using swin transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision 2021, Montreal, BC, Canada, 11–17 October 2021; pp. 1833–1844. [Google Scholar]
- Odena, A.; Dumoulin, V.; Olah, C. Deconvolution and checkerboard artifacts. Distill 2016, 1, e3. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).