Fine-Tuning a Large Language Model for the Classification of Diseases Caused by Environmental Pollution
Abstract
1. Introduction
1.1. Impact of Pollution on Health
1.2. Challenges for Early Diagnosis
1.3. Advances in Large Language Models
1.4. Challenges in Medical Text Classification
1.5. Research Objectives
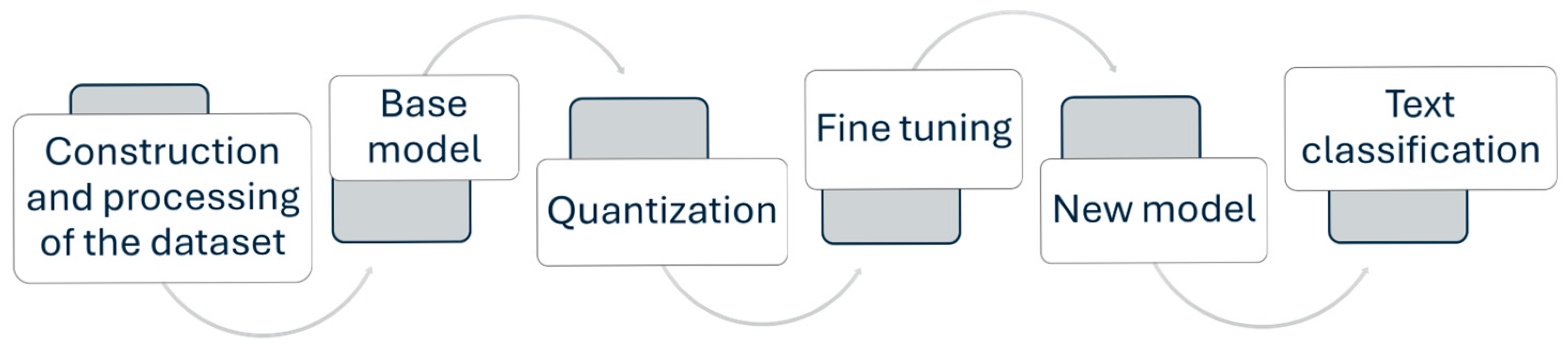
2. Materials and Methods
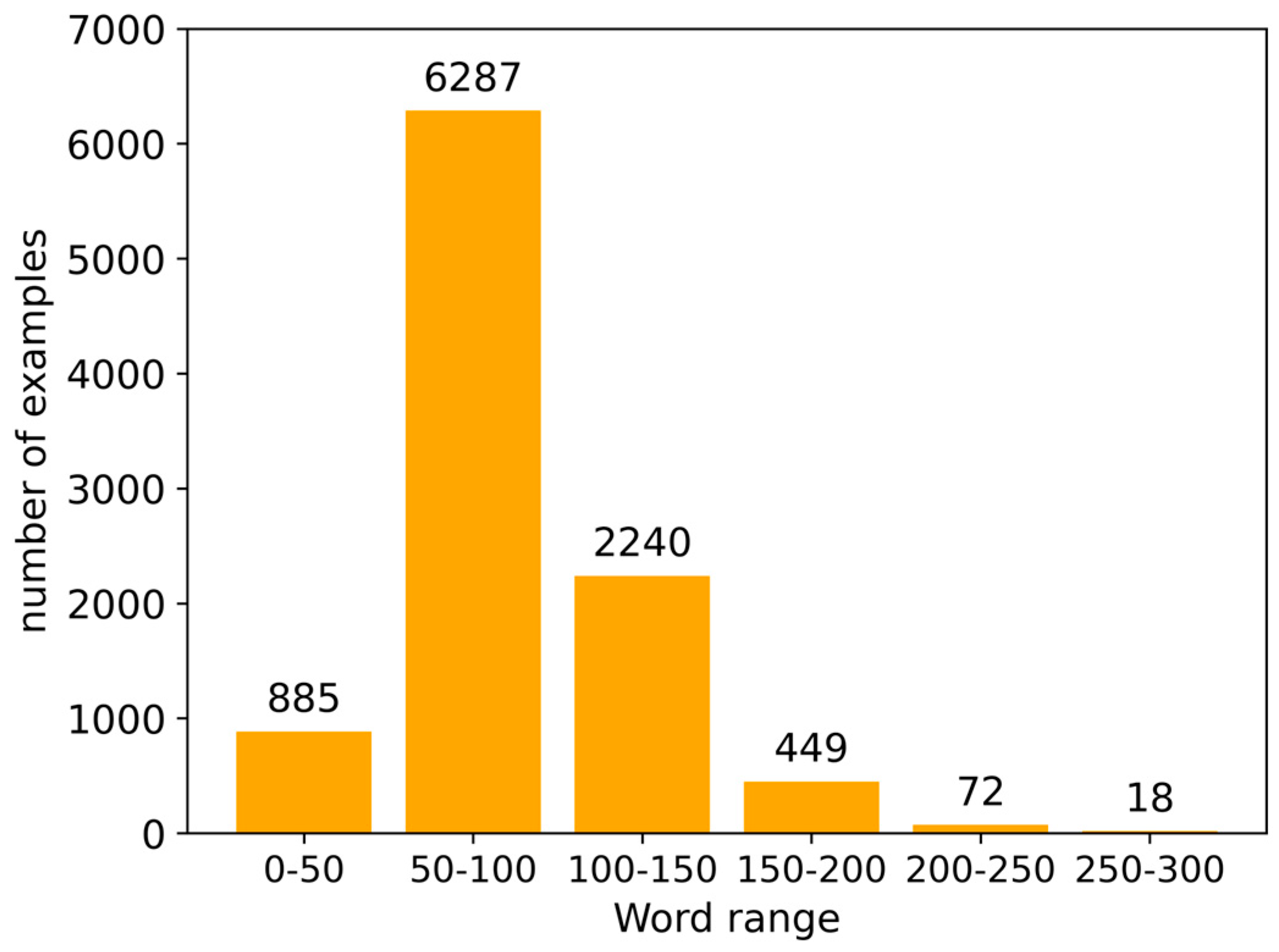
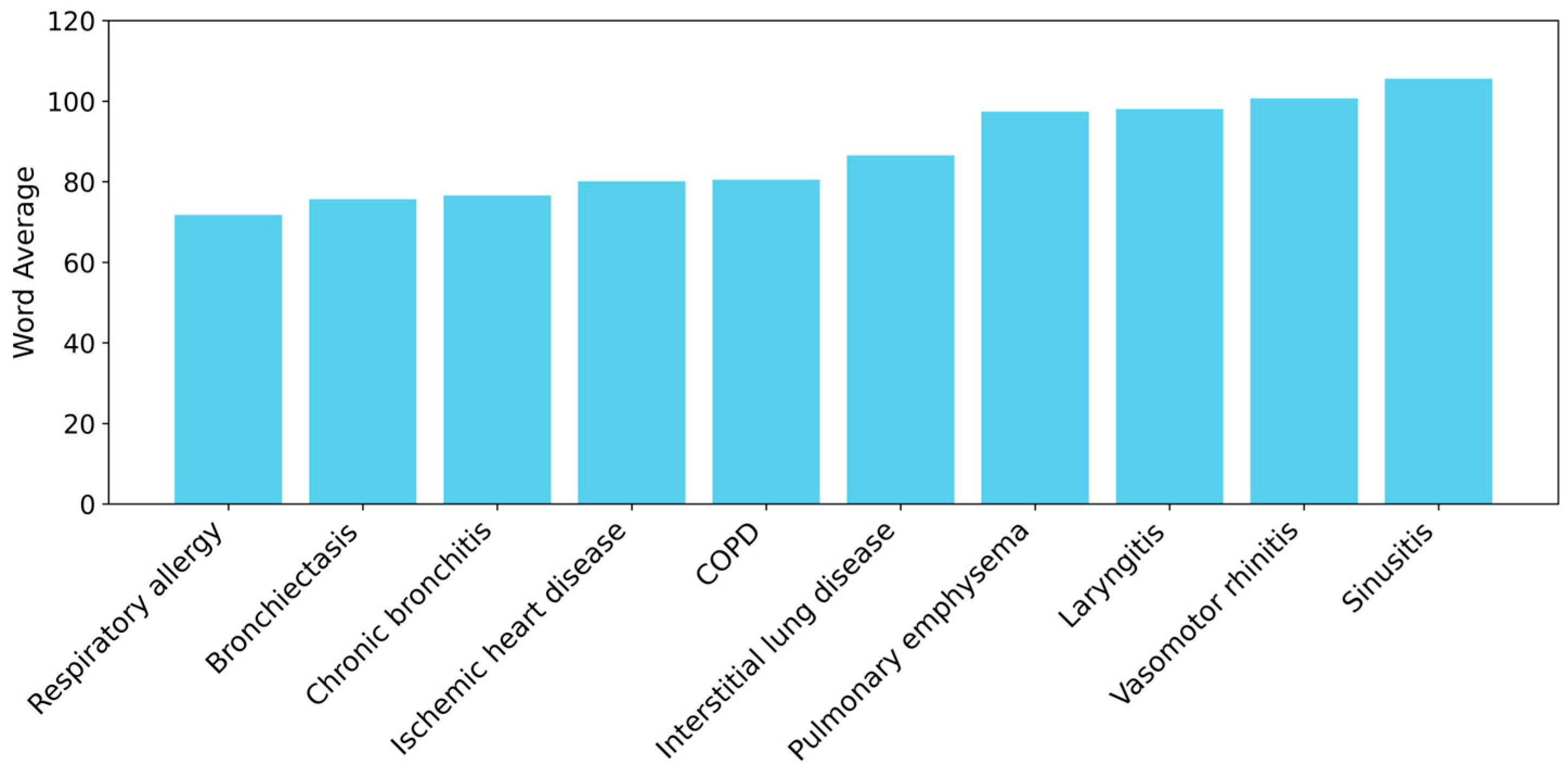
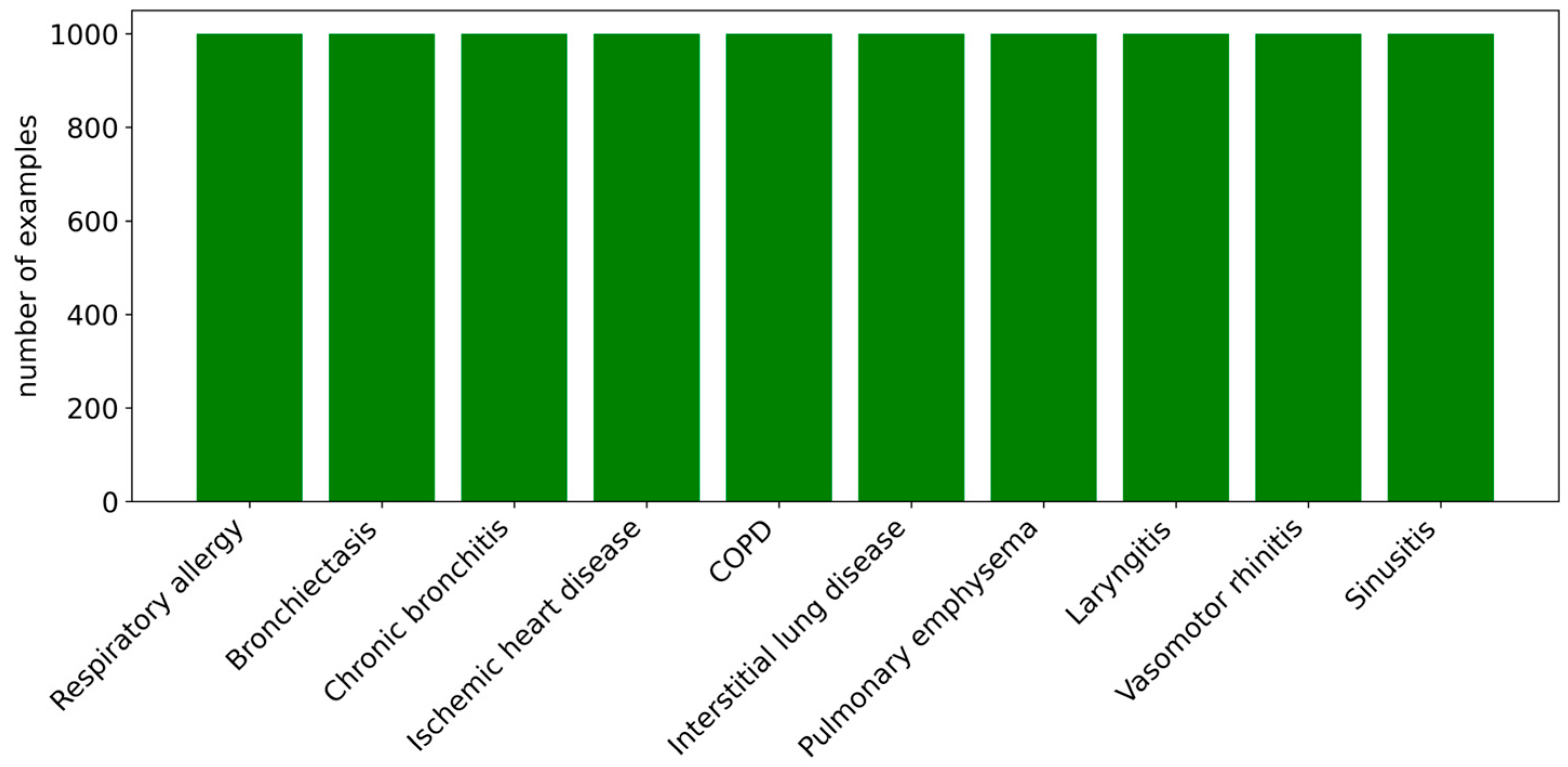
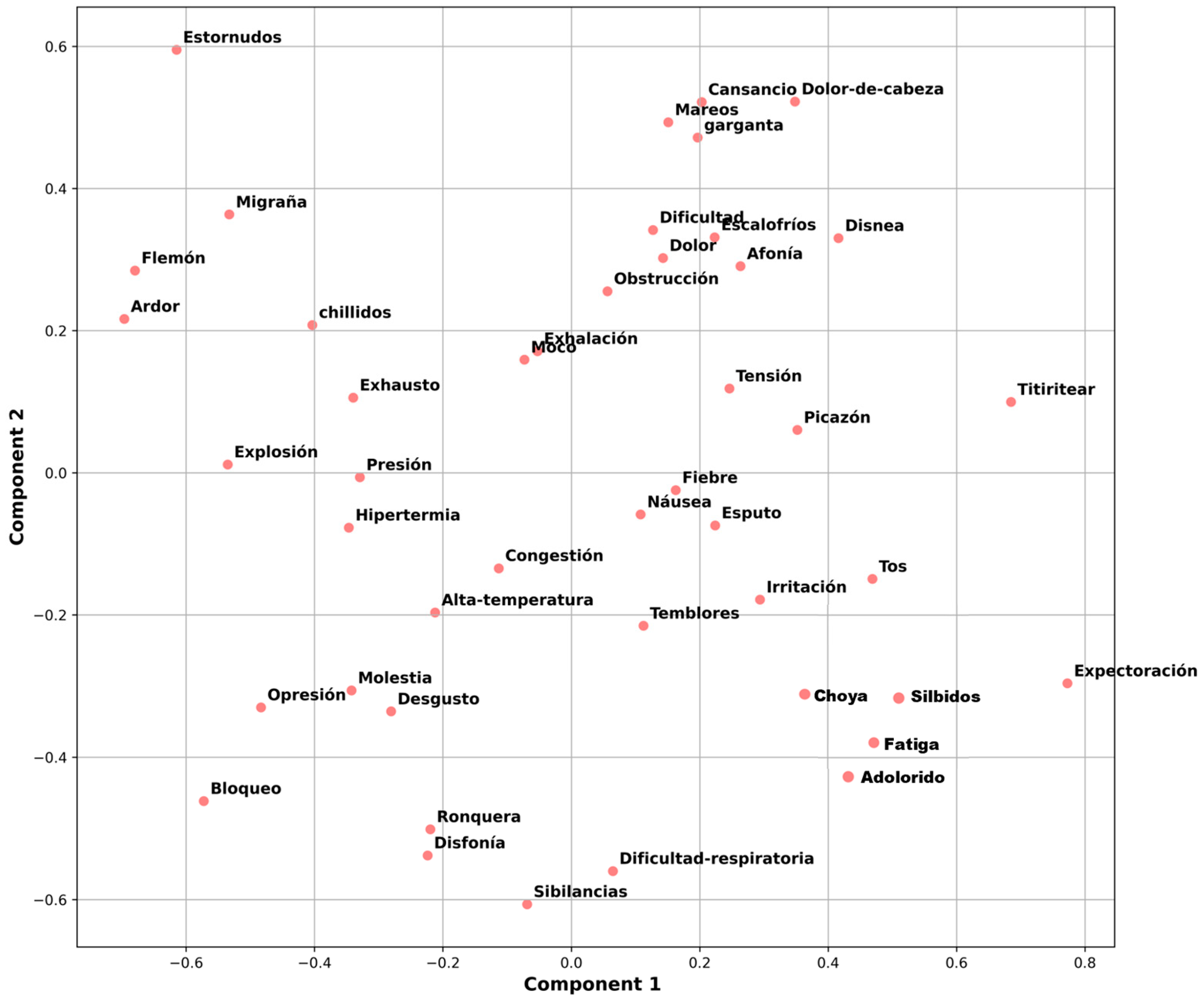
2.1. Construction and Processing of the Dataset
- Laryngitis;
- Bronchiectasis;
- Ischemic heart disease;
- Chronic bronchitis;
- Pulmonary emphysema;
- Sinusitis;
- Respiratory allergy;
- COPD (Chronic Obstructive Pulmonary Disease);
- Interstitial lung disease;
- Vasomotor rhinitis.
- Disease label: Each record in the dataset is associated with a label corresponding to a specific disease. Each category is represented by 1000 examples, resulting in a total of 10,000 examples in Spanish. Importantly, the dataset is balanced, ensuring that the model can make predictions without bias toward any particular class.
- Data augmentation: To enhance the robustness of the dataset, several augmentation techniques were applied, including the use of synonyms, translations, misspellings, and common slang expressions.
- Normalization. The text is adjusted to ensure consistency by removing unnecessary special characters, standardizing the use of uppercase and lowercase letters, and applying lemmatization (reducing words to their root form). This procedure reduces model complexity and enhances generalization.
- Noise removal. Irrelevant or repetitive elements that could interfere with model learning—such as excessive punctuation and stopwords (e.g., the, of, and)—were removed.
- Class balance verification. The dataset was evaluated to ensure that the classes were sufficiently balanced, thereby preventing bias toward categories with a larger number of examples.
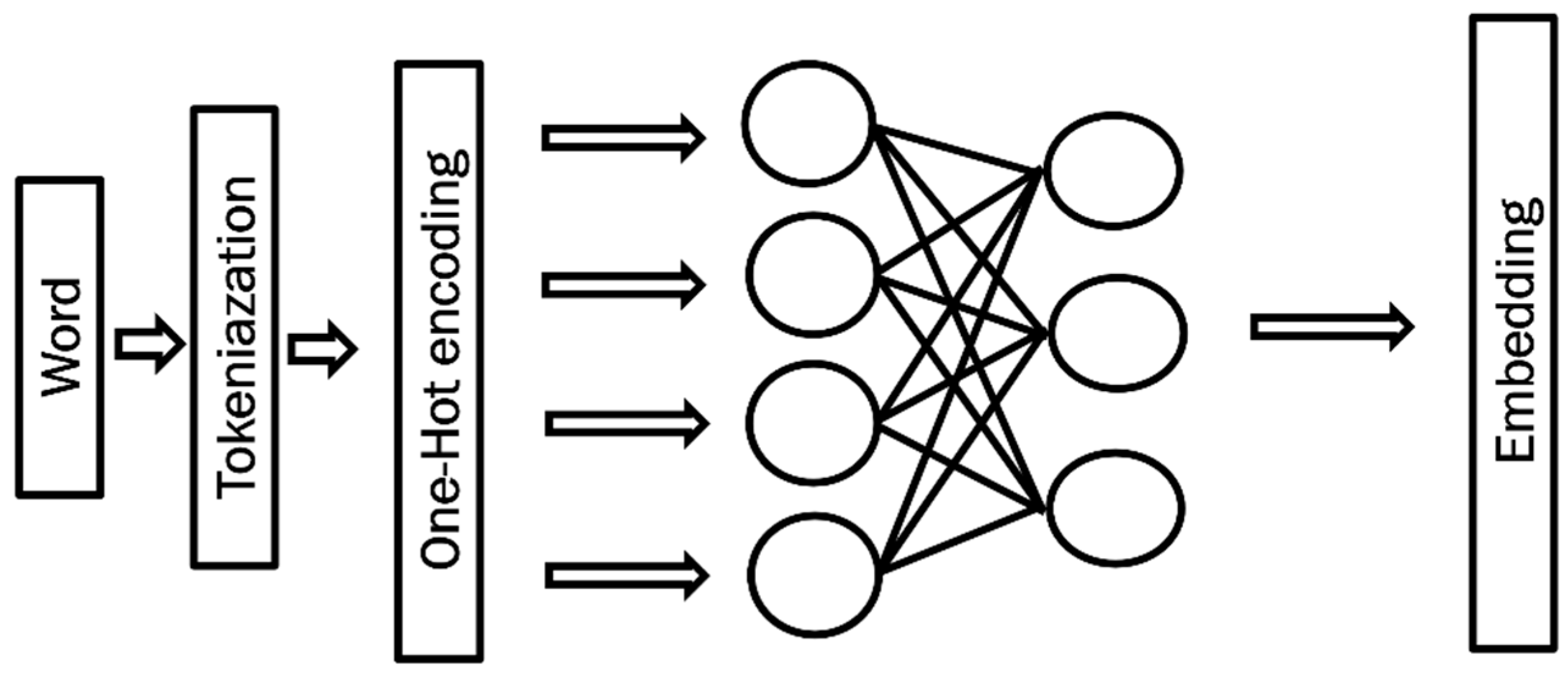
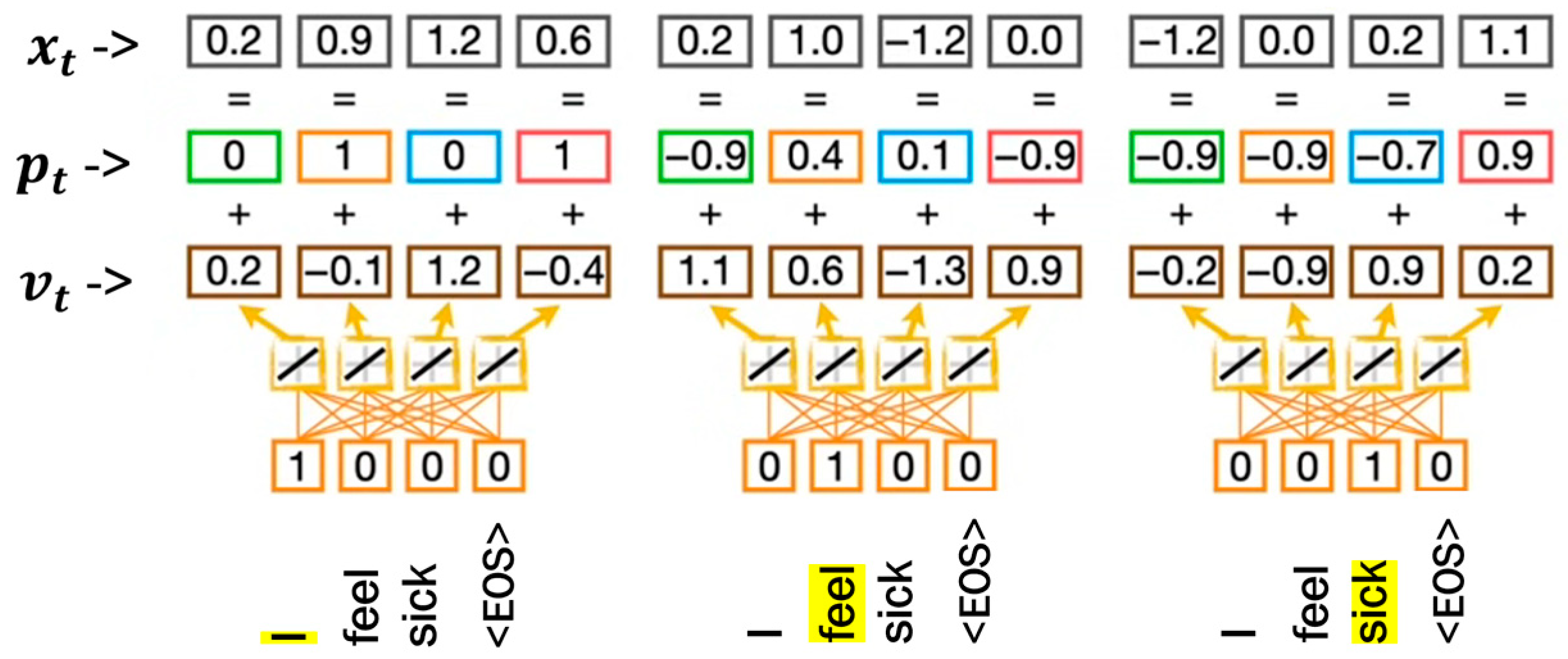
- Tokenization and embedding. The symptom reports were segmented into smaller units, such as words or subwords. Subsequently, each token was transformed into a high-dimensional numerical vector through embedding. These dense vector representations capture both semantic meaning and contextual relationships, enabling the model to interpret the underlying patterns in the reported symptoms.
2.2. Base Model
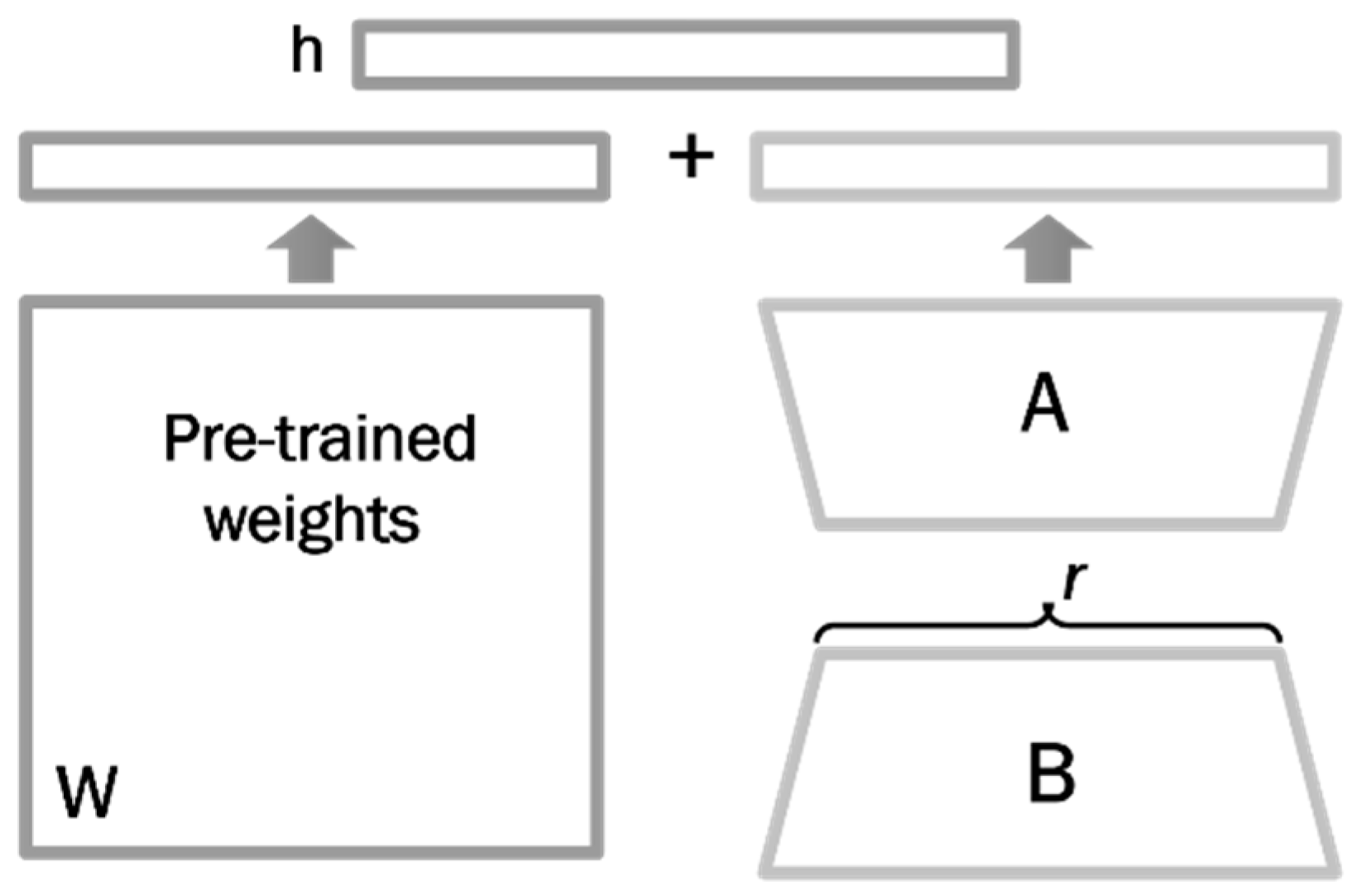
2.3. Quantization
2.4. LoRA
- Central Processing Unit (CPU): Intel Core i5 12th Gen;
- Graphics Processing Unit (GPU): NVIDIA GeForce RTX 4060 Ti;
- Operating System: Windows 10 Pro.
| Algorithm 1: Fine tuning pseudocode |
 |
3. Results
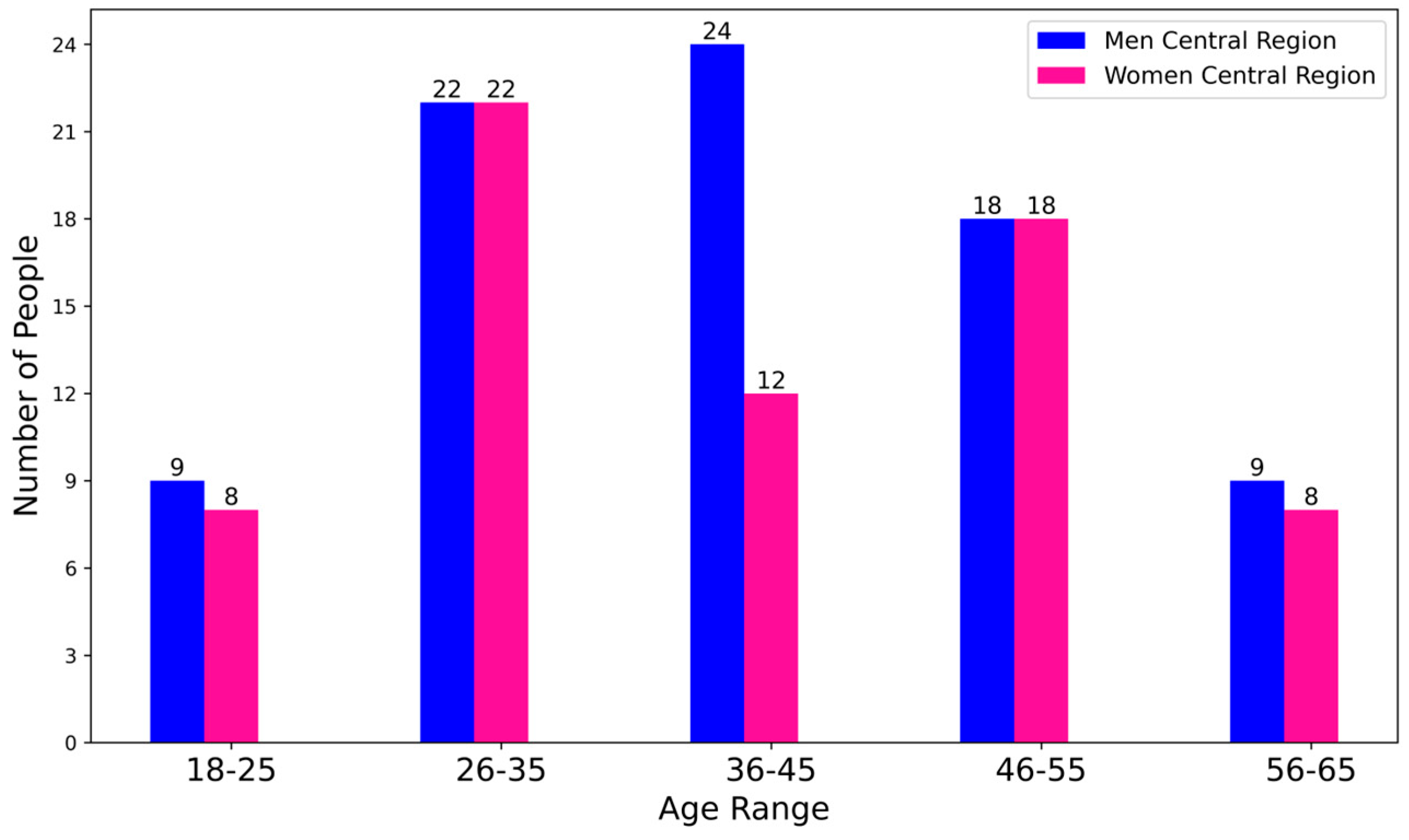
3.1. Dataset
- Controlled paraphrasing. New instances were generated by synonymously rewording the symptom descriptions, maintaining their original meaning while varying both structure and vocabulary. This procedure introduced variations in the same symptom set without altering its clinical essence.
- Variation in description length. Entries were modified by altering their length, either by adding supplementary details or by simplifying existing content, thereby capturing greater diversity in the expression of symptoms.
- Introduction of controlled typographical errors. Typographical and spelling errors were intentionally introduced to simulate human data-entry behavior. This strategy enhanced the model’s robustness by teaching it to handle common errors likely to appear in real-world data.
- Translation and back-translation. A translation strategy was employed in which texts were translated into other languages and subsequently back into the original language, thereby introducing additional variability in symptom descriptions while preserving key information.
3.2. Text Processing
3.3. Training
- Test set (20%): This subset contains instances not included during training, providing an unbiased assessment of the model’s final performance. Since the model has not been exposed to these data beforehand, it cannot adjust its parameters to them, thereby enabling the evaluation of its generalization capability and its effectiveness on unseen or real-world data that may be encountered in future applications.
- Validation set (20%): This subset was employed to evaluate the model’s performance throughout training and to guide the adjustment of hyperparameters, including learning rate and batch size. As a dataset distinct from the training set, it enabled assessment of the model on unseen data, thereby preventing overfitting and promoting effective generalization.
- Training set (60%): This subset was employed to train the language model and optimize its parameters. It comprises the majority of the examples and enables the model to learn the relationships between symptoms and their associated diseases. By exposing the model to a wide variety of instances, the training set allows the model to learn patterns and correlations within the data, thereby supporting accurate predictions and relevant responses when applied to new or unseen cases.
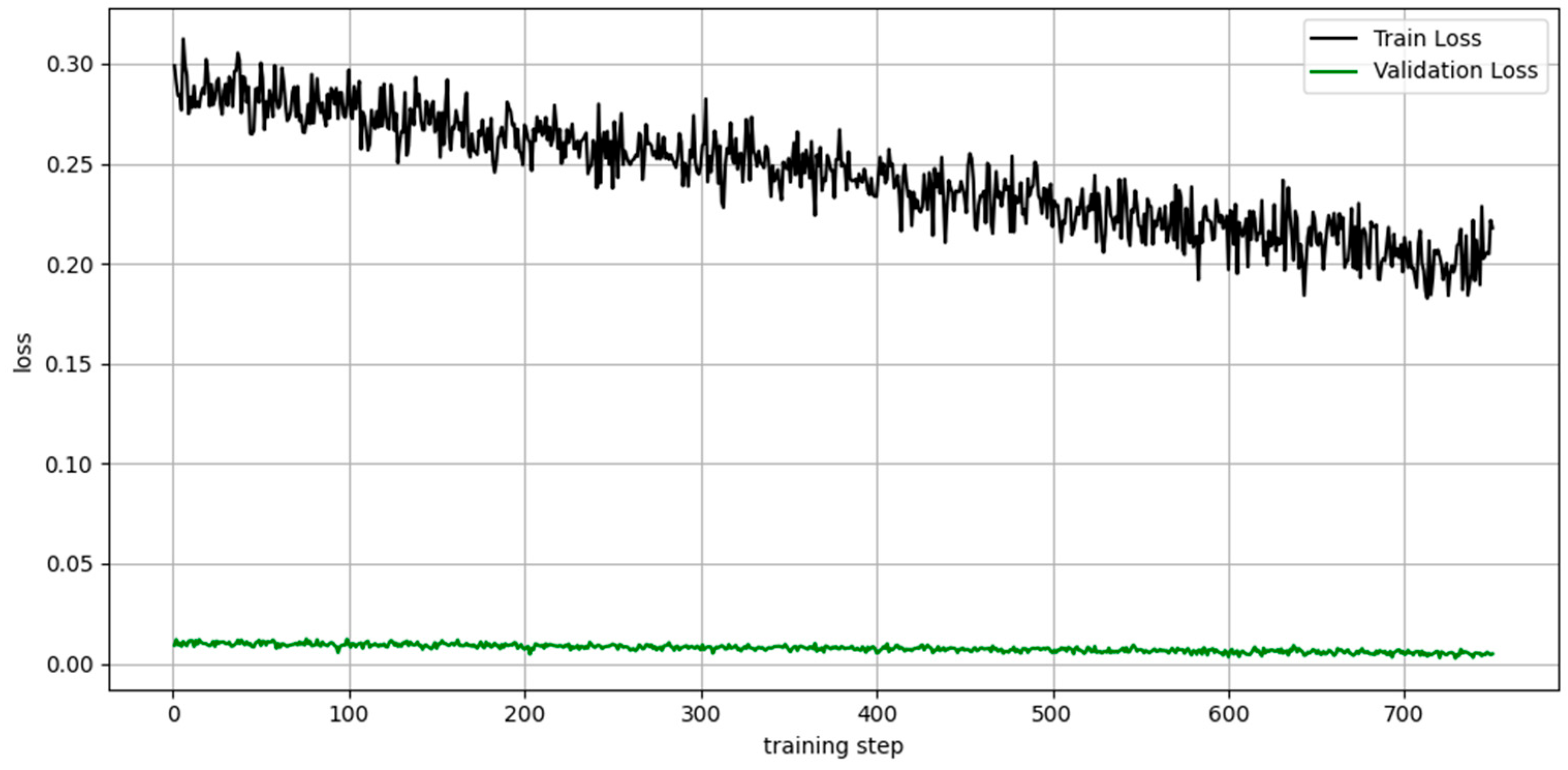
3.3.1. Evolution of the Loss During Training
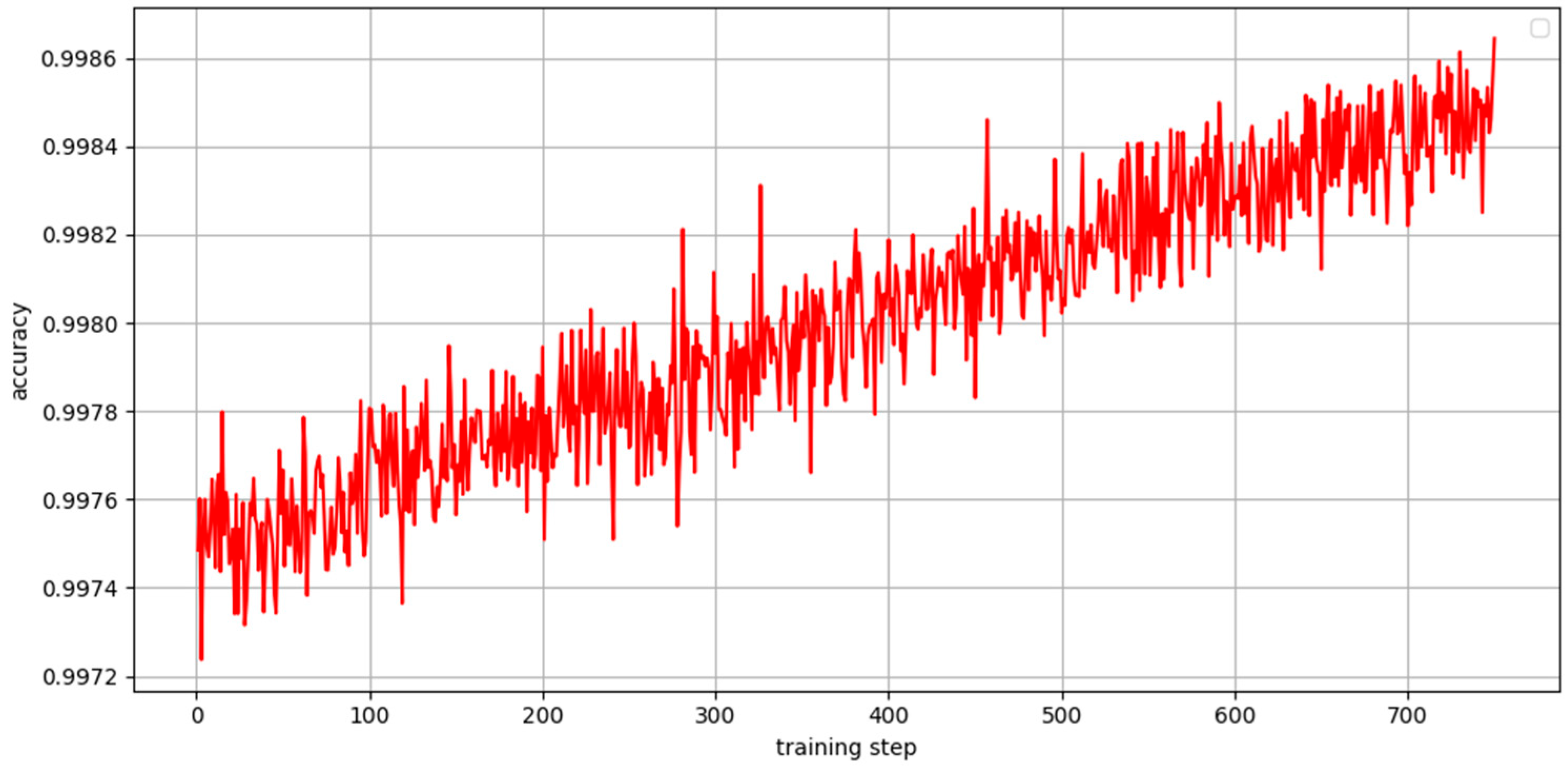
3.3.2. Evolution of Accuracy During Training
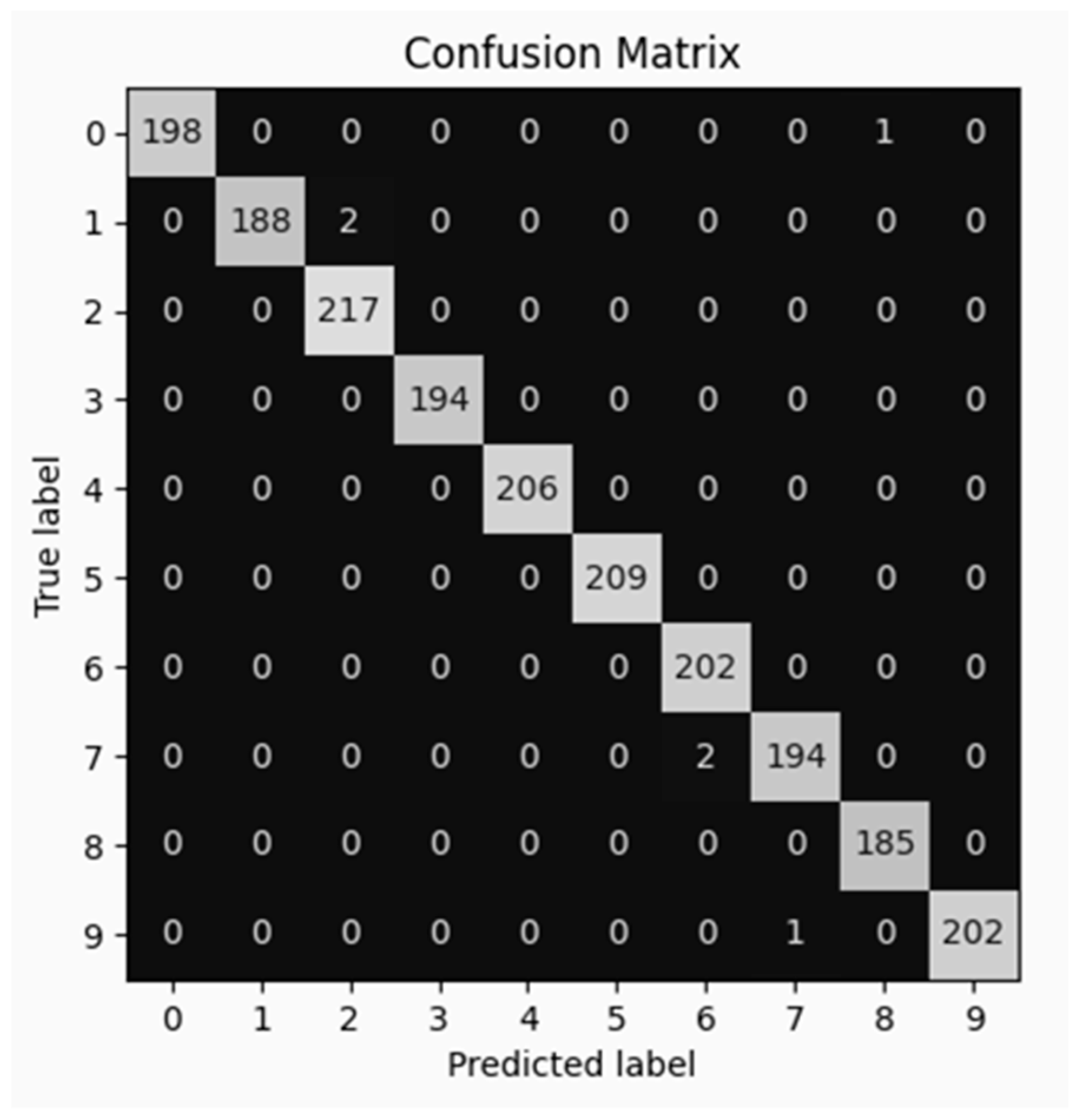
3.3.3. Confusion Matrix
3.3.4. F1 Score
- True positives (
- False positives (
- False Negatives (
- Precision (
- Recall (
- F1 score (
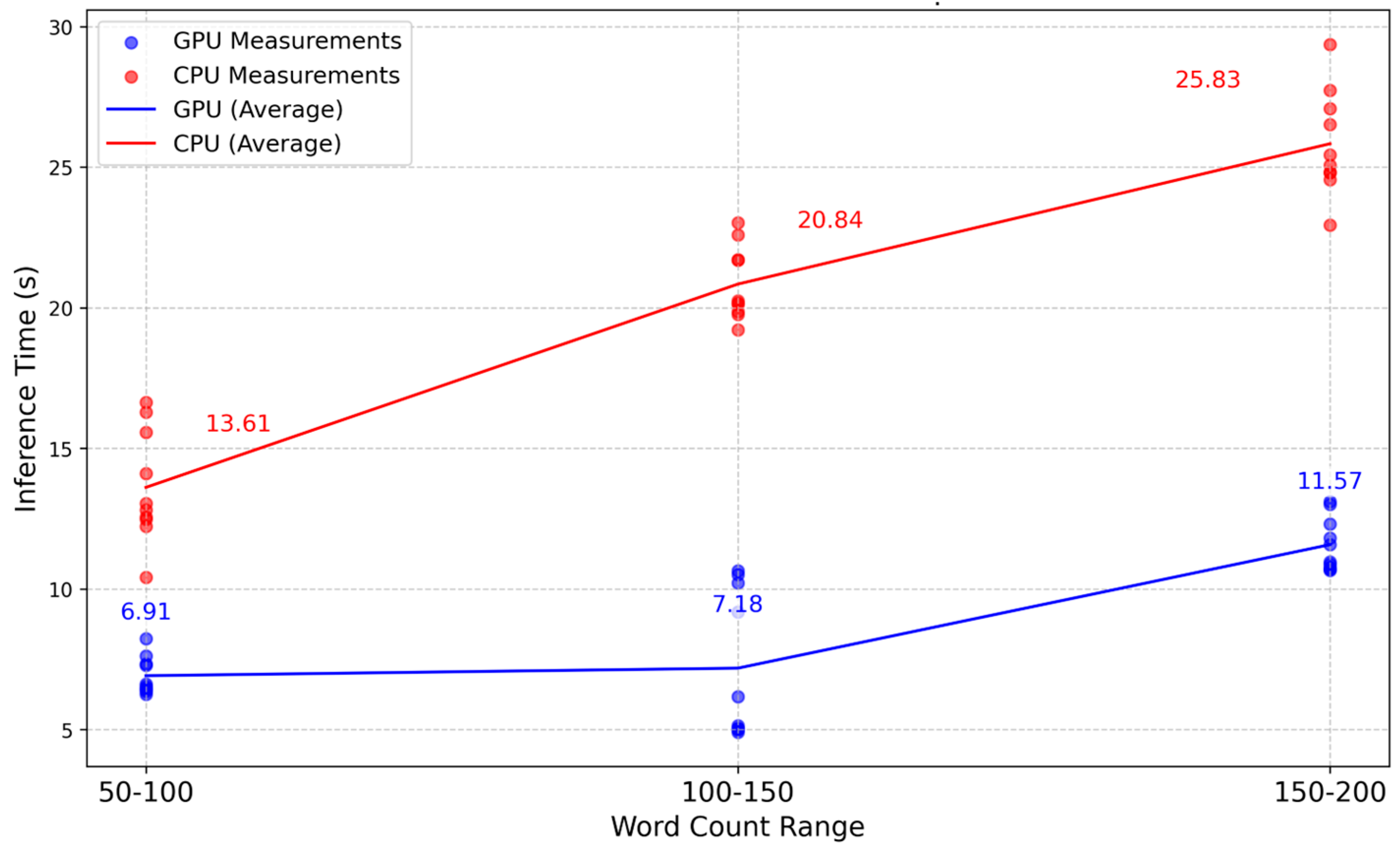
3.4. Inference Time
3.5. Comparison with the Base Model
3.6. Language
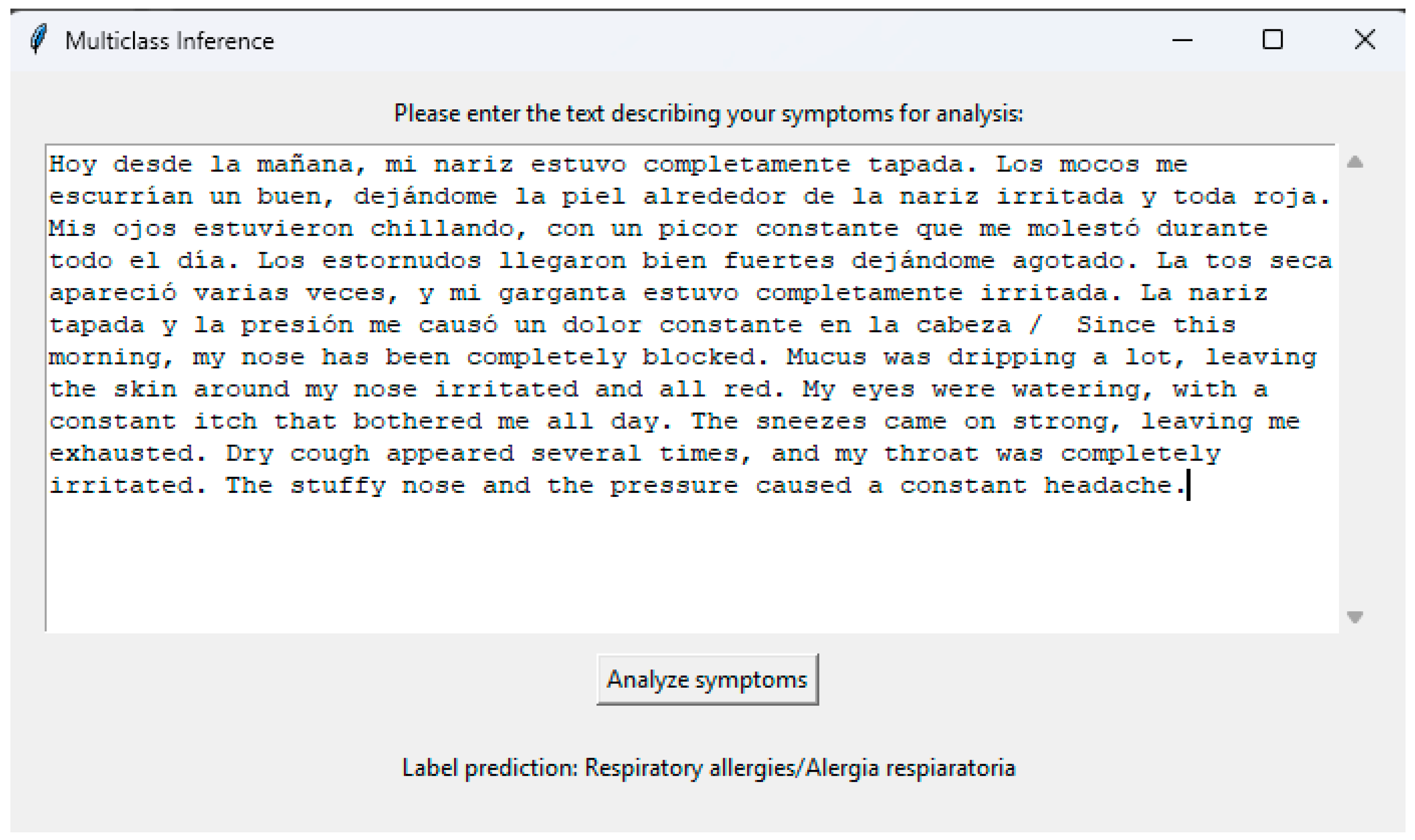
3.7. Graphical User Interface (GUI)
3.8. Qualitative Error Analysis
- Chronic bronchitis
- COPD (Chronic Obstructive Pulmonary Disease)
4. Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PM2.5 | Particulate Matter 2.5 micrometers |
| NO2 | Nitrogen Dioxide |
| O3 | Ozone |
| COPD | Chronic Obstructive Pulmonary Disease |
| AI | Artificial Intelligence |
| NLP | Natural Language Processing |
| LLM | Large Language Model |
| RNN | Recurrent Neural Network |
| GPU | Graphics Processing Unit |
| FFN | Feed-Forward Network |
| QLoRA | Quantized Low-Rank Adaptation |
| LoRA | Low-Rank Adaptation |
| CPU | Central Processing Unit |
| PCA | Principal Component Analysis |
| GUI | Graphical User Interface |
References
- Goldsmith, J.R. Environmental Pollution and Its Effects on Health; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Jurafsky, D.; Martin, J.H. Speech and Language Processing, 3rd ed.; Pearson: London, UK, 2020. [Google Scholar]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Al Nazi, Z.; Peng, W. Large language models in healthcare and medical domain: A review. Informatics 2024, 11, 57. [Google Scholar] [CrossRef]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Petroni, F.; Rocktäschel, T.; Lewis, P.; Bakhtin, A.; Wu, Y.; Miller, A.H.; Riedel, S. Language models as knowledge bases? arXiv 2019, arXiv:1909.01066. [Google Scholar] [CrossRef]
- Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M.A.; Lacroix, T.; Rozière, B.; Goyal, N.; Hambro, E.; Azhar, F.; et al. LLaMA: Open and efficient foundation language models. arXiv 2023, arXiv:2302.13971. [Google Scholar] [CrossRef]
- Radford, A.; Narasimhan, K.; Salimans, T.; Sutskever, I. Improving Language Understanding by Generative Pre-Training. 2018. Available online: https://cdn.openai.com/research-covers/language-unsupervised/language_understanding_paper.pdf (accessed on 17 July 2025).
- Chowdhery, A.; Narang, S.; Devlin, J.; Bosma, M.; Mishra, G.; Roberts, A.; Barham, P.; Chung, H.W.; Sutton, C.; Gehrmann, S.; et al. Palm: Scaling language modeling with pathways. arXiv 2022, arXiv:2204.02311. [Google Scholar] [CrossRef]
- Alsentzer, E.; Murphy, J.; Boag, W.; Weng, W.-H.; Jin, D.; Naumann, T.; McDermott, M. Publicly Available Clinical BERT Embeddings. In Proceedings of the 2nd Clinical Natural Language Processing Workshop, Minneapolis, MN, USA, 7 June 2019. [Google Scholar]
- Strubell, E.; Ganesh, A.; McCallum, A. Energy and Policy Considerations for Deep Learning in NLP. In Proceedings of the ACL 2019, Florence, Italy, 1–6 July 2019. [Google Scholar]
- Patterson, D.; Gonzalez, J.; Le, Q.; Liang, C.; Munguia, L.M.; Rothchild, D.; So, D. Carbon Emissions and Large Neural Network Training. arXiv 2021, arXiv:2104.10350. [Google Scholar] [CrossRef]
- Hu, E.J.; Shen, Y.; Wallis, P.; Allen-Zhu, Z.; Li, Y.; Wang, S.; Wang, L.; Chen, W.; Chen, Y.; Li, H. LoRA: Low-Rank Adaptation of Large Language Models. arXiv 2021, arXiv:2106.09685. [Google Scholar]
- Dettmers, T.; Lewis, M.; Belkada, Y.; Zettlemoyer, L.; Su, J. QLoRA: Efficient Finetuning of Quantized LLMs. arXiv 2023, arXiv:2305.14314. [Google Scholar] [CrossRef]
- Peng, Y.; Yan, S.; Lu, Z. Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets. In Proceedings of the 18th BioNLP Workshop and Shared Task, Florence, Italy, 1 August 2019; pp. 58–65. [Google Scholar] [CrossRef]
- Bousquet, J.; Khaltaev, N.; Cruz, A.A.; Denburg, J.; Fokkens, W.J.; Togias, A.; Zuberbier, T. Allergic diseases and asthma in the era of environmental pollution: A global perspective. World Allergy Organ. J. 2020, 13, 100118. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Dufter, P.; Schmitt, M.; Schütze, H. Position Information in Transformers: An Overview. Comput. Linguist. 2022, 48, 733–763. [Google Scholar] [CrossRef]
- Li, Y. Theoretical Analysis of Positional Encodings in Transformer Models: Impact on Expressiveness and Generalization. arXiv 2025, arXiv:2506.06398. [Google Scholar] [CrossRef]
- Zhou, D.; Shi, Y.; Kang, B.; Yu, W.; Jiang, Z.; Li, Y.; Jin, X.; Hou, Q.; Feng, J. Refiner: Refining self-attention for vision transformers. arXiv 2021, arXiv:2106.03714. [Google Scholar] [CrossRef]
- Al Nazi, Z.; Mashrur, F.R.; Islam, M.A.; Saha, S. Fibro-cosanet: Pulmonary fibrosis prognosis prediction using a convolutional self attention network. Phys. Med. Biol. 2021, 66, 225013. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Dong, L.; Wei, F.; Xu, K. Self-attention attribution: Interpreting information interactions inside transformer. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual, 2–9 February 2021; Volume 35, pp. 12963–12971. [Google Scholar]
| Features | 8B | 70B | 405B |
|---|---|---|---|
| FFN Dimension | 14,336 | 28,672 | 53,548 |
| Key/Value Heads | 8 | 8 | 8 |
| Layers | 32 | 80 | 126 |
| Model Dimension | 4096 | 8192 | 16,384 |
| Attention Heads | 32 | 64 | 128 |
| Vocabulary Size | 128,000 | ||
| Positional Embeddings | RoPE (θ = 500,000) | ||
| Hyperparameter | Value | Description |
|---|---|---|
| Learning_rate | 1 × 10−4 | Learning rate for the optimizer |
| per_device_train_batch_size | 8 | Batch size per device during training |
| LoRA rank (r) | 16 | Rank for the LoRA adaptation |
| LoRA alpha | 8 | Rank for the LoRA adaptation |
| per_device_eval_batch_size | 8 | Batch size per device during evaluation |
| LoRA_dropout | 0.05 | Dropout forregularization within LoRA |
| Max_LEN | 512 | Maximum token length for input truncation |
| Class | Precision | Recall | F1 Score |
|---|---|---|---|
| 0 Respiratory allergies | 1.0000 | 0.9950 | 0.9975 |
| 1 Bronchiectasis | 1.0000 | 0.9895 | 0.9947 |
| 2 Chronic bronchitis | 0.9857 | 1.0000 | 0.9928 |
| 3 ischemic heart disease | 1.0000 | 1.0000 | 1.0000 |
| 4 COPD | 1.0000 | 1.0000 | 1.0000 |
| 5 Interstitial lung disease | 1.0000 | 1.0000 | 1.0000 |
| 6 Pulmonary emphysema | 0.9902 | 1.0000 | 0.9951 |
| 7 Laryngitis | 0.9948 | 0.9898 | 0.9923 |
| 8 Vasomotor rhinitis | 0.9946 | 1.0000 | 0.9973 |
| 9 Sinusitis | 1.0000 | 0.9951 | 0.9975 |
| Class | Base Model F1 Score | F1 Score of the Fine-Tuned Model |
|---|---|---|
| 0 Respiratory allergies | 0.7245 | 0.9975 |
| 1 Bronchiectasis | 0.8031 | 0.9947 |
| 2 Chronic bronchitis | 0.7615 | 0.9928 |
| 3 ischemic heart disease | 0.7159 | 1.0000 |
| 4 COPD | 0.6698 | 1.0000 |
| 5 Interstitial lung disease | 0.7365 | 1.0000 |
| 6 Pulmonary emphysema | 0.6242 | 0.9951 |
| 7 Laryngitis | 0.7459 | 0.9923 |
| 8 Vasomotor rhinitis | 0.7629 | 0.9973 |
| 9 Sinusitis | 0.7595 | 0.9975 |
| Description | Classification | Analysis |
|---|---|---|
| Te escribo porke la neta ando bien jodido del pecho, Ai rato me falta el aire hasta pa ir a la tienda y me canzo bien rapido. la tos no me deja, sobre todo en las mañanas y siempre estoy escupiendo mocos güeros o amarillos bien feos. Abeces se me hace como un silbidito en el pecho, y asta me duele cuando respiro hondo. Ando todo cansado hasta pa subir un escalón y ya ni ganas de comer tengo. Se me ponen morados los labios y las uñas, y la neta me asusto, yo fumaba un buen antes pero ahorita ya ni un cigarro puedo | EPOC | porke—porque canzo—cansó abeces—a veces astá—hasta La neta—La Verdad Ando bien jodido—Estoy muy enfermo Un buen—mucho Mocos gueros—Flemas amariillas |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Angeles, J.F.; Rosales-Silva, A.J.; Vianney-Kinani, J.M.; Posadas-Durán, J.P.F.; Gallegos-Funes, F.J.; Velázquez-Lozada, E.; Miranda-González, A.A.; Uriostegui-Hernandez, D.; Estrada-Soubran, J.M. Fine-Tuning a Large Language Model for the Classification of Diseases Caused by Environmental Pollution. Appl. Sci. 2025, 15, 9772. https://doi.org/10.3390/app15179772
Hernández-Angeles JF, Rosales-Silva AJ, Vianney-Kinani JM, Posadas-Durán JPF, Gallegos-Funes FJ, Velázquez-Lozada E, Miranda-González AA, Uriostegui-Hernandez D, Estrada-Soubran JM. Fine-Tuning a Large Language Model for the Classification of Diseases Caused by Environmental Pollution. Applied Sciences. 2025; 15(17):9772. https://doi.org/10.3390/app15179772
Chicago/Turabian StyleHernández-Angeles, Julio Fernando, Alberto Jorge Rosales-Silva, Jean Marie Vianney-Kinani, Juan Pablo Francisco Posadas-Durán, Francisco Javier Gallegos-Funes, Erick Velázquez-Lozada, Armando Adrián Miranda-González, Dilan Uriostegui-Hernandez, and Juan Manuel Estrada-Soubran. 2025. "Fine-Tuning a Large Language Model for the Classification of Diseases Caused by Environmental Pollution" Applied Sciences 15, no. 17: 9772. https://doi.org/10.3390/app15179772
APA StyleHernández-Angeles, J. F., Rosales-Silva, A. J., Vianney-Kinani, J. M., Posadas-Durán, J. P. F., Gallegos-Funes, F. J., Velázquez-Lozada, E., Miranda-González, A. A., Uriostegui-Hernandez, D., & Estrada-Soubran, J. M. (2025). Fine-Tuning a Large Language Model for the Classification of Diseases Caused by Environmental Pollution. Applied Sciences, 15(17), 9772. https://doi.org/10.3390/app15179772








