3D Convolutional Neural Network Model for Detection of Major Depressive Disorder from Grey Matter Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
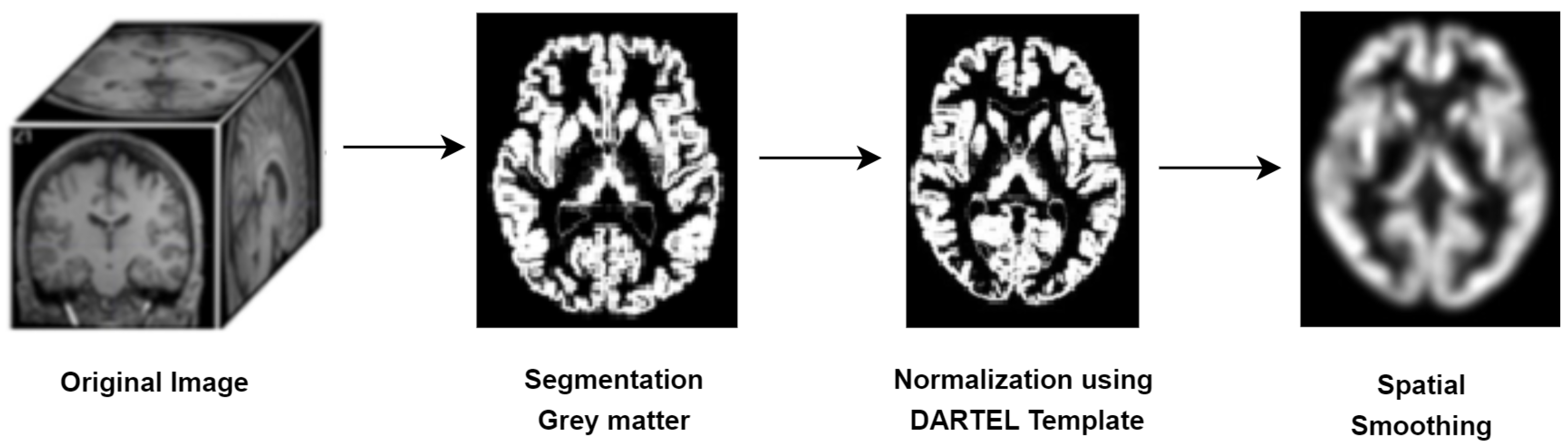
2.2. Data Preprocessing
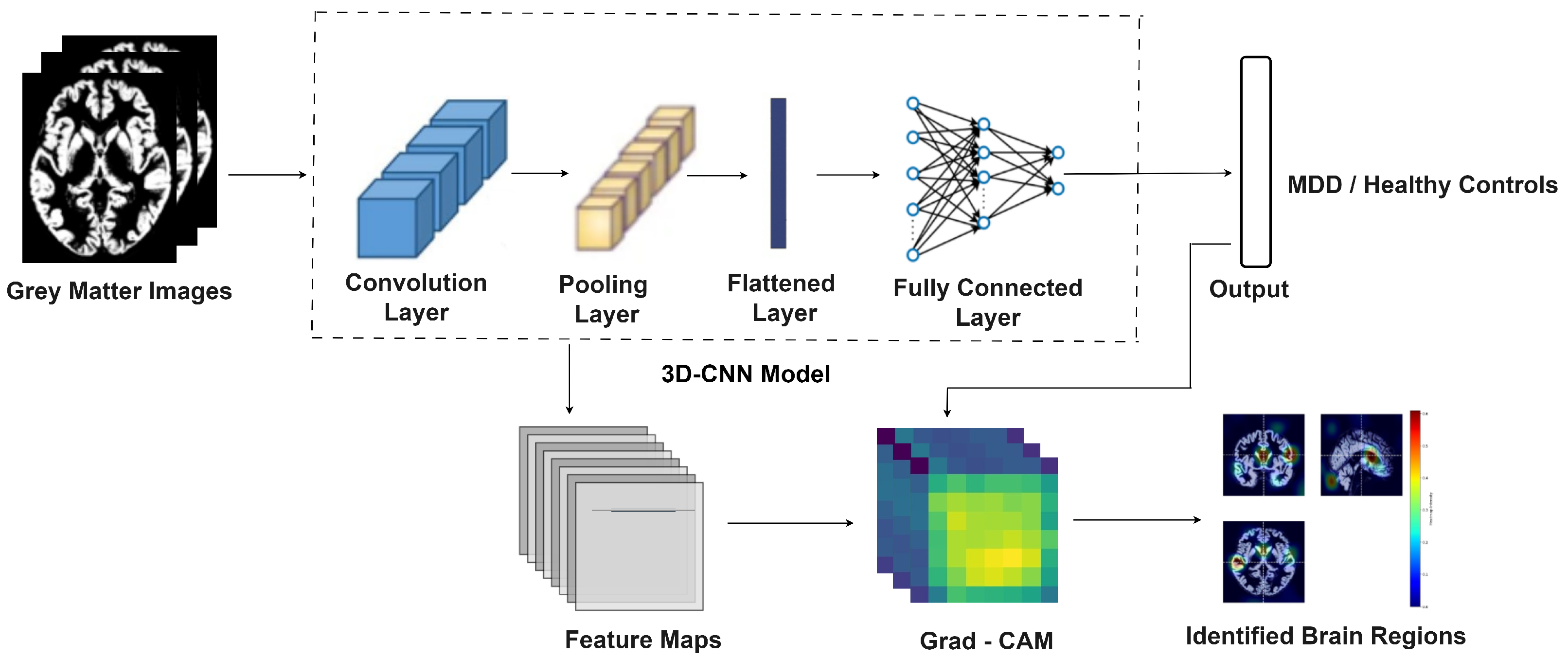
2.3. MDD-Net for Detection of Major Depressive Disorder
2.3.1. MDD-Net Model
2.3.2. Loss Function and Convergence
2.3.3. Hyperparameter Optimization
3. Results
3.1. Experimental Results
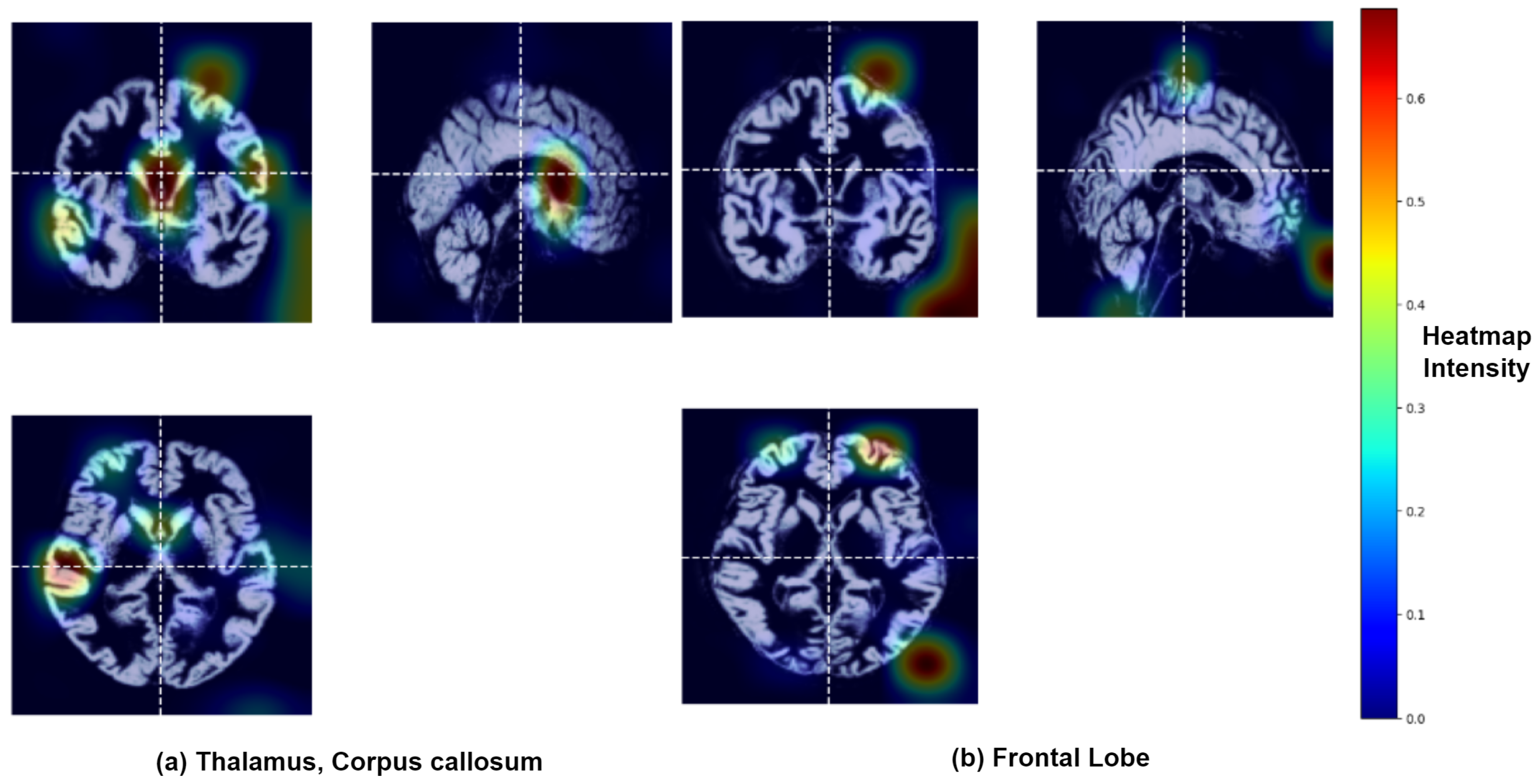
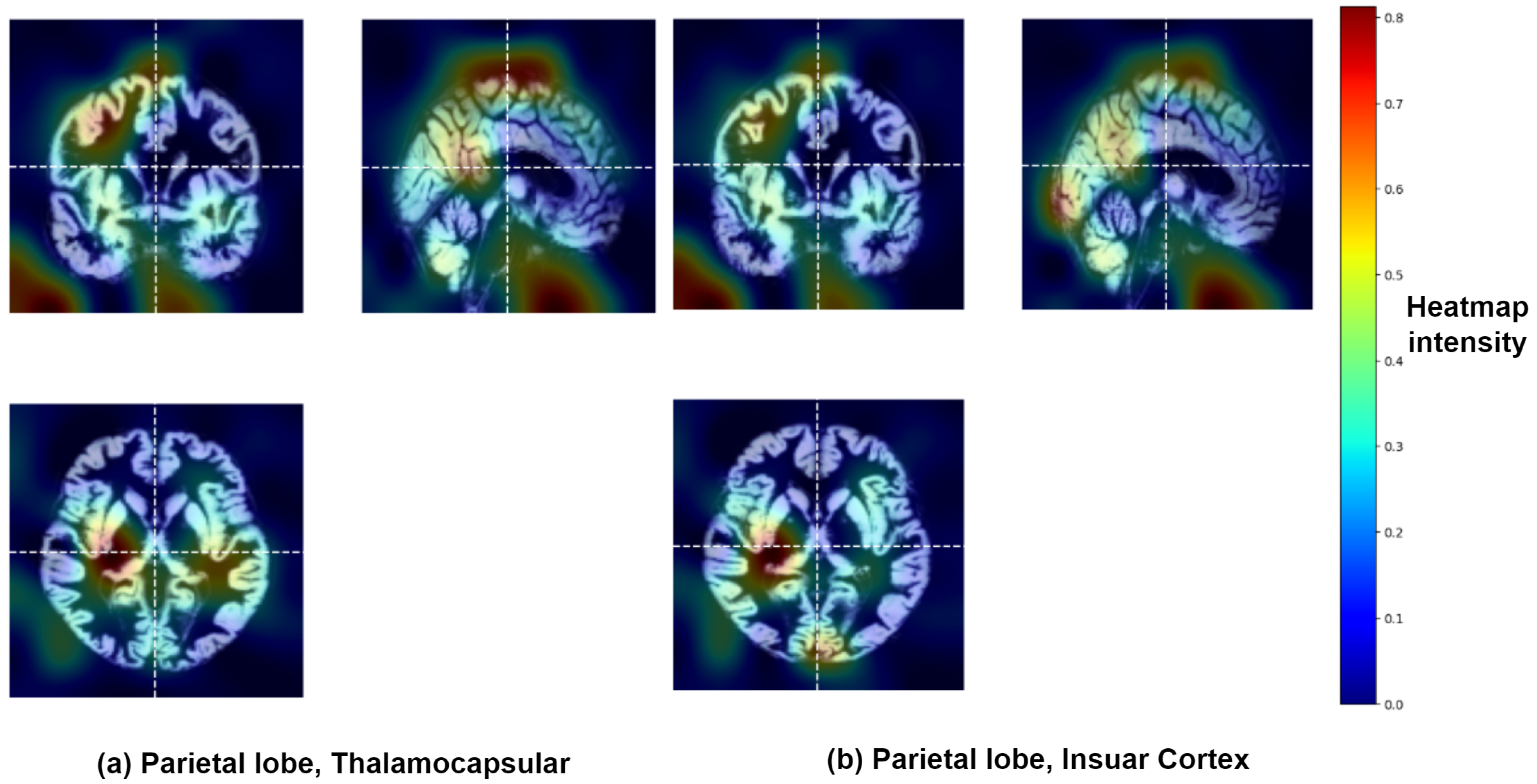
3.2. Identification of Brain Regions Affected by Major Depressive Disorder
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MRI | Magnetic resonance imaging |
| MDD | Major depressive disorder |
| HCs | Healthy controls |
| 3D CNN | 3D convolutional neural network |
| GM | Grey matter |
| Grad-CAM | Gradient-weighted class activation mapping |
References
- Otte, C.; Gold, S.M.; Penninx, B.W.; Pariante, C.M.; Etkin, A.; Fava, M.; Mohr, D.C.; Schatzberg, A.F. Major depressive disorder. Nat. Rev. Dis. Prim. 2016, 2, 16065. [Google Scholar] [CrossRef]
- Ren, Y.; Li, M.; Yang, C.; Jiang, W.; Wu, H.; Pan, R.; Yang, Z.; Wang, X.; Wang, W.; Wang, W.; et al. Suicidal risk is associated with hyper-connections in the frontal-parietal network in patients with depression. Transl. Psychiatry 2025, 15, 49. [Google Scholar] [CrossRef]
- Arnaud, A.; Brister, T.; Duckworth, K.; Foxworth, P.; Fulwider, T.; Suthoff, E.; Werneburg, B.; Aleksanderek, I.; Reinhart, M. Impact of Major Depressive Disorder on Comorbidities: A Systematic Literature Review. J. Clin. Psychiatry 2022, 83, 21r14328. [Google Scholar] [CrossRef]
- Reynolds, W.M.; Kobak, K.A. Reliability and validity of the Hamilton Depression Inventory: A paper-and-pencil version of the Hamilton Depression Rating Scale clinical interview. Psychol. Assess. 1995, 7, 472–483. [Google Scholar] [CrossRef]
- First, M.B.; Gaebel, W.; Maj, M.; Stein, D.J.; Kogan, C.S.; Saunders, J.B.; Poznyak, V.B.; Gureje, O.; Lewis-Fernández, R.; Maercker, A.; et al. An organization-and category-level comparison of diagnostic requirements for mental disorders in ICD-11 and DSM-5. World Psychiatry 2021, 20, 34–51. [Google Scholar] [CrossRef]
- Qiu, H.; Li, J. Major depressive disorder and magnetic resonance imaging: A mini-review of recent progress. Curr. Pharm. Des. 2018, 24, 2524–2529. [Google Scholar] [CrossRef] [PubMed]
- Thacker, N.; Jackson, A. Mathematical segmentation of grey matter, white matter and cerebral spinal fluid from MR image pairs. Br. J. Radiol. 2001, 74, 234–242. [Google Scholar] [CrossRef]
- Bedell, B.J.; Narayana, P.A. Volumetric analysis of white matter, gray matter, and CSF using fractional volume analysis. Magn. Reson. Med. 1998, 39, 961–969. [Google Scholar] [CrossRef]
- Girbau-Massana, D.; Garcia-Marti, G.; Marti-Bonmati, L.; Schwartz, R.G. Gray–white matter and cerebrospinal fluid volume differences in children with specific language impairment and/or reading disability. Neuropsychologia 2014, 56, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Zhang, Y.; Yang, Z.; Han, S.; Cheng, J. Reduced Brain Gray Matter Volume in Patients with First-Episode Major Depressive Disorder: A Quantitative Meta-Analysis. Front. Psychiatry 2021, 12, 671348. [Google Scholar] [CrossRef]
- Shad, M.U.; Muddasani, S.; Rao, U. Gray matter differences between healthy and depressed adolescents: A voxel-based morphometry study. J. Child Adolesc. Psychopharmacol. 2012, 22, 190–197. [Google Scholar] [CrossRef]
- Romeo, Z.; Biondi, M.; Oltedal, L.; Spironelli, C. The Dark and Gloomy Brain: Grey Matter Volume Alterations in Major Depressive Disorder–Fine-Grained Meta-Analyses. Depress. Anxiety 2024, 2024, 6673522. [Google Scholar] [CrossRef]
- Plis, S.; Hjelm, D.; Salakhutdinov, R.; Allen, E.; Bockholt, H.; Long, J.; Johnson, H.; Paulsen, J.; Turner, J.; Calhoun, V. Deep learning for neuroimaging: A validation study. Front. Neurosci. 2014, 8, 229. [Google Scholar] [CrossRef]
- Avberšek, L.; Repovš, G. Deep learning in neuroimaging data analysis: Applications, challenges, and solutions. Front. Neuroimaging 2022, 1, 981642. [Google Scholar] [CrossRef]
- Smucny, J.; Shi, G.; Davidson, I. Deep Learning in Neuroimaging: Overcoming Challenges with Emerging Approaches. Front. Psychiatry 2022, 13, 912600. [Google Scholar] [CrossRef]
- Wang, Y.; Gong, N.; Fu, C. Major depression disorder diagnosis and analysis based on structural magnetic resonance imaging and deep learning. J. Integr. Neurosci. 2021, 20, 977–984. [Google Scholar] [CrossRef]
- Lin, C.; Lee, S.H.; Huang, C.M.; Chen, G.Y.; Chang, W.; Liu, H.L.; Ng, S.H.; Lee, T.M.C.; Wu, S.C. Automatic diagnosis of late-life depression by 3D convolutional neural networks and cross-sample Entropy analysis from resting-state fMRI. Brain Imaging Behav. 2023, 17, 125–135. [Google Scholar] [CrossRef]
- Li, M.; Liu, M.; Kang, J.; Zhang, W.; Lu, S. Depression recognition method based on regional homogeneity features from emotional response fMRI using deep convolutional neural network. In Proceedings of the 2021 3rd International Conference on Intelligent Medicine and Image Processing, Tianjin, China, 23–26 April 2021; pp. 45–49. [Google Scholar]
- Lin, C.; Huang, C.; Chang, W.; Chang, Y.; Liu, H.; Ng, S.; Lin, H.; Lee, T.; Lee, S.; Wu, S. Predicting suicidality in late-life depression by 3D convolutional neural network and cross-sample entropy analysis of resting-state fMRI. Brain Behav. 2024, 14, e3348. [Google Scholar] [CrossRef]
- Belov, V.; Erwin-Grabner, T.; Aghajani, M.; Aleman, A.; Amod, A.R.; Basgoze, Z.; Benedetti, F.; Besteher, B.; Bülow, R.; Ching, C.R.; et al. Multi-site benchmark classification of major depressive disorder using machine learning on cortical and subcortical measures. Sci. Rep. 2024, 14, 1084. [Google Scholar] [CrossRef]
- Alotaibi, N.M.; Alhothali, A.M.; Ali, M.S. Multi-atlas ensemble graph neural network model for major depressive disorder detection using functional MRI data. Front. Comput. Neurosci. 2025, 19, 1537284. [Google Scholar] [CrossRef]
- Wu, W.; Yan, J.; Zhao, Y.; Sun, Q.; Zhang, H.; Cheng, J.; Liang, D.; Chen, Y.; Zhang, Z.; Li, Z.C. Multi-task learning for concurrent survival prediction and semi-supervised segmentation of gliomas in brain MRI. Displays 2023, 78, 102402. [Google Scholar] [CrossRef]
- Yan, J.; Zhao, Y.; Chen, Y.; Wang, W.; Duan, W.; Wang, L.; Zhang, S.; Ding, T.; Liu, L.; Sun, Q.; et al. Deep learning features from diffusion tensor imaging improve glioma stratification and identify risk groups with distinct molecular pathway activities. eBioMedicine 2021, 72, 103583. [Google Scholar] [CrossRef]
- Qu, X.; Xiong, Y.; Zhai, K.; Yang, X.; Yang, J. An Efficient Attention-Based Network for Screening Major Depressive Disorder with sMRI. In Proceedings of the 2023 29th International Conference on Mechatronics and Machine Vision in Practice (M2VIP), Queenstown, New Zealand, 21–24 November 2023; pp. 1–6. [Google Scholar]
- Wang, Q.; Li, L.; Qiao, L.; Liu, M. Adaptive multimodal neuroimage integration for major depression disorder detection. Front. Neuroinform. 2022, 16, 856175. [Google Scholar] [CrossRef]
- Yan, C.G.; Chen, X.; Li, L.; Castellanos, F.X.; Bai, T.J.; Bo, Q.J.; Cao, J.; Chen, G.M.; Chen, N.X.; Chen, W.; et al. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc. Natl. Acad. Sci. USA 2019, 116, 9078–9083. [Google Scholar] [CrossRef]
- Chen, X.; Lu, B.; Li, H.X.; Li, X.Y.; Wang, Y.W.; Castellanos, F.X.; Cao, L.P.; Chen, N.X.; Chen, W.; Cheng, Y.Q.; et al. The DIRECT consortium and the REST-meta-MDD project: Towards neuroimaging biomarkers of major depressive disorder. Psychoradiology 2022, 2, 32–42. [Google Scholar] [CrossRef]
- Shakya, A.; Biswas, M.; Pal, M. Classification of Radar data using Bayesian optimized two-dimensional Convolutional Neural Network. In Radar Remote Sensing; Elsevier: Amsterdam, The Netherlands, 2022; pp. 175–186. [Google Scholar]
- Fang, Y.; Potter, G.G.; Wu, D.; Zhu, H.; Liu, M. Addressing multi-site functional MRI heterogeneity through dual-expert collaborative learning for brain disease identification. Hum. Brain Mapp. 2023, 44, 4256–4271. [Google Scholar] [CrossRef]
- Wang, J.; Li, T.; Sun, Q.; Guo, Y.; Yu, J.; Yao, Z.; Hou, N.; Hu, B. Automatic Diagnosis of Major Depressive Disorder Using a High-and Low-Frequency Feature Fusion Framework. Brain Sci. 2023, 13, 1590. [Google Scholar] [CrossRef]
- Hong, J.; Huang, Y.; Ye, J.; Wang, J.; Xu, X.; Wu, Y.; Li, Y.; Zhao, J.; Li, R.; Kang, J.; et al. 3D FRN-ResNet: An Automated Major Depressive Disorder Structural Magnetic Resonance Imaging Data Identification Framework. Front. Aging Neurosci. 2022, 14, 912283. [Google Scholar] [CrossRef]
- Ramprasaath, R.; Selvaraju, M.; Das, A. Visual Explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 618–626. [Google Scholar]
- Samek, W.; Montavon, G.; Vedaldi, A.; Hansen, L.K.; Müller, K.R. Explainable AI: Interpreting, Explaining and Visualizing Deep Learning; Springer Nature: Berlin/Heidelberg, Germany, 2019; Volume 11700. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Computer Vision–ECCV 2014, Proceedings of the 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Proceedings, Part I 13; Springer: Berlin/Heidelberg, Germany, 2014; pp. 818–833. [Google Scholar]
- Binder, A.; Montavon, G.; Lapuschkin, S.; Müller, K.R.; Samek, W. Layer-wise relevance propagation for neural networks with local renormalization layers. In Artificial Neural Networks and Machine Learning–ICANN 2016, Proceedings of the 25th International Conference on Artificial Neural Networks, Barcelona, Spain, 6–9 September 2016; Proceedings, Part II 25; Springer: Berlin/Heidelberg, Germany, 2016; pp. 63–71. [Google Scholar]
- Chayer, C.; Freedman, M. Frontal lobe functions. Curr. Neurol. Neurosci. Rep. 2001, 1, 547–552. [Google Scholar] [CrossRef]
- Zhuo, C.; Li, G.; Lin, X.; Jiang, D.; Xu, Y.; Tian, H.; Wang, W.; Song, X. The rise and fall of MRI studies in major depressive disorder. Transl. Psychiatry 2019, 9, 335. [Google Scholar] [CrossRef]
- Zhang, F.F.; Peng, W.; Sweeney, J.A.; Jia, Z.Y.; Gong, Q.Y. Brain structure alterations in depression: Psychoradiological evidence. CNS Neurosci. Ther. 2018, 24, 994–1003. [Google Scholar] [CrossRef]
- Milner, B. Intellectual function of the temporal lobes. Psychol. Bull. 1954, 51, 42. [Google Scholar] [CrossRef]
- Lai, C.H. Gray matter volume in major depressive disorder: A meta-analysis of voxel-based morphometry studies. Psychiatry Res. Neuroimaging 2013, 211, 37–46. [Google Scholar] [CrossRef]
- Lorenzetti, V.; Allen, N.B.; Fornito, A.; Yücel, M. Structural brain abnormalities in major depressive disorder: A selective review of recent MRI studies. J. Affect. Disord. 2009, 117, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Coslett, H.B.; Schwartz, M.F. The parietal lobe and language. Handb. Clin. Neurol. 2018, 151, 365–375. [Google Scholar]
- Chen, Z.; Peng, W.; Sun, H.; Kuang, W.; Li, W.; Jia, Z.; Gong, Q. High-field magnetic resonance imaging of structural alterations in first-episode, drug-naive patients with major depressive disorder. Transl. Psychiatry 2016, 6, e942. [Google Scholar] [CrossRef] [PubMed]
- Kilic, F.; Kartal, F.; Erbay, M.F.; Karlidağ, R. Parietal Cortex Volume and Functions in Major Depression and Bipolar Disorder: A Cloud-Based Magnetic Resonans Imaging Study. Arch. Neuropsychiatry 2025, 61, 47. [Google Scholar]
- Klostermann, F.; Krugel, L.; Ehlen, F. Functional roles of the thalamus for language capacities. Front. Syst. Neurosci. 2013, 7, 32. [Google Scholar] [CrossRef]
- Ward, L.M. The thalamus: Gateway to the mind. Wiley Interdiscip. Rev. Cogn. Sci. 2013, 4, 609–622. [Google Scholar] [CrossRef]
- Zhang, K.; Zhu, Y.; Zhu, Y.; Wu, S.; Liu, H.; Zhang, W.; Xu, C.; Zhang, H.; Hayashi, T.; Tian, M. Molecular, functional, and structural imaging of major depressive disorder. Neurosci. Bull. 2016, 32, 273–285. [Google Scholar] [CrossRef]
- Nugent, A.C.; Davis, R.M.; Zarate, C.A., Jr.; Drevets, W.C. Reduced thalamic volumes in major depressive disorder. Psychiatry Res. Neuroimaging 2013, 213, 179–185. [Google Scholar] [CrossRef]
- Hinkley, L.; Marco, E.; Findlay, A.; Honma, S.; Jeremy, R.; Strominger, Z.; Bukshpun, P.; Wakahiro, M.; Brown, W.; Paul, L.; et al. The role of corpus callosum development in functional connectivity and cognitive processing. PLoS ONE 2012, 7, e39804. [Google Scholar] [CrossRef]
- Lee, S.; Pyun, S.; Choi, K.; Tae, W. Shape and Volumetric Differences in the Corpus Callosum Between Patients with Major Depressive Disorder and Healthy Controls. Psychiatry Investig. 2020, 17, 941–950. [Google Scholar] [CrossRef]
- Piras, F.; Vecchio, D.; Kurth, F.; Piras, F.; Banaj, N.; Ciullo, V.; Luders, E.; Spalletta, G. Corpus callosum morphology in major mental disorders: A magnetic resonance imaging study. Brain Commun. 2021, 3, fcab100. [Google Scholar] [CrossRef]
- Shura, R.D.; Hurley, R.A.; Taber, K.H. Insular cortex: Structural and functional neuroanatomy. J. Neuropsychiatry Clin. Neurosci. 2014, 26, 276–282. [Google Scholar] [CrossRef]
- Sliz, D.; Hayley, S. Major depressive disorder and alterations in insular cortical activity: A review of current functional magnetic imaging research. Front. Hum. Neurosci. 2012, 6, 323. [Google Scholar] [CrossRef]
- Peng, X.; Lin, P.; Wu, X.; Gong, R.; Yang, R.; Wang, J. Insular subdivisions functional connectivity dysfunction within major depressive disorder. J. Affect. Disord. 2018, 227, 280–288. [Google Scholar] [CrossRef]
- Takahashi, T.; Sasabayashi, D.; Yücel, M.; Whittle, S.; Suzuki, M.; Pantelis, C.; Malhi, G.S.; Allen, N.B. Gross anatomical features of the insular cortex in affective disorders. Front. Psychiatry 2024, 15, 1482990. [Google Scholar] [CrossRef]


| MDD | Healthy Controls | |
|---|---|---|
| No. of Subjects | 1276 | 1104 |
| Male | 463 | 462 |
| Female | 813 | 642 |
| Average Age | 36 years Range (14–80) | 36 years Range (12–82) |
| Layer (Type) | Output Shape | Parameters |
|---|---|---|
| Input Layer | (B, 1, 121, 145, 121) | 0 |
| Block 1 | ||
| Conv3D (8 filters, 3 × 3 × 3) | (B, 8, 121, 145, 121) | 224 (216 + 8) |
| Conv3D (8 filters, 3 × 3 × 3) | (B, 8, 121, 145, 121) | 1736 (1728 + 8) |
| MaxPool3D (2 × 2 × 2) | (B, 8, 60, 72, 60) | 0 |
| Block 2 | ||
| Conv3D (16 filters, 3 × 3 × 3) | (B, 16, 60, 72, 60) | 3472 (3456 + 16) |
| Conv3D (16 filters, 3 × 3 × 3) | (B, 16, 60, 72, 60) | 6928 (6912 + 16) |
| MaxPool3D (2 × 2 × 2) | (B, 16, 30, 36, 30) | 0 |
| Block 3 | ||
| Conv3D (32 filters, 3 × 3 × 3) | (B, 32, 30, 36, 30) | 13,856 (13,824 + 32) |
| Conv3D (32 filters, 3 × 3 × 3) | (B, 32, 30, 36, 30) | 27,680 (27,648 + 32) |
| MaxPool3D (2 × 2 × 2) | (B, 32, 15, 18, 15) | 0 |
| Block 4 | ||
| Conv3D (64 filters, 3 × 3 × 3) | (B, 64, 15, 18, 15) | 55,360 (55,296 + 64) |
| Conv3D (64 filters, 3 × 3 × 3) | (B, 64, 15, 18, 15) | 110,656 (110,592 + 64) |
| MaxPool3D (2 × 2 × 2) | (B, 64, 7, 9, 7) | 0 |
| Block 5 | ||
| Conv3D (128 filters, 3 × 3 × 3) | (B, 128, 7, 9, 7) | 221,312 (221,184 + 128) |
| Conv3D (128 filters, 3 × 3 × 3) | (B, 128, 7, 9, 7) | 442,496 (442,368 + 128) |
| MaxPool3D (2 × 2 × 2) | (B, 128, 3, 4, 3) | 0 |
| Block 6 | ||
| Conv3D (256 filters, 3 × 3 × 3) | (B, 256, 3, 4, 3) | 884,992 (884,736 + 256) |
| Conv3D (256 filters, 3 × 3 × 3) | (B, 256, 3, 4, 3) | 1,769,728 (1,769,472 + 256) |
| MaxPool3D (2 × 2 × 2) | (B, 256, 1, 2, 1) | 0 |
| Fully Connected Layers | ||
| Fully connected 1 (512 → 128) | (B, 128) | 65,664 (65,536 + 128) |
| BatchNorm1D (128) | (B, 128) | 256 (128 + 128) |
| Dropout | (B, 128) | 0 |
| Fully connected 2 (128 → 64) | (B, 64) | 8256 (8192 + 64) |
| Fully connected 3 (64 → 2) | (B, 2) | 130 (128 + 2) |
| Activation function | Rectified linear unit |
| Weight initialization | Kaiming initialization |
| Optimizer | AdamW |
| Model depth | 4 |
| Dropout rate | 0.7 |
| No. of epoch | 50 |
| Learning Rate | 0.001 |
| Loss function | Custom Categorical Cross-Entropy Loss |
| Batch size | 16 |
| Config | Model Depth | Optimizer | Learning Rate () | Loss Function Formulation |
|---|---|---|---|---|
| Config-A | 4 | Adam | 0.01 | |
| Config-B | 4 | AdamW | 0.001 | |
| Config-C | 4 | AdamW | 0.001 | |
| Config-D | 6 | AdamW | 0.001 | |
| Config-E | 6 | Adam | 0.0001 | |
| Config-F | 6 | AdamW | 0.001 |
| Config | Accuracy | Sensitivity | Specificity | F1 Score | AUROC | AUPRC |
|---|---|---|---|---|---|---|
| Config-A | 66.95% | 0.93 | 0.36 | 0.75 | 0.74 | 0.78 |
| Config-B | 72.26% | 0.83 | 0.59 | 0.76 | 0.80 | 0.81 |
| Config-C | 52.38% | 0.13 | 0.99 | 0.22 | 0.77 | 0.77 |
| Config-D | 71.43% | 0.75 | 0.67 | 0.74 | 0.76 | 0.76 |
| Config-E | 72.00% | 0.67 | 0.76 | 0.72 | 0.80 | 0.81 |
| Config-F | 53.78% | 0.13 | 0.98 | 0.24 | 0.75 | 0.77 |
| SI No. | Authors | Sample Size | Model Used | Accuracy |
|---|---|---|---|---|
| 1. | Yuqi et al. [29] | MDD = 441, HC = 395 | DFH | 57.7% |
| 2. | Wang et al. [30] | MDD = 54, HC = 62 | Ensemble model | 72.4% |
| 3. | Lin et al. [17] | MDD = 49, HC = 28 | 3D CNN | 85% |
| 4. | Hong et al. [31] | MDD = 34, HC = 34 | 3D FRN-ResNet | 86.7% |
| 5. | Proposed | MDD = 1276, HC = 1104 | 3D CNN | 72.26% |
| SI No. | Brain Regions | Functionalities |
|---|---|---|
| 1. | Frontal Lobe | Planning, decision-making, problem-solving, voluntary motor control, behavior, emotional regulation, speech and hearing |
| 2. | Parietal Lobe | Integration of somatosensory information like touch, pain, temperature, proprioception, spatial orientation, perception, aspects of language and mathematical processing |
| 3. | Temporal Lobe | Primary auditory processing, language comprehension, encoding and retrieval of declarative memory, visual object and face recognition and emotional processing |
| 4. | Thalamus | Regulation of consciousness, memory, emotion, alertness, sleep-wake cycles, relaying sensory and motor signals to the cerebral cortex |
| 5. | Insular Cortex | Involved in interceptive awareness, taste processing, automatic control, pain perception and emotional regulation |
| 6. | Corpus Callosum | Communication and coordination of motor, sensory and cognitive functions |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
A. R., B.; Adiga, A.; Mahanand, B.S.; DIRECT Consortium. 3D Convolutional Neural Network Model for Detection of Major Depressive Disorder from Grey Matter Images. Appl. Sci. 2025, 15, 10312. https://doi.org/10.3390/app151910312
A. R. B, Adiga A, Mahanand BS, DIRECT Consortium. 3D Convolutional Neural Network Model for Detection of Major Depressive Disorder from Grey Matter Images. Applied Sciences. 2025; 15(19):10312. https://doi.org/10.3390/app151910312
Chicago/Turabian StyleA. R., Bindiya, Aditya Adiga, B. S. Mahanand, and DIRECT Consortium. 2025. "3D Convolutional Neural Network Model for Detection of Major Depressive Disorder from Grey Matter Images" Applied Sciences 15, no. 19: 10312. https://doi.org/10.3390/app151910312
APA StyleA. R., B., Adiga, A., Mahanand, B. S., & DIRECT Consortium. (2025). 3D Convolutional Neural Network Model for Detection of Major Depressive Disorder from Grey Matter Images. Applied Sciences, 15(19), 10312. https://doi.org/10.3390/app151910312






