Exploring Three Methods for Sampling Oral Microbiota in Older People: A Comparative Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Participants
- -
- Presence of cognitive complaints;
- -
- Presence of at least 1 natural tooth (to be able to use the paper point method on all patients).
- -
- Incompetent to follow or to perform the given sampling instructions correctly (e.g., due to severe cognitive problems and/or physical problems);
- -
- Incapacitated by law.
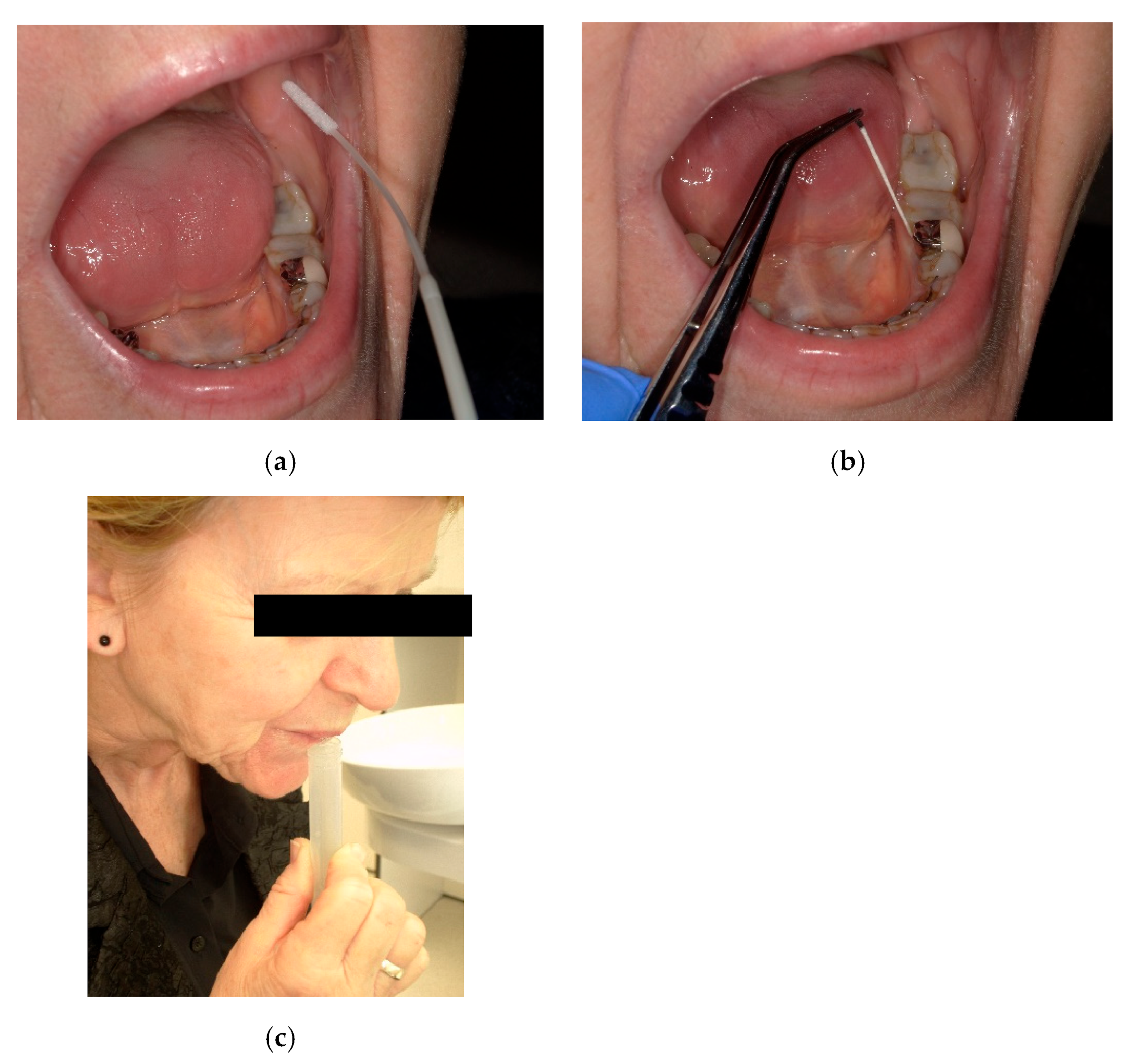
2.3. Sampling Methods
2.4. Oral Microbiota
2.5. Microbiological Q-PCR Test
2.6. Bacterial DNA Extraction
| 0 CFU | − | Negative |
| 1–10 CFU | + | Minimal amount |
| 10–100 CFU | ++ | Low amount |
| 100–1000 CFU | +++ | Moderate amount |
| 1000–10,000 CFU | ++++ | Moderately high amount |
| 10,000–100,000 CFU | +++++ | High amount |
| 100,000–>1,000,000 CFU | ++++++ | Very high amount |
2.7. The Ease of Use and Cost of the Three Different Sampling Methods
- -
- Cost of sample material (expensive (>€5) −, fair deal (€2–€5) +/−, cheap (<€2) +);
- -
- Cost of laboratory analysis (price in euros);
- -
- Participant comfort (good +, average +/−, poor −);
- -
- Sampling time length (short + (<30 s), average +/− (30 s–2 min), long − (>2 min));
- -
- Ease of sampling (for the practitioner) (easy +, average +/−, difficult −);
- -
- Accessibility of the sample location (good +, average +/−, poor −);
2.8. Sample Size Calculation
2.9. Statistical Analysis
3. Results
3.1. Characteristics of the Participants
3.2. Comparison of the Different Sampling Methods
3.3. Comparing Sampling Methods for Bacterial Detection
3.4. The Association Between Bacterial Match and Participant Characteristics
3.5. Characteristics of the Sampling Methods
4. Discussion
Limitations and Strengths of This Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Deo, P.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [PubMed]
- Barboza-Solís, C.; Acuña-Amador, L.A. The Oral Microbiota: A Literature Review for Updating Professionals in Dentistry. Part I. Odovtos Int. J. Dent. Sci. 2020, 22, 59–68. [Google Scholar] [CrossRef]
- Kilian, M.; Chapple, I.L.C.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.L.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef]
- Pruntel, S.M.; van Munster, B.C.; de Vries, J.J.; Vissink, A.; Visser, A. Oral Health as a Risk Factor for Alzheimer Disease. J. Prev. Alzheimers Dis. 2024, 11, 249–258. [Google Scholar] [CrossRef]
- Herrera, D.; Sanz, M.; Shapira, L.; Brotons, C.; Chapple, I.; Frese, T.; Graziani, F.; Hobbs, F.D.R.; Huck, O.; Hummers, E.; et al. Periodontal diseases and cardiovascular diseases, diabetes, and respiratory diseases: Summary of the consensus report by the European Federation of Periodontology and WONCA Europe. Eur. J. Gen. Pract. 2024, 30, 2320120. [Google Scholar] [CrossRef]
- Yano, Y.; Hua, X.; Wan, Y.; Suman, S.; Zhu, B.; Dagnall, C.L.; Hutchinson, A.; Jones, K.; Hicks, B.D.; Shi, J.; et al. Comparison of Oral Microbiota Collected Using Multiple Methods and Recommendations for New Epidemiologic Studies. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Boutaga, K.; Savelkoul, P.H.; Winkel, E.G.; van Winkelhoff, A.J. Comparison of Subgingival Bacterial Sampling with Oral Lavage for Detection and Quantification of Periodontal Pathogens by Real-Time Polymerase Chain Reaction. J. Periodontol. 2007, 78, 79–86. [Google Scholar] [CrossRef]
- Belstrøm, D.; Sembler-Møller, M.L.; Grande, M.A.; Kirkby, N.; Cotton, S.L.; Paster, B.J.; Holmstrup, P.; Yilmaz, Ö. Microbial profile comparisons of saliva, pooled and site-specific subgingival samples in periodontitis patients. PLoS ONE 2017, 12, e0182992. [Google Scholar] [CrossRef]
- Smola, S.; Rettenberger, G.; Simmet, T.; Burysek, L. Comparison of sample collection methods for the PCR detection of oral anaerobic pathogens. Lett. Appl. Microbiol. 2003, 36, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Maki, K.A.; Kazmi, N.; Barb, J.J.; Ames, N. The Oral and Gut Bacterial Microbiomes: Similarities, Differences, and Connections. Biol. Res. Nurs. 2020, 23, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Bang, E.; Oh, S.; Ju, U.; Chang, H.E.; Hong, J.-S.; Baek, H.-J.; Kim, K.-S.; Lee, H.-J.; Park, K.U. Factors influencing oral microbiome analysis: From saliva sampling methods to next-generation sequencing platforms. Sci. Rep. 2023, 13, 10086. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Liu, J.; Huang, S.; Wang, X.-Y.; Chen, X.; Liu, G.-H.; Ye, K.; Song, W.; Masters, C.L.; Wang, J.; et al. Antiageing strategy for neurodegenerative diseases: From mechanisms to clinical advances. Signal Transduct. Target. Ther. 2025, 10, 76. [Google Scholar] [CrossRef]
- Plachokova, A.S.; Gjaltema, J.; Hagens, E.R.C.; Hashemi, Z.; Knüppe, T.B.A.; Kootstra, T.J.M.; Visser, A.; Bloem, B.R. Periodontitis: A Plausible Modifiable Risk Factor for Neurodegenerative Diseases? A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 4504. [Google Scholar] [CrossRef]
- Arweiler, N.B.; Netuschil, L. The Oral Microbiota. Adv. Exp. Med. Biol. 2016, 902, 45–60. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef]
- D’ambrosio, F.; Santella, B.; Di Palo, M.P.; Giordano, F.; Giudice, R.L. Characterization of the Oral Microbiome in Wearers of Fixed and Removable Implant or Non-Implant-Supported Prostheses in Healthy and Pathological Oral Conditions: A Narrative Review. Microorganisms 2023, 11, 1041. [Google Scholar] [CrossRef]
- Mukai, Y.; Torii, M.; Urushibara, Y.; Kawai, T.; Takahashi, Y.; Maeda, N.; Ohkubo, C.; Ohshima, T. Analysis of plaque microbiota and salivary proteins adhering to dental materials. J. Oral Biosci. 2020, 62, 182–188. [Google Scholar] [CrossRef]
- Singh, H.; Maharaj, R.G.; Naidu, R. Oral health among the elderly in 7 Latin American and Caribbean cities, 1999-2000: A cross-sectional study. BMC Oral Health 2015, 15, 46. [Google Scholar] [CrossRef]
- Wang, T.-F.; Huang, C.-M.; Chou, C.; Yu, S. Effect of oral health education programs for caregivers on oral hygiene of the elderly: A systemic review and meta-analysis. Int. J. Nurs. Stud. 2015, 52, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Richardson, B.N.; Noh, H.I.; Webster, C.I.; Zhang, W.; Kim, S.; Yang, I.; Bai, J. Oral Microbiome, Mental Health, and Sleep Outcomes During the COVID-19 Pandemic: An Observational Study in Chinese and Korean American Immigrants. OMICS J. Integr. Biol. 2023, 27, 180–190. [Google Scholar] [CrossRef]
- Caselli, E.; Fabbri, C.; D’aCcolti, M.; Soffritti, I.; Bassi, C.; Mazzacane, S.; Franchi, M. Defining the oral microbiome by whole-genome sequencing and resistome analysis: The complexity of the healthy picture. BMC Microbiol. 2020, 20, 120. [Google Scholar] [CrossRef]
- Tomás, I.; Regueira-Iglesias, A.; López, M.; Arias-Bujanda, N.; Novoa, L.; Balsa-Castro, C.; Tomás, M. Quantification by qpcr of pathobionts in chronic periodontitis: Development of predictive models of disease severity at site-specific level. Front. Microbiol. 2017, 8, 1443. [Google Scholar] [CrossRef]
- Castillo, Y.; Delgadillo, N.A.; Neuta, Y.; Iniesta, M.; Sanz, M.; Herrera, D.; Pianeta, R.; Lafaurie, G.I.; Castillo, D.M. Design and validation of a quantitative polymerase chain reaction test for the identification and quantification of uncultivable bacteria associated with periodontitis. Arch. Oral Biol. 2023, 154, 105758. [Google Scholar] [CrossRef]
| Participants, n = 28 (%) | p-Value | |
|---|---|---|
| Gender; female | 13 (46%) | |
| Gender; male | 15 (54%) | 0.705 |
| Age, year, mean (SD) | 77.5 (5.3) | |
| Smoking | 17 (60.7) | |
| No medication | 0 (0.0) | |
| 1–4 medications | 15 (53.6) | |
| Polypharmacy | 13 (46.4) | |
| Antibiotic use (<6 months) | 14 (50.0) | |
| Oral status of the participants | ||
| Pocket depth, mean (SD) | 6.3 (1.0) | |
| Number of teeth, mean (SD) | 22.0 (5.4) | |
| Number of carious teeth per participant, mean (SD) | 4 (2.1) | |
| Mucosal pathology | 7 (8.5) | |
| A maxillary partial denture | 4 (14.3) | |
| A mandibular partial denture | 3 (10.7) | |
| A complete maxillary denture | 2 (7.1) | |
| A complete mandibular denture | 0 (0.0) | |
| Participant | Number of Teeth | Pocket Depth | Aa | Pg | Tf | Td | Pi | Fn | Pm | Pn | Cg | Cr | En | Match (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 OF | − | − | +++ | +++ | − | +++ | + | +++ | + | + | − | |||
| 1 PP | 13 | 7 mm | − | − | +++++ | +++++ | − | +++++ | ++++ | ++++++ | ++++ | + | +++ | 91 |
| 1 Swab | − | − | +++ | +++ | − | +++ | ++ | +++ | +++ | + | + | |||
| 2 OF | − | +++ | +++ | +++ | − | +++ | + | ++++ | ++ | ++ | + | |||
| 2 PP | 28 | 6 mm | − | +++ | +++ | +++ | − | ++++ | ++ | ++++ | ++++ | ++ | ++ | 100 |
| 2 Swab | − | +++ | +++ | +++ | − | +++ | + | +++ | + | ++ | + | |||
| 3 OF | − | − | +++ | +++ | − | ++ | + | − | + | ++ | − | |||
| 3 PP | 26 | 7 mm | − | − | ++++ | ++++ | − | ++++ | +++ | +++++ | ++++ | +++ | ++ | 82 |
| 3 Swab | − | − | ++ | ++ | − | ++ | − | − | + | + | + | |||
| 4 OF | − | +++ | +++ | ++++ | − | +++ | +++ | +++ | ++ | ++ | ++ | |||
| 4 PP | 22 | 6 mm | − | ++ | +++ | +++ | − | ++++ | +++ | ++++ | +++ | ++ | ++ | 100 |
| 4 Swab | − | +++ | +++ | +++ | − | +++ | +++ | ++++ | ++ | ++ | ++ | |||
| 5 OF | − | − | ++++ | +++ | − | +++ | ++ | ++++ | ++ | +++ | − | |||
| 5 PP | 26 | 7 mm | − | − | +++++ | ++++ | − | +++++ | ++++ | +++++ | ++++ | +++++ | − | 100 |
| 5 Swab | − | − | +++ | ++ | − | ++ | ++ | +++ | + | ++ | − | |||
| 6 OF | − | − | ++++ | ++++ | − | +++ | +++ | − | ++ | +++ | +++ | |||
| 6 PP | 26 | 7 mm | − | − | ++++ | +++++ | − | +++++ | +++ | − | +++ | ++++ | +++ | 100 |
| 6 Swab | − | − | +++ | +++ | − | +++ | ++ | − | ++ | +++ | ++ | |||
| 7 OF | − | +++ | +++ | +++ | − | +++ | ++ | +++ | +/− | ++ | ++ | |||
| 7 PP | 27 | 8 mm | − | +++++ | +++++ | ++++++ | − | +++++ | ++++ | +++++ | ++++ | ++++ | ++++ | 100 |
| 7 Swab | − | ++ | ++ | +++ | − | ++ | ++ | ++ | +/− | + | ++ | |||
| 8OF | − | +++ | +++ | +++ | +++ | +++ | ++ | ++++ | + | ++ | + | |||
| 8 PP | 25 | 7 mm | − | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | +++++ | ++ | +++ | +++ | 100 |
| 8 Swab | − | ++ | ++ | +++ | ++ | ++ | ++ | ++++ | +/− | ++ | ++ | |||
| 9 OF | − | +++ | +++ | − | − | +++ | +++ | +++ | + | ++ | +++ | |||
| 9 PP | 17 | 6 mm | − | +++ | +++ | − | − | ++++ | +++ | +++++ | +++ | +++ | +++ | 100 |
| 9 Swab | − | ++ | ++ | − | − | + | ++ | ++ | + | + | ++ | |||
| 10 OF | − | − | +++ | +++ | − | +++ | ++ | − | + | +++ | − | |||
| 10 PP | 27 | 7 mm | − | − | +++ | +++ | − | +++++ | +++ | +++++ | +++ | ++ | − | 91 |
| 10 Swab | − | − | ++ | ++ | − | ++ | + | +++ | +/− | + | − | |||
| 11 OF | − | +++ | +++ | ++++ | +++ | +++ | + | ++++ | + | +++ | ++ | |||
| 11 PP | 17 | 7 mm | − | +++ | ++++ | +++++ | ++++ | ++++ | ++ | +++++ | +++ | +++++ | +++ | 100 |
| 11 Swab | − | ++ | ++ | +++ | ++ | +++ | − | +++ | +/− | ++ | ++ | |||
| 12 OF | − | − | +++ | +++ | +++ | +++ | ++ | ++++ | + | +++ | + | |||
| 12 PP | 25 | 5 mm | − | − | +++ | +++ | +++ | ++++ | ++ | ++++ | +++ | +++++ | + | 100 |
| 12 Swab | − | − | ++ | +++ | ++ | ++ | + | ++ | + | ++ | + | |||
| 13 OF | − | − | ++++ | ++++ | − | ++++ | +++ | − | − | +++ | ++ | |||
| 13 PP | 27 | 7 mm | − | − | +++++ | ++++ | +++ | +++++ | ++++ | +++ | ++ | ++++++ | +++ | 82 |
| 13 Swab | − | − | ++ | ++ | − | ++ | ++ | − | − | + | +/− | |||
| 14 OF | − | − | ++ | +++ | − | +++ | +++ | ++++ | + | ++ | − | |||
| 14 PP | 26 | 5 mm | − | − | ++ | +++ | − | ++++ | ++ | +++ | ++ | +++ | − | 100 |
| 14 Swab | − | − | ++ | ++ | − | ++++ | +++ | ++++ | +++ | +++ | − | |||
| 15 OF | − | − | − | +++ | +++ | ++++ | +++ | +++++ | ++ | ++++ | +++ | |||
| 15 PP | 25 | 7 mm | − | − | − | ++++ | ++++ | +++++ | +++ | ++++ | +++ | ++++ | ++ | 91 |
| 15 Swab | − | − | − | +++ | +++ | ++ | ++ | − | +/− | ++ | ++ | |||
| 16 OF | − | +++ | +++ | ++++ | − | +++ | ++ | − | − | +++ | − | |||
| 16 PP | 18 | 5 mm | − | +++ | ++++ | ++++ | − | +++++ | ++++ | ++++ | ++ | +++ | − | 91 |
| 16 Swab | − | +++ | ++ | +++ | − | ++ | +/- | +++ | − | ++ | − | |||
| 17 OF | − | ++ | +++ | +++ | − | +++ | +++ | +++ | − | ++ | +/− | |||
| 17 PP | 21 | 7 mm | − | + | ++++ | +++++ | − | +++++ | ++++ | +++++ | +++ | ++++ | ++ | 73 |
| 17 Swab | − | + | − | ++ | − | ++ | − | ++ | + | + | + | |||
| 18 OF | − | +++ | ++++ | ++++ | ++++ | +++ | +++ | ++ | ++ | +++ | + | |||
| 18 PP | 25 | 8 mm | − | +++++ | +++++ | ++++++ | ++++ | +++++ | +++ | +++ | +++ | +++++ | +++ | 100 |
| 18 Swab | − | ++ | ++ | +++ | ++ | ++ | + | ++ | +/− | ++ | + | |||
| 19 OF | − | − | − | − | − | ++ | + | − | + | − | − | |||
| 19 PP | 11 | 5 mm | − | − | − | − | − | ++++ | ++ | − | +++ | +/− | − | 100 |
| 19 Swab | − | − | − | − | − | +/− | − | − | +/− | − | − | |||
| 20 OF | − | ++ | +++ | − | − | +++ | ++ | +++ | + | +++ | − | |||
| 20 PP | 15 | 5 mm | − | ++ | +++ | − | − | +++++ | +++ | ++++ | +++ | ++++ | − | 100 |
| 20 Swab | − | ++ | ++ | − | − | ++ | + | ++ | +/− | + | − | |||
| 21 OF | ++ | +++ | − | ++++ | ++++ | +++ | + | − | ++ | ++ | + | |||
| 21 PP | 22 | 7 mm | +++++ | +++++ | − | ++++++ | +++++ | ++++++ | +++++ | − | ++++ | +++++ | ++ | 100 |
| 21 Swab | + | +++ | − | ++++ | +++ | ++ | ++ | − | + | ++ | + | |||
| 22 OF | − | − | ++ | − | − | + | +/− | ++ | ++ | + | − | |||
| 22 PP | 24 | 5 mm | − | − | +++ | − | − | +++ | +/− | +++ | +++ | ++ | − | 100 |
| 22 Swab | − | − | ++ | − | − | + | +/− | ++ | + | + | − | |||
| 23 OF | − | +++ | ++++ | +++ | +++ | ++++ | ++++ | ++++ | +++ | +++ | + | |||
| 23 PP | 16 | 7 mm | − | ++++ | ++++ | +++++ | ++++ | +++++ | ++++ | ++++++ | ++++ | ++++ | ++ | 100 |
| 23 Swab | − | +++ | +++ | +++ | +++ | ++ | ++ | +++ | + | ++ | +/- | |||
| 24 OF | − | − | ++ | +++ | − | +++ | + | − | + | ++ | − | |||
| 24 PP | 22 | 5 mm | − | ++ | ++ | ++ | − | ++++ | ++++ | +++ | ++ | ++ | − | 91 |
| 24 Swab | − | − | ++ | ++ | − | ++ | + | − | + | + | − | |||
| 25 OF | − | +++ | +++ | ++++ | +++ | ++ | + | − | + | ++ | +/- | |||
| 25 PP | 28 | 5 mm | − | +++ | +++ | ++++ | +++ | ++++ | ++ | ++++ | ++ | ++ | + | 91 |
| 25 Swab | − | ++ | ++ | +++ | ++ | ++ | + | +++ | +/− | + | +/- | |||
| 26 OF | − | +++ | ++++ | − | − | ++++ | +++ | ++++ | ++ | ++ | − | |||
| 26 PP | 24 | 5 mm | − | ++ | +++ | − | − | +++ | +++ | +++ | ++ | +++ | − | 100 |
| 26 Swab | − | +++ | +++ | − | − | ++ | ++ | +++ | ++ | + | − | |||
| 27 OF | − | +++ | ++++ | ++++ | ++++ | +++ | ++ | ++++ | + | ++ | +++ | |||
| 27 PP | 16 | 7 mm | − | ++++ | +++++ | +++++ | ++++ | +++++ | ++++ | ++++++ | +++ | ++++ | +++ | 91 |
| 27 Swab | − | ++++ | ++++ | +++++ | ++++ | +++ | +++ | − | +/− | +++ | ++++ | |||
| 28 OF | − | − | +++ | +++ | − | ++ | ++ | +++ | + | ++ | − | |||
| 28 PP | 10 | 7 mm | − | − | +++++ | +++++ | − | +++++ | ++++ | +++++ | ++++ | +++++ | − | 100 |
| 28 Swab | − | − | +++ | ++ | − | ++ | ++ | +++ | + | + | − |
| Bacteria Species | p-Value |
|---|---|
| Aggregatibacter actinomycetemcomitans | 1.000 |
| Porphyromonas gingivalis | 0.368 |
| Tannerella forsythia | 0.368 |
| Treponema denticola | 1.000 |
| Fusobacterium nucleatum | 0.368 |
| Prevotella intermedia | 1.000 |
| Parvimonas micra | 0.018 |
| Prevotalla nigrescens | 0.021 |
| Eubacterium nodatum | 0.097 |
| Eikenella corrodens | 0.368 |
| Campylobacter concisus | 0.135 |
| Bacteria Match (%) | Number of Participants | Number Smoking (%) | Polypharmacy (%) | Antibiotic Use (%) |
|---|---|---|---|---|
| 100 | 18 | 10 (59) | 9 (69) | 9 (64) |
| 91 | 7 | 5 (29) | 2 (15) | 4 (29) |
| 82 | 2 | 2 (12) | 1 (8) | 0 (0) |
| 73 | 1 | 0 (0) | 1 (8) | 1 (7) |
| Cost of Sample Material Anno 2024 in The Netherlands (€) | Cost of Laboratory Analysis Anno 2024 | Participant Comfort | Time of Sampling | Ease of Sampling (for the Practitioner) | Accessibility of the Sample Location | |
|---|---|---|---|---|---|---|
| Swab | (0.03) + | 75 euro | + | ++ | ++ | ++ |
| Paper point | (0.12) + | 75 euro | − | − | − | − |
| Oral fluid collection | (0.15) + | 75 euro | ++ | − | − | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pruntel, S.M.; Valk, H.d.; Vissink, A.; Visser, A. Exploring Three Methods for Sampling Oral Microbiota in Older People: A Comparative Study. Appl. Sci. 2025, 15, 10297. https://doi.org/10.3390/app151810297
Pruntel SM, Valk Hd, Vissink A, Visser A. Exploring Three Methods for Sampling Oral Microbiota in Older People: A Comparative Study. Applied Sciences. 2025; 15(18):10297. https://doi.org/10.3390/app151810297
Chicago/Turabian StylePruntel, Sanne M., Hanneke de Valk, Arjan Vissink, and Anita Visser. 2025. "Exploring Three Methods for Sampling Oral Microbiota in Older People: A Comparative Study" Applied Sciences 15, no. 18: 10297. https://doi.org/10.3390/app151810297
APA StylePruntel, S. M., Valk, H. d., Vissink, A., & Visser, A. (2025). Exploring Three Methods for Sampling Oral Microbiota in Older People: A Comparative Study. Applied Sciences, 15(18), 10297. https://doi.org/10.3390/app151810297








