Classification of Blackcurrant Genotypes by Ploidy Levels on Stomata Microscopic Images with Deep Learning: Convolutional Neural Networks and Vision Transformers
Abstract
1. Introduction
- Computerized classification of ploidy levels based on microscopic images of stomata, whereas other researchers focus on detection, segmentation and measuring stomata. To the best of our knowledge, no other research group has yet applied artificial intelligence to ploidy-level classification.
- The approach of training a model on one set and testing it on a second, different dataset for ploidy-level classification, as opposed to our previous research [14], where both training and testing was performed on subsets of a single dataset.
- Application of Vision Transformers to stomata microscopic images, as other works do not apply ViTs to any stomata-related issues.
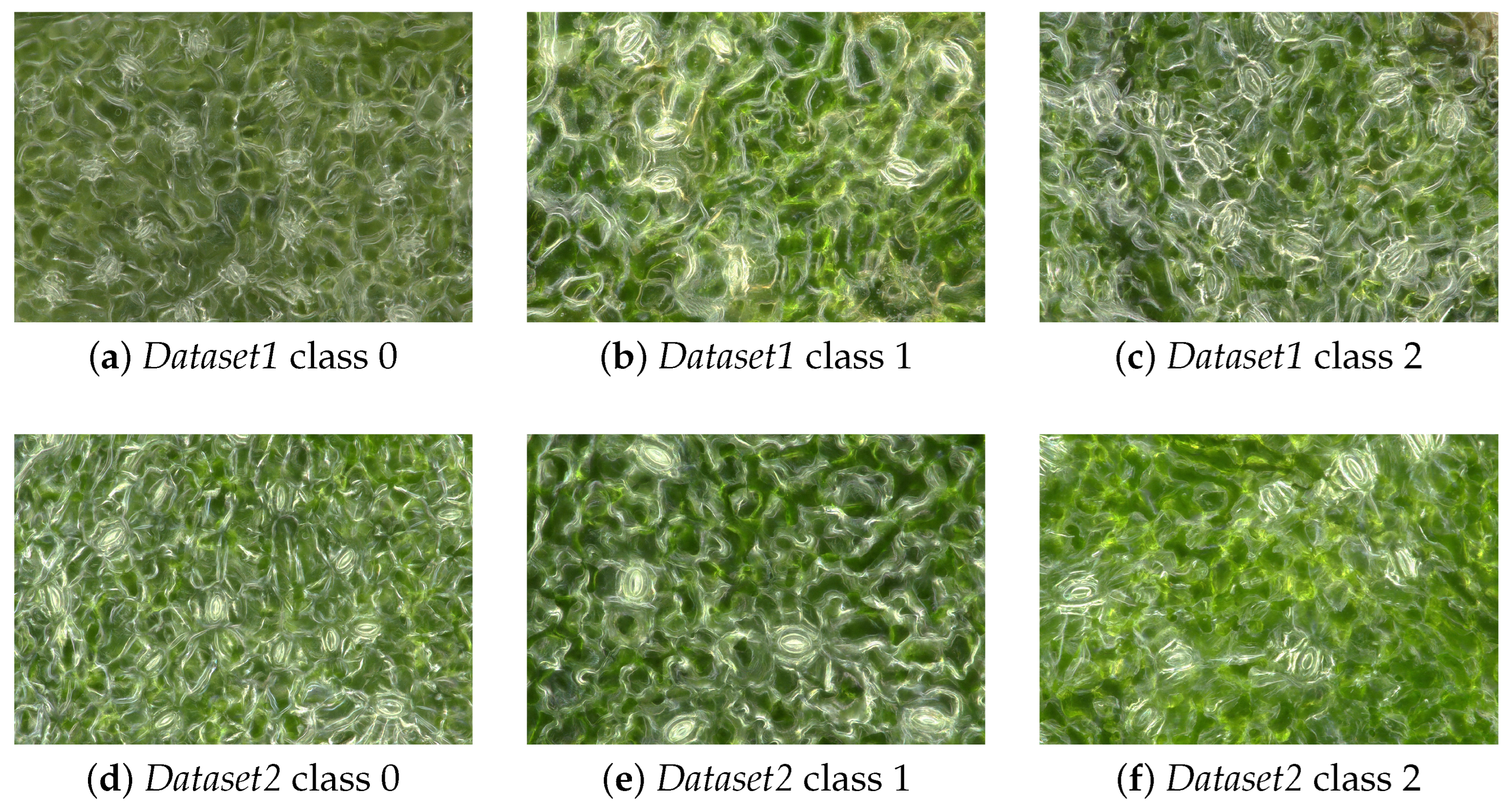
2. Datasets
2.1. Dataset1
2.2. Dataset2
3. Methods
3.1. Convolutional Neural Networks
3.2. Transformers
4. Experiments and Results
4.1. ResNet Experimental Setup
4.2. ViT Experimental Setup
4.3. Model Evaluation Metrics
4.4. Experiments on Raw Datasets
4.4.1. Experiments on Two Classes
4.4.2. Experiments on Augmented Datasets
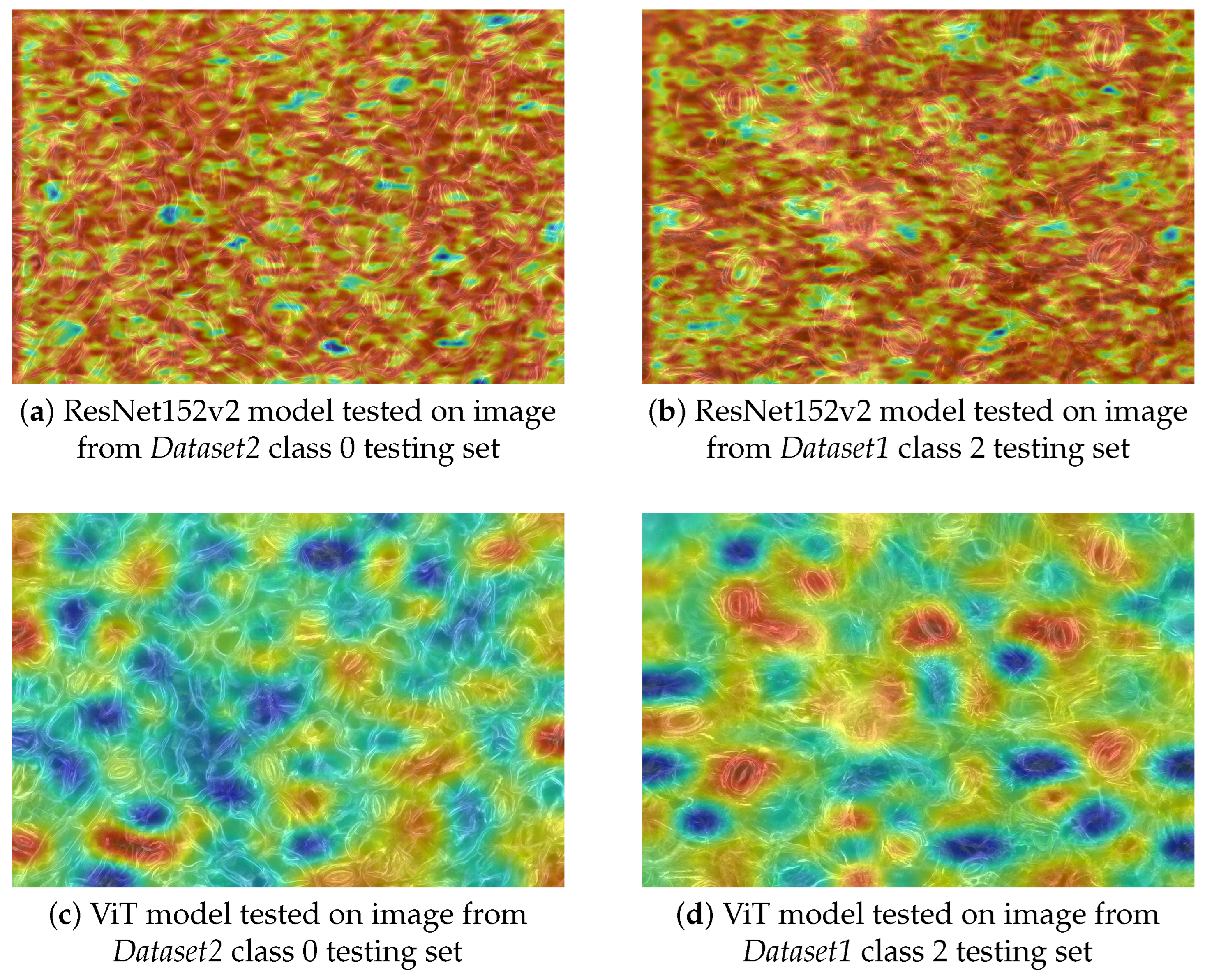
4.4.3. Gradient-Weighted Class Activation Mapping
5. Conclusions
6. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casson, S.A.; Hetherington, A.M. Environmental regulation of stomatal development. Curr. Opin. Plant Biol. 2010, 13, 90–95. [Google Scholar] [CrossRef]
- Mtileni, M.; Venter, N.; Glennon, K. Ploidy differences affect leaf functional traits, but not water stress responses in a mountain endemic plant population. S. Afr. J. Bot. 2021, 138, 76–83. [Google Scholar] [CrossRef]
- Van Laere, K.; França, S.C.; Vansteenkiste, H.; Van Huylenbroeck, J.; Steppe, K.; Van Labeke, M.C. Influence of ploidy level on morphology, growth and drought susceptibility in Spathiphyllum wallisii. Acta Physiol. Plant. 2010, 33, 1149–1156. [Google Scholar] [CrossRef]
- Sai, N.; Bockman, J.P.; Chen, H.; Watson-Haigh, N.; Xu, B.; Feng, X.; Piechatzek, A.; Shen, C.; Gilliham, M. StomaAI: An efficient and user-friendly tool for measurement of stomatal pores and density using deep computer vision. New Phytol. 2023, 238, 904–915. [Google Scholar] [CrossRef]
- Wu, T.L.; Chen, P.Y.; Du, X.; Wu, H.; Ou, J.Y.; Zheng, P.X.; Wu, Y.L.; Wang, R.S.; Hsu, T.C.; Lin, C.Y.; et al. StomaVision: Stomatal trait analysis through deep learning. bioRxiv 2024. [Google Scholar] [CrossRef]
- Fetter, K.C.; Eberhardt, S.; Barclay, R.S.; Wing, S.; Keller, S.R. StomataCounter: A neural network for automatic stomata identification and counting. New Phytol. 2019, 223, 1671–1681. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Jayakody, H.; Liu, S.; Whitty, M.; Petrie, P. Microscope image based fully automated stomata detection and pore measurement method for grapevines. Plant Methods 2017, 13, 94. [Google Scholar] [CrossRef]
- Andayani, U.; Sumantri, I.B.; Pahala, A.; Muchtar, M.A. The implementation of deep learning using Convolutional Neural Network to classify based on stomata microscopic image of curcuma herbal plants. IOP Conf. Ser. Mater. Sci. Eng. 2020, 851, 012035. [Google Scholar] [CrossRef]
- Casado-García, A.; del Canto, A.; Sanz-Saez, A.; Pérez-López, U.; Bilbao-Kareaga, A.; Fritschi, F.B.; Miranda-Apodaca, J.; Muñoz-Rueda, A.; Sillero-Martínez, A.; Yoldi-Achalandabaso, A.; et al. LabelStoma: A tool for stomata detection based on the YOLO algorithm. Comput. Electron. Agric. 2020, 178, 105751. [Google Scholar] [CrossRef]
- Sun, Z.; Song, Y.; Li, Q.; Cai, J.; Wang, X.; Zhou, Q.; Huang, M.; Jiang, D. An integrated method for tracking and monitoring stomata dynamics from microscope videos. Plant Phenomics 2021, 2021, 9835961. [Google Scholar] [CrossRef]
- Wacker, T.S.; Smith, A.G.; Jensen, S.M.; Pflüger, T.; Hertz, V.G.; Rosenqvist, E.; Liu, F.; Dresbøll, D.B. Stomata morphology measurement with interactive machine learning: Accuracy, speed, and biological relevance? Plant Methods 2025, 21, 95. [Google Scholar] [CrossRef]
- Razzaq, A.; Shahid, S.; Akram, M.; Ashraf, M.; Iqbal, S.; Hussain, A.; Azam Zia, M.; Qadri, S.; Saher, N.; Shahzad, F.; et al. Stomatal state identification and classification in quinoa microscopic imprints through deep learning. Complexity 2021, 2021, 9938013. [Google Scholar] [CrossRef]
- Konopka, A.; Struniawski, K.; Kozera, R.; Ortenzi, L.; Marasek-Ciołakowska, A.; Machlańska, A. Deep Learning Classification of Blackcurrant Genotypes by Ploidy Levels on Stomata Microscopic Images. In Computational Science—ICCS 2025 Workshops; Springer Nature: Cham, Switzerland, 2025; pp. 135–148. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. arXiv 2017, arXiv:1706.03762. [Google Scholar] [CrossRef]
- Li, X.; Guo, S.; Gong, L.; Lan, Y. An automatic plant leaf stoma detection method based on YOLOv5. IET Image Process. 2022, 17, 67–76. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Podwyszyńska, M.; Pluta, S. In vitro tetraploid induction of the blackcurrant (Ribes nigrum L.) and preliminary phenotypic observations. Zemdirb. Agric. 2019, 106, 151–158. [Google Scholar] [CrossRef]
- Haykin, S. Neural Networks and Learning Machines, 3rd ed.; Pearson: Bloomington, MN, USA, 2008. [Google Scholar]
- Rosenblatt, F. The Perceptron—A Perceiving and Recognizing Automaton; Report 85-460-1; Cornell Aeronautical Laboratory: Buffalo, NY, USA, 1957. [Google Scholar]
- Konopka, A.; Struniawski, K.; Kozera, R. Performance analysis of Residual Neural Networks in soil bacteria microscopic image classification. In Proceedings of the 37th Annual European Simulation and Modelling Conference, ESM 2023, Toulouse, France, 24–26 October 2023; pp. 144–148. [Google Scholar]
- Slavutskii, L.; Lazareva, N.; Portnov, M.; Slavutskaya, E. Neural net without deep learning: Signal approximation by multilayer perceptron. In Proceedings of the 2nd International Conference on Computer Applications for Management and Sustainable Development of Production and Industry (CMSD-II-2022), SPIE, Dushanbe, Tajikistan, 21–23 December 2022; p. 11. [Google Scholar] [CrossRef]
- Wang, L.; Meng, Z. Multichannel two-dimensional Convolutional Neural Network based on interactive features and group strategy for chinese sentiment analysis. Sensors 2022, 22, 714. [Google Scholar] [CrossRef]
- Konopka, A.; Kozera, R.; Sas-Paszt, L.; Trzcinski, P.; Lisek, A. Identification of the selected soil bacteria genera based on their geometric and dispersion features. PLoS ONE 2023, 18, e0293362. [Google Scholar] [CrossRef]
- Konopka, A.; Kozera, R.; Sas-Paszt, L.; Trzciński, P. Automated imaging and machine learning for soil bacteria classification: Challenges and insights. Eng. Appl. Artif. Intell. 2025, 159, 111369. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Tseng, C.C.; Lee, S.L.; Xiao, B.Y. Image classification using Convolutional Neural Networks with different convolution operations. In Proceedings of the 2020 IEEE International Conference on Consumer Electronics—Taiwan (ICCE-Taiwan), Taoyuan, Taiwan, 28–30 September 2020; pp. 1–2. [Google Scholar] [CrossRef]
- LeCun, Y.; Jackel, L.D.; Bottou, L.; Cortes, C.; Denker, J.S.; Drucker, H.; Guyon, I.; Muller, U.A.; Sackinger, E.; Simard, P.; et al. Learning algorithms for classification: A comparison on handwritten digit recognition. In Neural Networks; World Scientific: Pohang, Republic of Korea, 1995; pp. 261–276. [Google Scholar] [CrossRef]
- Vang-Mata, R. Multilayer Perceptrons: Theory and Applications; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2020. [Google Scholar]
- Zhao, L.; Zhang, Z. A improved pooling method for Convolutional Neural Networks. Sci. Rep. 2024, 14, 1589. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Borawar, L.; Kaur, R. ResNet: Solving Vanishing Gradient in Deep Networks. In Proceedings of International Conference on Recent Trends in Computing; Springer Nature: Singapore, 2023; pp. 235–247. [Google Scholar] [CrossRef]
- Philipp, G.; Song, D.; Carbonell, J.G. Gradients explode—Deep networks are shallow—ResNet explained. In Proceedings of the 6th International Conference on Learning Representations, ICLR 2018—Workshop Track Proceedings, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in Deep Residual Networks. arXiv 2016, arXiv:1603.05027. [Google Scholar] [CrossRef]
- Varshney, M.; Singh, P. Optimizing nonlinear activation function for convolutional neural networks. Signal Image Video Process 2021, 15, 1323–1330. [Google Scholar] [CrossRef]
- Azar, G.A.; Emami, M.; Fletcher, A.; Rangan, S. Learning embedding representations in high dimensions. In Proceedings of the 2024 58th Annual Conference on Information Sciences and Systems (CISS), Princeton, NJ, USA, 13–15 March 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Zheng, C.; Gao, Y.; Shi, H.; Huang, M.; Li, J.; Xiong, J.; Ren, X.; Ng, M.; Jiang, X.; Li, Z.; et al. DAPE: Data-Adaptive Positional Encoding for length extrapolation. Adv. Neural Inf. Process. Syst. 2024, 37, 26659–26700. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Shah, S.M.A.H.; Khan, M.Q.; Ghadi, Y.Y.; Jan, S.U.; Mzoughi, O.; Hamdi, M. A hybrid neuro-fuzzy approach for heterogeneous patch encoding in ViTs using contrastive embeddings and deep knowledge dispersion. IEEE Access 2023, 11, 83171–83186. [Google Scholar] [CrossRef]
- Keras. 2015. Available online: https://keras.io (accessed on 30 September 2025).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020; pp. 38–45. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems 32; Curran Associates, Inc.: Red Hook, NY, USA, 2019; pp. 8024–8035. [Google Scholar]
- Clark, A. Pillow (PIL Fork) Documentation. 2015. Available online: https://buildmedia.readthedocs.org/media/pdf/pillow/latest/pillow.pdf (accessed on 30 September 2025).
- Marcel, S.; Rodriguez, Y. Torchvision the machine-vision package of torch. In Proceedings of the 18th ACM International Conference on Multimedia, Firenze, Italy, 25–29 October 2010; MM ’10. ACM: New York, NY, USA; pp. 1485–1488. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. arXiv 2016, arXiv:1610.02391. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking model scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. arXiv 2022, arXiv:2201.03545. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using shifted windows. arXiv 2021, arXiv:2103.14030. [Google Scholar] [CrossRef]
- Zhu, L.; Liao, B.; Zhang, Q.; Wang, X.; Liu, W.; Wang, X. Vision Mamba: Efficient visual representation learning with bidirectional state space model. arXiv 2024, arXiv:2401.09417. [Google Scholar] [CrossRef]
| Tr | Te | mod | acc | 0_p | 0_r | 0_f1 | 1_p | 1_r | 1_f1 | 2_p | 2_r | 2_f1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | vit-b | 0.57 | 0.83 | 0.72 | 0.77 | 0.82 | 0.12 | 0.21 | 0.44 | 0.88 | 0.59 |
| 2 | 1 | vit-b | 0.52 | 0.49 | 0.99 | 0.66 | 1 | 0 | 0.01 | 0.57 | 0.56 | 0.57 |
| 1 | 2 | vit-l | 0.56 | 0.90 | 0.59 | 0.71 | 0.96 | 0.16 | 0.28 | 0.43 | 0.93 | 0.59 |
| 2 | 1 | vit-l | 0.59 | 0.72 | 0.96 | 0.82 | 1 | 0 | 0.01 | 0.48 | 0.79 | 0.60 |
| 1 | 2 | RN | 0.68 | 0.72 | 0.66 | 0.69 | 0.63 | 0.91 | 0.75 | 0.72 | 0.47 | 0.57 |
| 2 | 1 | RN | 0.54 | 0.91 | 0.64 | 0.75 | 0 | 0 | 0 | 0.42 | 0.97 | 0.59 |
| Tr | Te | mod | acc | 0_p | 0_r | 0_f1 | 2_p | 2_r | 2_f1 |
|---|---|---|---|---|---|---|---|---|---|
| 1_02 | 2_02 | vit-b | 0.81 | 0.88 | 0.71 | 0.78 | 0.76 | 0.90 | 0.82 |
| 2_02 | 1_02 | vit-b | 0.88 | 0.84 | 0.93 | 0.88 | 0.92 | 0.82 | 0.87 |
| 1_02 | 2_02 | vit-l | 0.82 | 0.93 | 0.69 | 0.79 | 0.75 | 0.95 | 0.84 |
| 2_02 | 1_02 | vit-l | 0.82 | 0.74 | 0.99 | 0.85 | 0.98 | 0.66 | 0.79 |
| 1_02 | 2_02 | RN | 0.79 | 0.88 | 0.68 | 0.76 | 0.74 | 0.90 | 0.81 |
| 2_02 | 1_02 | RN | 0.80 | 0.94 | 0.63 | 0.76 | 0.72 | 0.96 | 0.83 |
| Tr | Te | mod | acc | 0_p | 0_r | 0_f1 | 1_p | 1_r | 1_f1 | 2_p | 2_r | 2_f1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1_a | 2 | vit-b | 0.59 | 0.91 | 0.71 | 0.80 | 0.91 | 0.14 | 0.25 | 0.45 | 0.92 | 0.60 |
| 2_a | 1 | vit-b | 0.60 | 0.69 | 0.91 | 0.78 | 1 | 0.07 | 0.13 | 0.52 | 0.83 | 0.64 |
| 1_a_02 | 2_02 | vit-b | 0.73 | 0.93 | 0.49 | 0.64 | - | - | - | 0.65 | 0.97 | 0.78 |
| 2_a_02 | 1_02 | vit-b | 0.80 | 0.72 | 0.97 | 0.83 | - | - | - | 0.95 | 0.63 | 0.76 |
| 1_a | 2 | vit-l | 0.53 | 0.94 | 0.44 | 0.60 | 0.90 | 0.19 | 0.31 | 0.42 | 0.97 | 0.58 |
| 2_a | 1 | vit-l | 0.59 | 0.69 | 0.89 | 0.78 | 1 | 0.08 | 0.14 | 0.50 | 0.82 | 0.62 |
| 1_a_02 | 2_02 | vit-l | 0.78 | 0.91 | 0.62 | 0.73 | - | - | - | 0.71 | 0.94 | 0.81 |
| 2_a_02 | 1_02 | vit-l | 0.88 | 0.85 | 0.91 | 0.88 | - | - | - | 0.90 | 0.84 | 0.87 |
| 1_a | 2 | RN | 0.60 | 0.52 | 0.90 | 0.66 | 0.68 | 0.44 | 0.54 | 0.73 | 0.47 | 0.57 |
| 2_a | 1 | RN | 0.59 | 0.72 | 0.82 | 0.77 | 1 | 0.07 | 0.14 | 0.49 | 0.87 | 0.62 |
| 1_a_02 | 2_02 | RN | 0.78 | 0.91 | 0.63 | 0.74 | - | - | - | 0.72 | 0.94 | 0.81 |
| 2_a_02 | 1_02 | RN | 0.85 | 0.87 | 0.83 | 0.85 | - | - | - | 0.84 | 0.88 | 0.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konopka, A.; Kozera, R.; Marasek-Ciołakowska, A.; Machlańska, A. Classification of Blackcurrant Genotypes by Ploidy Levels on Stomata Microscopic Images with Deep Learning: Convolutional Neural Networks and Vision Transformers. Appl. Sci. 2025, 15, 10735. https://doi.org/10.3390/app151910735
Konopka A, Kozera R, Marasek-Ciołakowska A, Machlańska A. Classification of Blackcurrant Genotypes by Ploidy Levels on Stomata Microscopic Images with Deep Learning: Convolutional Neural Networks and Vision Transformers. Applied Sciences. 2025; 15(19):10735. https://doi.org/10.3390/app151910735
Chicago/Turabian StyleKonopka, Aleksandra, Ryszard Kozera, Agnieszka Marasek-Ciołakowska, and Aleksandra Machlańska. 2025. "Classification of Blackcurrant Genotypes by Ploidy Levels on Stomata Microscopic Images with Deep Learning: Convolutional Neural Networks and Vision Transformers" Applied Sciences 15, no. 19: 10735. https://doi.org/10.3390/app151910735
APA StyleKonopka, A., Kozera, R., Marasek-Ciołakowska, A., & Machlańska, A. (2025). Classification of Blackcurrant Genotypes by Ploidy Levels on Stomata Microscopic Images with Deep Learning: Convolutional Neural Networks and Vision Transformers. Applied Sciences, 15(19), 10735. https://doi.org/10.3390/app151910735





