Lung Opacity Segmentation in Chest CT Images Using Multi-Head and Multi-Channel U-Nets with Partially Supervised Learning
Abstract
1. Introduction
2. Materials and Methods
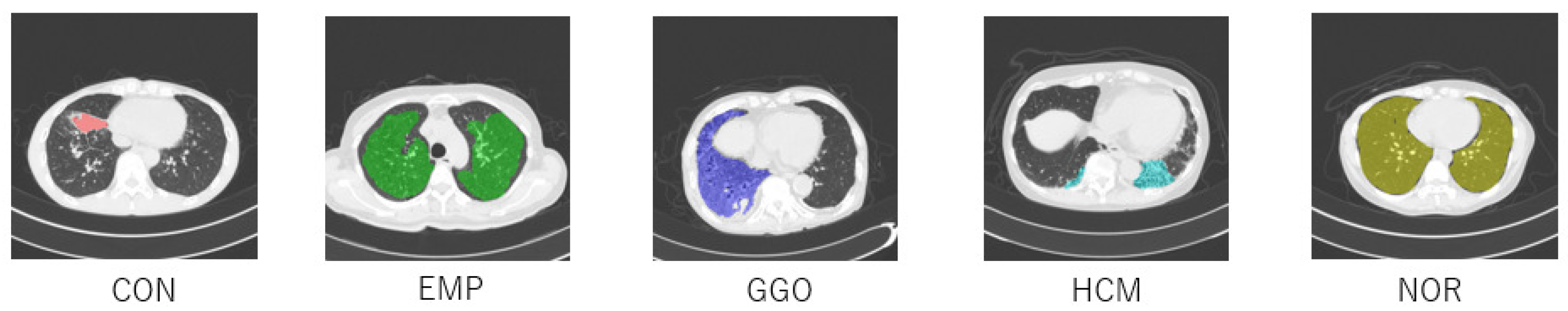
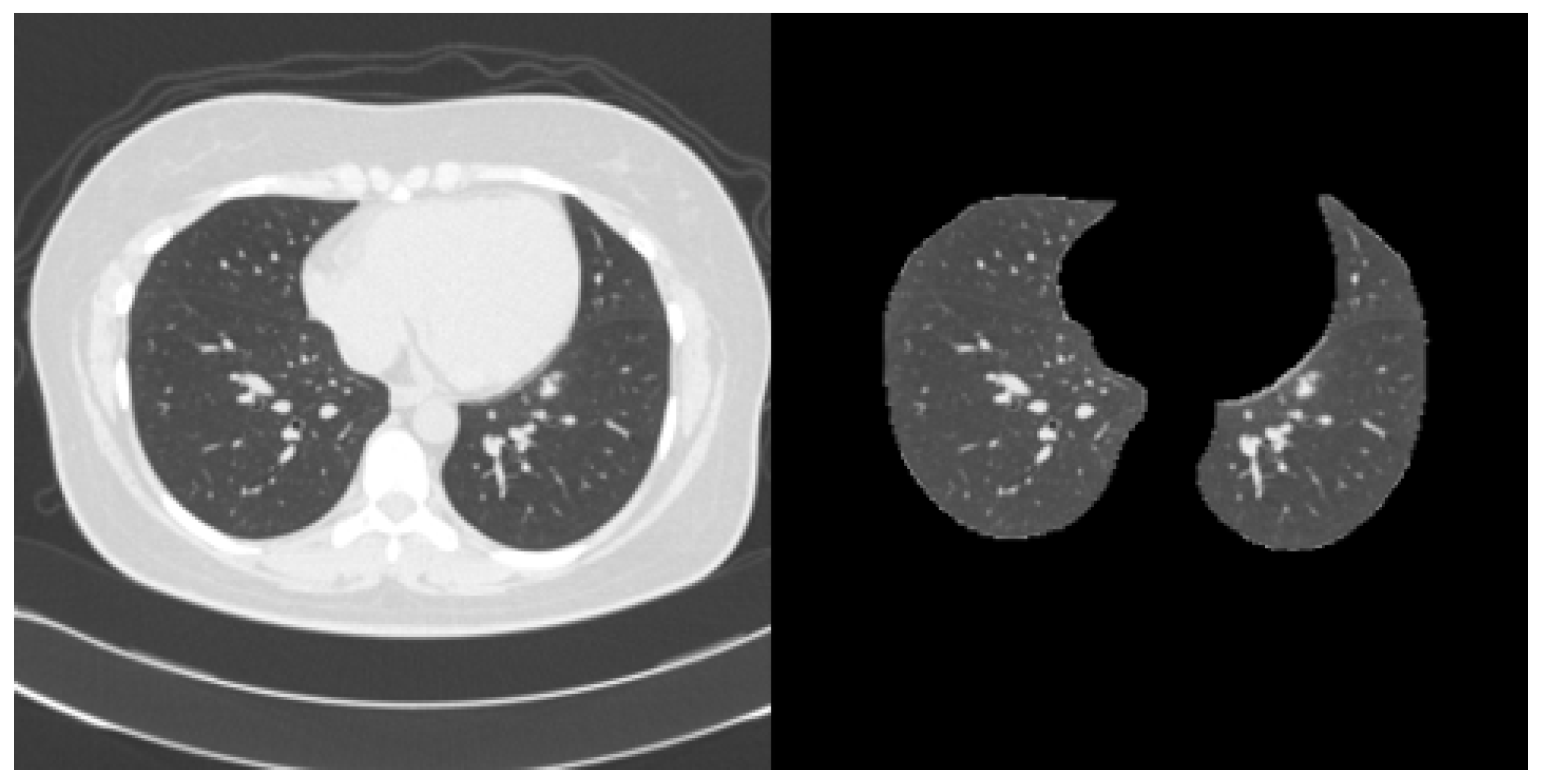
2.1. Dataset
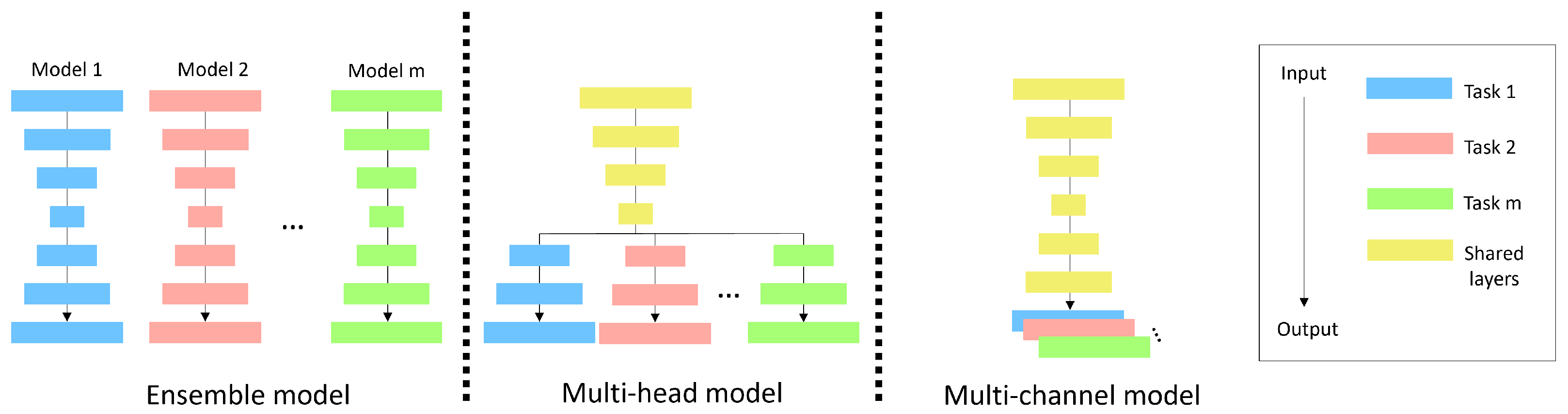
2.2. Methods
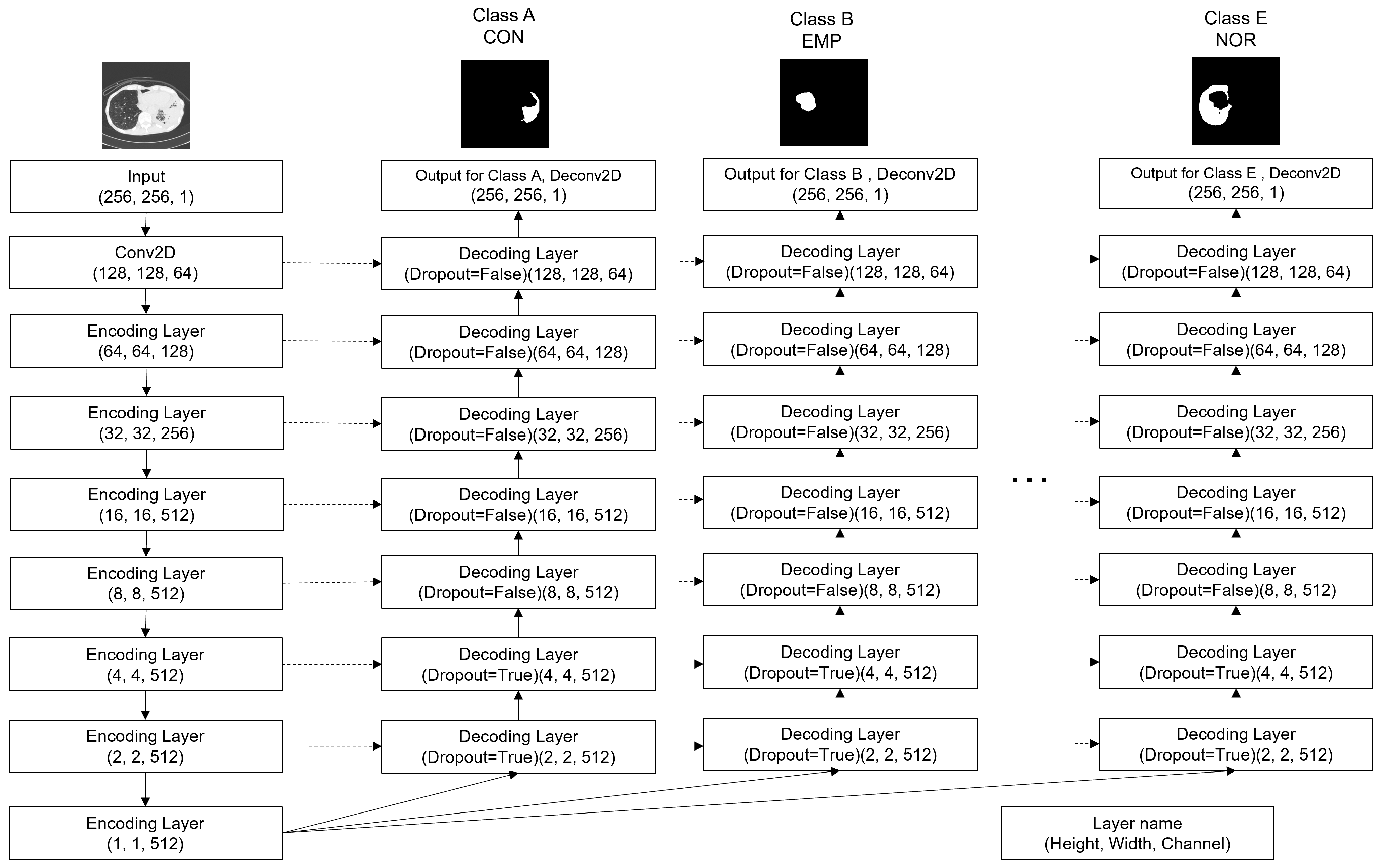
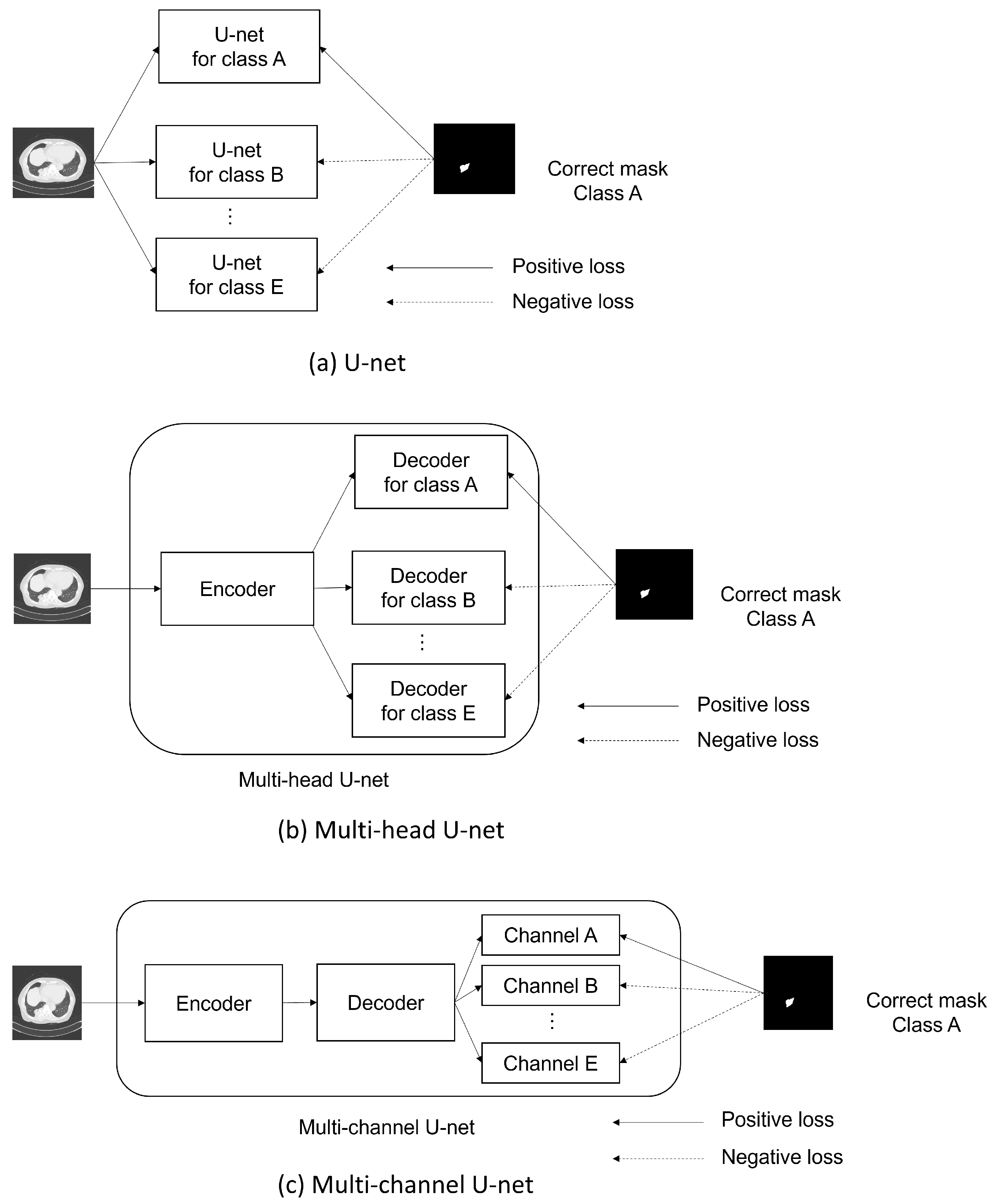
2.3. Multi-Head U-Net
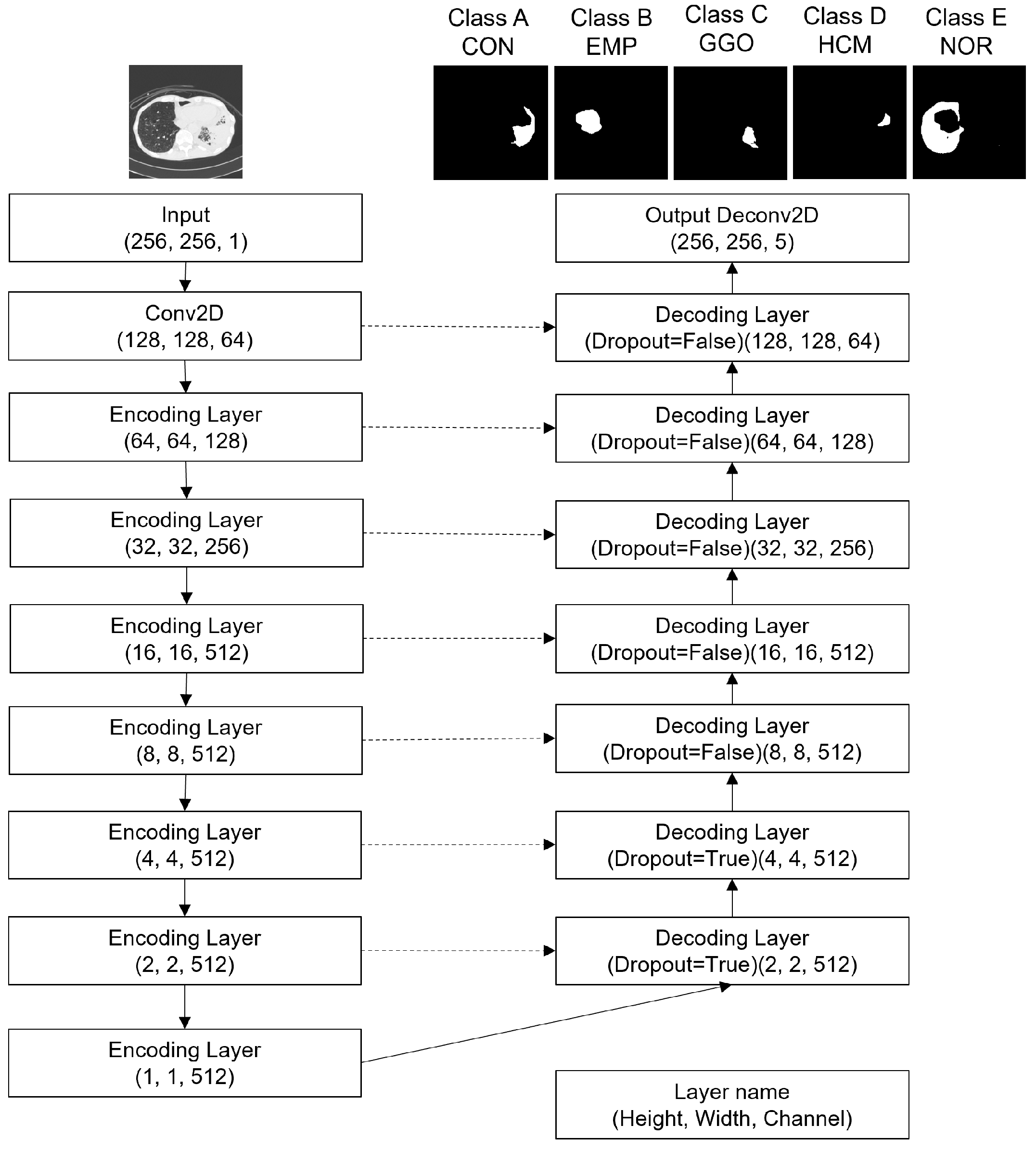
2.4. Multi-Channel U-Net
2.5. Partially Supervised Learning (Two-Loss Learning)
3. Experiments
3.1. Training Procedure
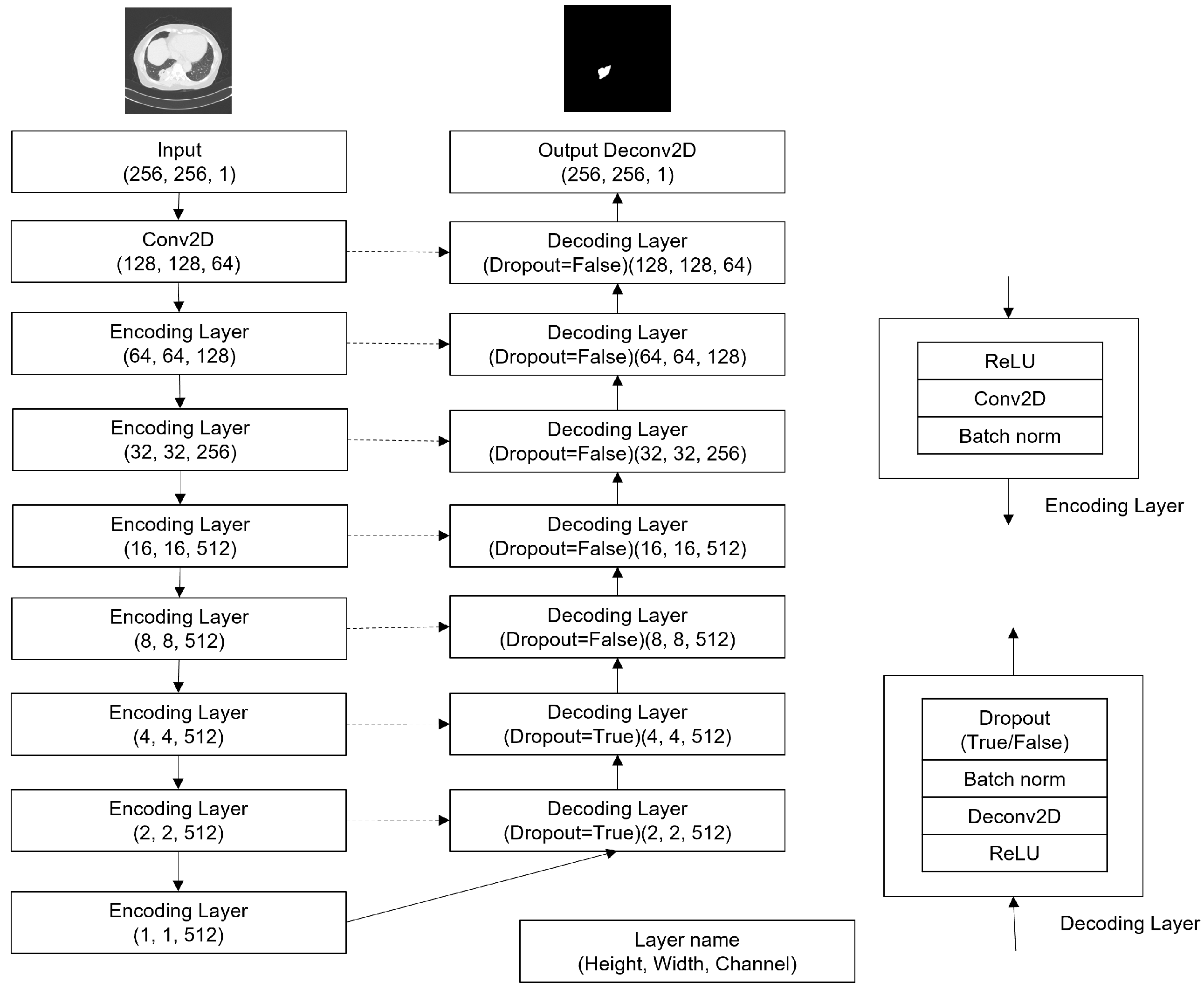
3.1.1. U-Net
- 1.
- Five U-nets (Figure 4) are created with random weight initialization.
- 2.
- Five U-nets are independently trained on the partially annotated images of each target class for 100 epochs using the positive loss function.
- 1.
- Five U-nets are created with random weight initialization.
- 2
- For training data of a single target class, each U-net executes one epoch of two-loss learning, changing the target class one by one (a total of five epochs is implemented in this step).
- 3.
- Step 2 is repeated 100 times.
3.1.2. Multi-Head and Multi-Channel U-Net
- 1.
- 2.
- The model is trained on the partially annotated images for 100 epochs using the positive loss function. Note that the weight update is implemented only on the layers that are related to the class of the input image (multi-head: encoder only; multi-channel: both encoder and decoder).
- 1.
- A model is created with random weight initialization.
- 2.
- Then, the model is trained on the partially annotated images. In detail, for training data of a single target class, the model executes one epoch of two-loss learning, changing the learning target class one by one (a total of five epochs is implemented in this step).
- 3.
- Step 2 is repeated 100 times.
3.2. Testing Procedure
3.2.1. Prediction Ensemble
3.2.2. Evaluation Metric
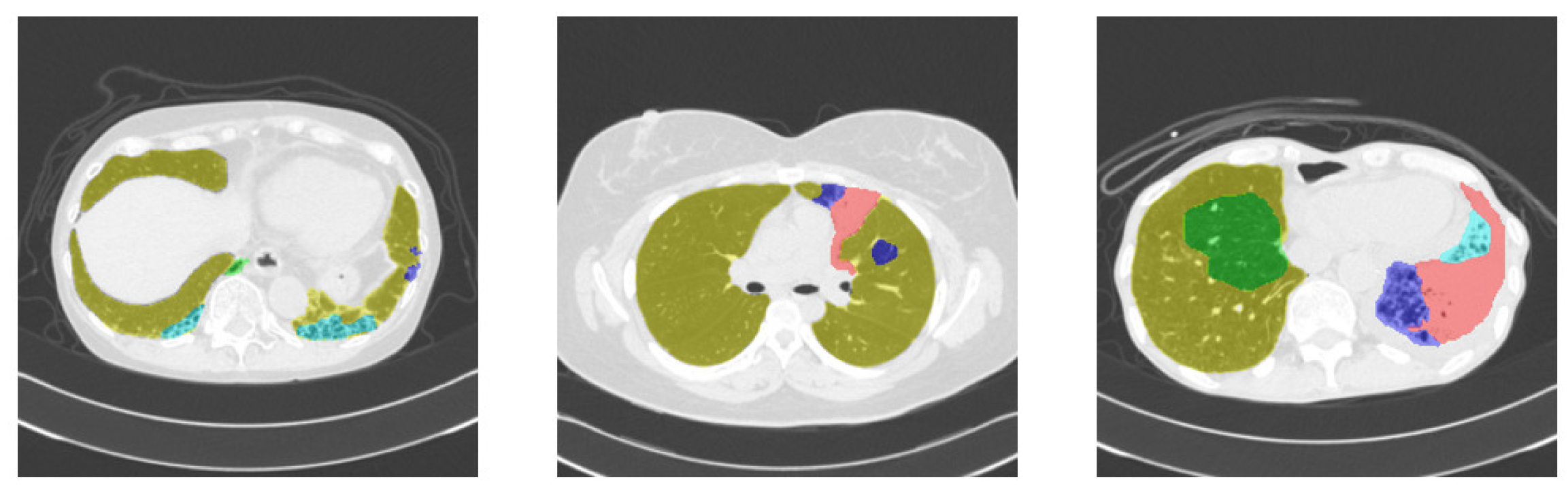
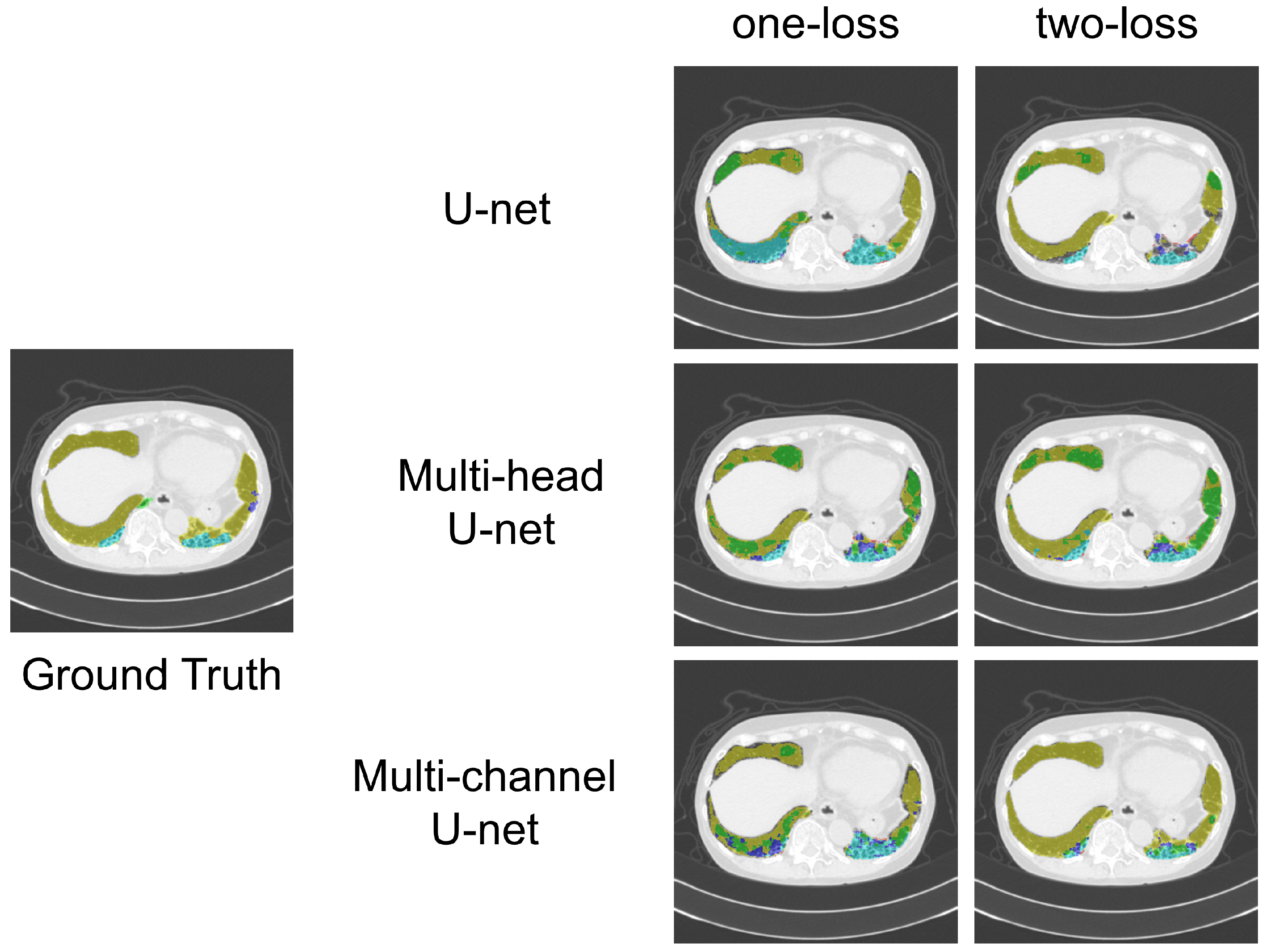
3.3. Results
- Similarities: Both the multi-head and multi-channel U-nets were designed to share a common goal: To efficiently encode opacity features for all classes by using a shared encoder.
- Differences and rationale for choosing the multi-channel U-net: The key differences between the two models lie in their decoders and the way loss functions are backpropagated.Multi-head U-net: This design uses separate decoders for each class, which enables the creation of segmentation maps specialized for each class. However, the backpropagation of loss functions from other classes does not reach a given decoder, limiting the flow of information.Multi-channel U-net: This design employs a shared decoder. While this may not create decoders specifically tailored to each class, it allows the backpropagation of loss functions from all the classes to the decoder layers.
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeasmin, M.N.; Al Amin, M.; Joti, T.J.; Aung, Z.; Azim, M.A. Advances of AI in image-based computer-aided diagnosis: A review. Array 2024, 23, 100357. [Google Scholar] [CrossRef]
- Fujita, H. AI-based computer-aided diagnosis (AI-CAD): The latest review to read first. Radiol. Phys. Technol. 2020, 13, 6–19. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Ye, T.; Yang, Y.; Liu, G.; Zhu, L. Segmamba: Long-range sequential modeling mamba for 3d medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; Springer: Cham, Switzerland, 2024; pp. 578–588. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Rani, V.; Kumar, M.; Gupta, A.; Sachdeva, M.; Mittal, A.; Kumar, K. Self-supervised learning for medical image analysis: A comprehensive review. Evol. Syst. 2024, 15, 1607–1633. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Gerdprasert, T.; Mabu, S. Pseudo-LabelingWith Contrastive Perturbation Using CNN & ViT for Chest X-ray Classification. In Proceedings of the 2023 IEEE 13th International Workshop on Computational Intelligence and Applications (IWCIA), Hiroshima, Japan, 11–12 November 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 65–69. [Google Scholar]
- Nguyen, Q.H.; Hoang, D.A.; Pham, H.V. Combination of 2D and 3D nnU-Net for ground glass opacity segmentation in CT images of Post-COVID-19 patients. Comput. Biol. Med. 2025, 195, 110376. [Google Scholar] [CrossRef] [PubMed]
- Akila Agnes, S.; Arun Solomon, A.; Karthick, K. Wavelet U-Net++ for accurate lung nodule segmentation in CT scans: Improving early detection and diagnosis of lung cancer. Biomed. Signal Process. Control 2024, 87, 105509. [Google Scholar] [CrossRef]
- Hoang-Thi, T.N.; Vakalopoulou, M.; Christodoulidis, S.; Paragios, N.; Revel, M.P.; Chassagnon, G. Deep learning for lung disease segmentation on CT: Which reconstruction kernel should be used? Diagn. Interv. Imaging 2021, 102, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Hamada, T.; Mabu, S.; Ikebe, S. Segmentation of Diffuse Lung Diseases in Computed Tomography Images Using Partially Supervised Learning: A Model Construction and Learning for Feature Extraction Considering Lung Opacities. In Proceedings of the Thirtieth International Symposium on Artificial Life and Robotics, Beppu, Japan, 22–24 January 2025. [Google Scholar]
- Liu, H.; Xu, Z.; Gao, R.; Li, H.; Wang, J.; Chabin, G.; Oguz, I.; Grbic, S. COSST: Multi-Organ Segmentation With Partially Labeled Datasets Using Comprehensive Supervisions and Self-Training. IEEE Trans. Med. Imaging 2024, 43, 1995–2009. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, J.; Xia, Y.; Shen, C. Learning From Partially Labeled Data for Multi-Organ and Tumor Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 14905–14919. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Kido, S.; Mabu, S.; Yanagawa, M.; Tomiyama, N.; Sato, Y. Segmentation of Diffuse Lung Abnormality Patterns on Computed Tomography Images using Partially Supervised Learning. Adv. Biomed. Eng. 2022, 11, 25–36. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015; Conference Track Proceedings. Bengio, Y., LeCun, Y., Eds.; 2015. [Google Scholar]
- Hofmanninger, J.; Prayer, F.; Pan, J.; Röhrich, S.; Prosch, H.; Langs, G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur. Radiol. Exp. 2020, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Liu, Q.; Cui, C.; Asad, Z.; Yang, H.; Huo, Y. Single Dynamic Network for Multi-label Renal Pathology Image Segmentation. In Proceedings of the 5th International Conference on Medical Imaging with Deep Learning, Zurich, Switzerland, 6–8 July 2022; Konukoglu, E., Menze, B., Venkataraman, A., Baumgartner, C., Dou, Q., Albarqouni, S., Eds.; Proceedings of Machine Learning Research. PMLR: Cambridge, MA, USA, 2022; Volume 172, pp. 304–314. [Google Scholar]
- Ye, Y.; Xie, Y.; Zhang, J.; Chen, Z.; Xia, Y. UniSeg: A Prompt-Driven Universal Segmentation Model as Well as A Strong Representation Learner. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2023, Vancouver, BC, Canada, 8–12 October 2023; Greenspan, H., Madabhushi, A., Mousavi, P., Salcudean, S., Duncan, J., Syeda-Mahmood, T., Taylor, R., Eds.; Springer: Cham, Switzerland, 2023; pp. 508–518. [Google Scholar]
| Model Name | Number of Weights | One-Loss Learning | Two-Loss Learning |
|---|---|---|---|
| U-net | 165,849,930 | ||
| Multi-head U-net | 62,667,653 | ||
| Multi-channel U-net | 28,276,869 |
| Number of Shared Decoding Layers | Dice Coefficient |
|---|---|
| 0 (Multi-head U-net) | 0. |
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 (Multi-channel U-net) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mabu, S.; Hamada, T.; Ikebe, S.; Kido, S. Lung Opacity Segmentation in Chest CT Images Using Multi-Head and Multi-Channel U-Nets with Partially Supervised Learning. Appl. Sci. 2025, 15, 10373. https://doi.org/10.3390/app151910373
Mabu S, Hamada T, Ikebe S, Kido S. Lung Opacity Segmentation in Chest CT Images Using Multi-Head and Multi-Channel U-Nets with Partially Supervised Learning. Applied Sciences. 2025; 15(19):10373. https://doi.org/10.3390/app151910373
Chicago/Turabian StyleMabu, Shingo, Takuya Hamada, Satoru Ikebe, and Shoji Kido. 2025. "Lung Opacity Segmentation in Chest CT Images Using Multi-Head and Multi-Channel U-Nets with Partially Supervised Learning" Applied Sciences 15, no. 19: 10373. https://doi.org/10.3390/app151910373
APA StyleMabu, S., Hamada, T., Ikebe, S., & Kido, S. (2025). Lung Opacity Segmentation in Chest CT Images Using Multi-Head and Multi-Channel U-Nets with Partially Supervised Learning. Applied Sciences, 15(19), 10373. https://doi.org/10.3390/app151910373






