Very First Application of Compact Benchtop NMR Spectrometers to Complex Biofluid Analysis and Metabolite Tracking for Future Metabolomics Studies: A Retrospective Decennial Report from November 2014
Abstract
1. Introduction
2. Materials and Methods
2.1. Urine Sample Collection
2.2. Urine Sample Storage, Processing and Preparation
2.3. LF 1H NMR Measurements at an Operating Frequency of 60 MHz
Signal-to-Noise (STN) Ratio Values
2.4. MF NMR Measurements (Operating Frequency 400 MHz)
2.5. Assignment of 1H NMR Resonances of Human Urine Samples
3. Results
3.1. Very First LF 60 MHz 1H NMR Spectra of Human Urine
3.2. Prima Facie Benchtop 1H NMR Analysis of Urine Collected from Patients with NPC1 Disease and Their Heterozygous Carrier Controls
4. Discussion of Early LF Benchtop NMR Experiments Applied to NPC1 Disease Urine
Compact LF 1H NMR Monitoring of LSDs and Associated Gastric Permeability Complications in Human Urine
5. Developments, Updates and Future Perspectives of the Metabolomics Applications and Potential of LF Benchtop NMR Analysis
5.1. Benchtop NMR-Based Metabolomics Analysis of Type 2 Diabetes (T2D) in Humans
5.2. Value of Compact NMR Analysis of Biofluids for the Diagnosis of Tuberculosis (TB)
5.3. Benchtop NMR Spectroscopic Applications to Chronic Kidney Disease Diagnosis in Cats
5.4. Benchtop NMR-Based Metabolomics Investigation of Colitis in Mice
5.5. Neonatal Sepsis in Humans
5.6. Benchtop NMR Estimation of Inflammatory and Cardiovascular Disease Markers in Human Blood Plasma and Serum
6. Limitations of Our Original 2014 Benchtop NMR Study, and Longitudinal Improvements in the Technology Since That Time
6.1. Water Resonance Suppression in Biofluid Samples for Compact 1H NMR Analysis
6.2. Dealing with Resonance Superimposition Challenges at LF Operating Frequencies
6.3. Potential Confounding Bioanalytical Effects of Added 2H2O
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Percival, B.C.; Grootveld, M.; Gibson, M.; Osman, Y.; Molinari, M.; Jafari, F.; Sahota, T.; Martin, M.; Casanova, F.; Mather, M.L.; et al. Low-field, benchtop NMR spectroscopy as a potential tool for point-of-care diagnostics of metabolic conditions: Validation, protocols and computational models. High Throughput 2019, 8, 2. [Google Scholar] [CrossRef]
- Grootveld, M.; Percival, B.C.; Gibson, M.; Osman, Y.; Edgar, M.; Molinari, M.; Mather, M.L.; Casanova, F.; Wilson, P.B. Progress in low-field benchtop NMR spectroscopy in chemical and biochemical analysis. Anal. Chim. Acta 2019, 1067, 11–30. [Google Scholar] [CrossRef]
- Leenders, J.; Grootveld, M.; Percival, B.C.; Gibson, M.; Casanova, F.; Wilson, P.B. Benchtop low frequency 60 MHz NMR analysis of urine: A comparative metabolomics investigation. Metabolites 2020, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Edgar, M.; Percival, B.C.; Gibson, M.; Jafari, F.; Grootveld, M. Low-field benchtop NMR spectroscopy as a potential non-stationary tool for point-of-care urinary metabolite tracking in diabetic conditions. Diabetes Res. Clin. Pract. 2021, 171, 108554. [Google Scholar] [CrossRef] [PubMed]
- Percival, B.C.; Wann, A.; Taylor, S.; Edgar, M.; Gibson, M.; Grootveld, M. Metabolomics distinction of cigarette smokers from non-smokers using non-stationary benchtop NMR analysis of human saliva. In Oral Health Care—An Important Issue of the Modern Society; Ardelean, L., Ed.; Dentistry; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- Grootveld, M.; Page, G.; Bhogadia, M.; Edgar, M. Updates and original case studies focused on the NMR-linked metabolomics analysis of human oral fluids Part I: Emerging platforms and perspectives. Appl. Sci. 2022, 12, 1235. [Google Scholar] [CrossRef]
- Edgar, M.; Kuhn, S.; Page, G.; Grootveld, M. Computational simulation of 1H NMR profiles of complex biofluid analyte mixtures at differential operating frequencies: Applications to low-field benchtop spectra. Magn. Reson. Chem. 2022, 60, 1097. [Google Scholar] [CrossRef]
- Mierisová, Š.; Ala-Korpela, M. MR spectroscopy quantitation: A review of frequency domain methods. NMR Biomed. 2001, 14, 247–259. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0—The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D6008–D6117. [Google Scholar] [CrossRef]
- Ruiz-Rodado, V.; Luque-Baena, R.M.; te Vruchte, D.; Probert, F.; Lachmann, R.H.; Hendriksz, C.J.; Wraith, J.E.; Imrie, J.; Elizondo, D.; Sillence, D.; et al. 1H NMR-linked urinary metabolic profiling of Niemann-Pick class C1 (NPC1) disease: Identification of potential new biomarkers using correlated component regression (CCR) and genetic algorithm (GA) analysis strategies. Curr. Metabolomics 2014, 2, 88–121. [Google Scholar] [CrossRef]
- Gouilleux, B.; Charrier, B.; Akoka, S.; Felpin, F.X.; Rodriguez-Zubiri, M.; Giraudeau, P. Ultrafast 2D NMR on a benchtop spectrometer: Applications and perspectives. TrAC Trends Anal. Chem. 2016, 83, 65–75. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. NMR Metabolomics Methods for investigating disease. Anal. Chem. 2023, 95, 83–99. [Google Scholar] [CrossRef]
- Lhoste, C.; Lorandel, B.; Praud, C.; Marchand, A.; Mishra, R.; Dey, A.; Bernard, A.; Dumez, J.N.; Giraudeau, P. Ultrafast 2D NMR for the analysis of complex mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 2022, 130–131, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Silwood, C.J.L.; Lynch, E.; Claxson, A.W.D.; Grootveld, M.C. (1)H and (13)C NMR spectroscopic analysis of human saliva. J. Dent. Res. 2002, 81, 422–427. [Google Scholar] [CrossRef]
- Available online: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&ved=2ahUKEwi1q-7F9dKOAxWRQEEAHZnMINMQFnoECBAQAQ&url=https%3A%2F%2Fmagritek.com%2F2018%2F07%2F15%2F1h-and-13c-peak-assignments-of-quinine-using-1d-and-2d-nmr-methods-part-3%2F&usg=AOvVaw0Lo4lTbby-Blu4_79XdOix&opi=89978449 (accessed on 22 July 2025).
- Leal, A.F.; Espejo-Mojica, A.J.; Sánchez, O.F.; Ramírez, C.M.; Reyes, L.H.; Cruz, J.C.; Alméciga-Díaz, C.J. Lysosomal storage diseases: Current therapies and future alternatives. J. Mol. Med. 2020, 98, 931–946. [Google Scholar] [CrossRef]
- Vinet, B.; Panzini, B.; Boucher, M.; Massicotte, J. Automated enzymatic assay for the determination of sucrose in serum and urine and Its use as a marker of gastric damage. Clin. Chem. 1998, 44, 2369–2371. [Google Scholar] [CrossRef]
- Sutherland, L.R.; Verhoef, M.; Wallace, J.L.; Van Rosendaal, G.; Crutcher, R.; Meddings, J.B. A simple, non-invasive marker of gastric damage: Sucrose permeability. Lancet 1994, 343, 998–1000. [Google Scholar] [CrossRef] [PubMed]
- Meddings, J.B.; Sutherland, L.R.; Byles, N.I.; Wallace, J.L. Sucrose: A novel permeability marker for gastroduodenal disease. Gastroenterology 1993, 104, 1619–1626. [Google Scholar] [CrossRef]
- Macura, B.; Kiecka, A.; Szczepanik, M. Intestinal permeability disturbances: Causes, diseases and therapy. Clin. Exp. Med. 2024, 24, 232. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, R.J.; Bodamer, O.A.; Watson, M.S.; Wilcox, W.R. Lysosomal storage diseases: Diagnostic confirmation and management of presymptomatic individuals. Genet. Med. 2011, 13, 457–484. [Google Scholar] [CrossRef]
- Zhang, Z.; Yue, P.; Lu, T.; Wang, Y.; Wei, Y.; Wei, X. Role of lysosomes in physiological activities, diseases, and therapy. J. Hematol. Oncol. 2021, 14, 79. [Google Scholar] [CrossRef] [PubMed]
- Carubbi, F.; Barbato, A.; Burlina, A.B.; Francini, F.; Mignani, R.; Pegoraro, E.; Landini, L.; De Danieli, G.; Bruni, S.; Strazzullo, P.; et al. Nutrition in adult patients with selected lysosomal storage diseases. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.; Pastores, G.M. Hepatic manifestations of lysosomal storage disorders: Differential diagnosis, investigations, and treatment, current and upcoming. EMJ 2021, 6, 70–79. [Google Scholar]
- Poorthuis, B. Lysosomal Storage Disorders. In Inherited Metabolic Disease in Adults: A Clinical Guide; Hollak, C.E.M., Lachmann, R., Eds.; online edn.; Oxford Monographs on Medical Genetics: New York, NY, USA, 2016; Oxford Academic: Oxford, UK, 2016. [Google Scholar] [CrossRef]
- Suhre, K.; Wallaschofski, H.; Raffler, J.; Nele, F.; Haring, R.; Michael, K.; Wasner, C.; Krebs, A.; Kronenberg, F.; Chang, D.; et al. A genome-wide association study of metabolic traits in human urine. Nat. Genet. 2011, 43, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Moreno, P.; Rodriguez, I.; Izquierdo-Garcia, J.L. Benchtop NMR-based metabolomics: First steps for biomedical application. Metabolites 2023, 13, 614. [Google Scholar] [CrossRef]
- Percival, B.C.; Leenders, J.; Grootveld, M. ‘State-of-the-Art’ metabolomics investigations of type 2 diabetes. In Obesity and Diabetes: New Surgical and Nonsurgical Approaches; Faintuch, J., Ed.; Springer Nature Publications Ltd.: London, UK, 2020; pp. 265–282. [Google Scholar]
- Stolz, M.; Schlawne, C.; Hoffmann, J.; Hartmann, V.; Marini, I.; Fritsche, A.; Peter, A.; Bakchoul, T.; Schick, F. Feasibility of precise and reliable glucose quantification in human whole blood samples by 1 tesla benchtop NMR. NMR Biomed 2020, 33, e4358. [Google Scholar] [CrossRef]
- Reddy, N.; Verma, N.; Dungan, K. Monitoring technologies-continuous glucose monitoring, mobile technology, biomarkers of glycemic control. In Endotext [Internet] 2023; Feingold, K.R., Ahmed, S.F., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar] [PubMed]
- Izquierdo-Garcia, J.L.; Comella-Del-Barrio, P.; Campos-Olivas, R.; Villar-Hernandez, R.; Prat-Aymerich, C.; De Souza-Galvao, M.L.; Jimenez-Fuentes, M.A.; Ruiz-Manzano, J.; Stojanovic, Z.; Gonzalez, A.; et al. Discovery and validation of an NMR-based metabolomic profile in urine as TB biomarker. Sci. Rep. 2020, 10, 22317. [Google Scholar] [CrossRef]
- Comella-Del-Barrio, P.; Izquierdo-Garcia, J.L.; Gautier, J.; Doresca, M.J.C.; Campos-Olivas, R.; Santiveri, C.M.; Muriel-Moreno, B.; Prat-Aymerich, C.; Abellana, R.; Perez-Porcuna, T.M.; et al. Urine NMR-based TB metabolic fingerprinting for the diagnosis of TB in children. Sci. Rep. 2021, 11, 12006. [Google Scholar] [CrossRef]
- Ruiz-Cabello, J.; Sevilla, I.A.; Olaizola, E.; Bezos, J.; Miguel-Coello, A.B.; Munoz-Mendoza, M.; Beraza, M.; Garrido, J.M.; Izquierdo-Garcia, J.L. Benchtop nuclear magnetic resonance-based metabolomic approach for the diagnosis of bovine tuberculosis. Transbound Emerg. Dis. 2021, 69, e859–e870. [Google Scholar] [CrossRef]
- Finch, N.; Percival, B.; Hunter, E.; Blagg, R.J.; Blackwell, E.; Sagar, J.; Ahmad, Z.; Chang, M.W.; Hunt, J.A.; Mather, M.L.; et al. Preliminary demonstration of benchtop NMR metabolic profiling of feline urine: Chronic kidney disease as a case study. BMC Res. Notes 2021, 14, 469. [Google Scholar] [CrossRef]
- Grootveld, M. Metabolic Profiling: Disease and Xenobiotics; Royal Society of Chemistry Publications: Cambridge, UK, 2014; Issues in Toxicology Series. [Google Scholar]
- Haslauer, K.E.; Hemmler, D.; Schmitt-Kopplin, P.; Heinzmann, S.S. Guidelines for the use of deuterium oxide (D2O) in 1H NMR metabolomics. Anal. Chem. 2019, 91, 11063–11069. [Google Scholar] [CrossRef]
- Song, Z.; Ohnishi, Y.; Osada, S.; Gan, L.; Jiang, J.; Hu, Z.; Kumeta, H.; Kumaki, Y.; Yokoi, Y.; Nakamura, K.; et al. Application of benchtop NMR for metabolomics study using feces of mice with DSS-Induced colitis. Metabolites 2023, 13, 611. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fleischmann-Struzek, C.; Goldfarb, D.M.; Schlattmann, P.; Schlapbach, L.J.; Reinhart, K.; Kissoon, N. The global burden of paediatric and neonatal sepsis: A systematic review. Lancet Respir. Med. 2018, 6, 223–230. [Google Scholar] [CrossRef]
- Stocchero, M.; Cannet, C.; Napoli, C.; Demetrio, E.; Baraldi, E.; Giordano, G. Low-Field Benchtop NMR to Discover Early-Onset Sepsis: A Proof of Concept. Metabolites 2023, 13, 1029. [Google Scholar] [CrossRef]
- Nitschke, P.; Lodge, S.; Hall, D.; Schaefer, H.; Spraul, M.; Embade, N.; Millet, O.; Holmes, E.; Wist, J.; Nicholson, J.K. Direct low field J-edited diffusional proton NMR spectroscopic measurement of COVID-19 inflammatory biomarkers in human serum. Analyst 2022, 147, 4213–4221. [Google Scholar] [CrossRef]
- Wist, J.; Nitschke, P.; Conde, R.; de Diego, A.; Bizkarguenaga, M.; Lodge, S.; Hall, D.; Chai, Z.; Wang, W.; Kowlessur, S.; et al. Benchtop proton NMR spectroscopy for high-throughput lipoprotein quantification in human serum and plasma. Anal. Chem. 2025, 97, 6399–6409. [Google Scholar] [CrossRef] [PubMed]
- Giraudeau, P.; Silvestre, V.; Akoka, S. Optimizing water suppression for quantitative NMR-based metabolomics: A tutorial review. Metabolomics 2015, 11, 1041–1055. [Google Scholar] [CrossRef]
- Hoult, D.I. Solvent peak saturation with single phase and quadrature fourier transformation. J. Magn. Reson. 1976, 21, 337–347. [Google Scholar] [CrossRef]
- Gouilleux, B.; Charrier, B.; Akoka, S.; Giraudeau, P. Gradient-based solvent suppression methods on a benchtop spectrometer. Magn. Reson. Chem. 2017, 55, 91–98. [Google Scholar] [CrossRef]
- Gouilleux, B.; Farjon, J.; Giraudeau, P. Gradient-based pulse sequences for benchtop NMR spectroscopy. J. Magn. Reson. 2020, 319, 106810. [Google Scholar] [CrossRef] [PubMed]
- Mckay, R.T. How the 1D-NOESY suppresses solvent signal in metabonomics NMR spectroscopy: An examination of the pulse sequence components and evolution. Concepts Magn. Reson. Part A 2011, 38A, 197–220. [Google Scholar] [CrossRef]
- Simpson, A.J.; Brown, S.A. Purge NMR: Effective and easy solvent suppression. J. Magn. Reson. 2005, 175, 340–346. [Google Scholar] [CrossRef]
- Soong, R.; Wolff, W.; Pellizzari, J.; Downey, K.; Chen, S.; Biswas, R.G.; Bastawrous, M.; Goerling, B.; Busse, V.; Busse, F.; et al. Water suppression 101 for benchtop NMR–An accessible guide and primer including fully interactive training videos. J. Magn. Reson. Open 2024, 19, 100150. [Google Scholar] [CrossRef]
- Ogg, R.J.; Kingsley, P.B.; Taylor, J.S. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR Spectroscopy. J. Magn. Reson. B. 1994, 104, 1–10. [Google Scholar] [CrossRef]
- Gouilleux, B.; Rouger, L.; Giraudeau, P. Chapter Two—Ultrafast 2D NMR: Methods and Applications. In Annual Reports on NMR Spectroscopy; Webb, G.A., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 93, pp. 75–144. [Google Scholar]
- Karamanos, T.K.; Kalverda, A.P.; Thompson, G.S.; Radford, S.E. Mechanisms of amyloid formation revealed by solution NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2015, 88–89, 86–104. [Google Scholar] [CrossRef]
- Tenenhaus, M.; Tenenhaus, A.; Groenen, P.J. Regularized generalized canonical correlation analysis: A framework for sequential multiblock component methods. Psychometrika 2017, 82, 737–777. [Google Scholar] [CrossRef] [PubMed]
- Giraudeau, P.; Felpin, F.X. Flow reactors integrated with in-line monitoring using benchtop NMR spectroscopy. React. Chem. Eng. 2018, 3, 399–413. [Google Scholar] [CrossRef]
- Castaing-Cordier, T.; Bouillaud, D.; Bowyer, P.; Gonçalves, O.; Giraudeau, P.; Farjon, J. Highly Resolved Pure-Shift Spectra on a Compact NMR Spectrometer. ChemPhysChem 2019, 20, 736–744. [Google Scholar] [CrossRef] [PubMed]
- McCarney, E.R.; Dykstra, R.; Galvosas, P. Evaluation of benchtop NMR Diffusion Ordered Spectroscopy for small molecule mixture analysis. Magn. Reson. Imaging 2019, 56, 103–109. [Google Scholar] [CrossRef]
- Schmid, N.; Bruderer, S.; Paruzzo, F.; Fischetti, G.; Toscano, G.; Graf, D.; Fey, M.; Henrici, A.; Ziebart, V.; Heitmann, B.; et al. Deconvolution of 1D NMR spectra: A deep learning-based approach. J. Magn. Reson. 2023, 347, 107357. [Google Scholar] [CrossRef]
- Matviychuk, Y.; Yeo, J.; Holland, D.J. A field-invariant method for quantitative analysis with benchtop NMR. J. Magn. Reson. 2019, 298, 35–47. [Google Scholar] [CrossRef]
- Bruker Analytik GmbH. Bruker Topspin Software, Version 4.0.3. NMRSIM (1999), Part No. H9171 (version 6.1.1). Bruker Analytik GmbH: Berlin, Germany, 1999.
- Oxford Instruments X-Pulse. Available online: https://nmr.oxinst.com/x-pulse (accessed on 10 May 2025).
- Magritek Spinsolve 60. Available online: https://magritek.com/products/benchtop-nmrspectrometer-spinsolve/spinsolve-60/ (accessed on 2 May 2025).
- Magritek Spinsolve 80. Available online: https://magritek.com/products/benchtop-nmrspectrometer-spinsolve/spinsolve-80/ (accessed on 2 May 2025).
- Magritek Spinsolve 90. Available online: https://magritek.com/products/benchtop-nmrspectrometer-spinsolve/spinsolve-90-mhz/ (accessed on 2 May 2025).
- Bruker Fourier 80. Available online: https://www.bruker.com/en/products-and-solutions/mr/nmr/fourier80.html (accessed on 9 May 2025).
- Nanalysis 60 MHz. Available online: https://www.nanalysis.com/60mhz (accessed on 17 May 2025).
- Nanalysis 100 MHz. Available online: https://www.nanalysis.com/100mhz (accessed on 17 May 2025).
- ThermoFisher picoSpin 45. Available online: https://www.thermofisher.com/order/catalog/product/912A0911 (accessed on 17 May 2025).
- ThermoFisher picoSpin 80. Available online: https://www.thermofisher.com/order/catalog/product/912A0913 (accessed on 17 May 2025).
- Q Magnetics QM-125. Available online: https://www.qmagnetics.com/ (accessed on 17 May 2025).
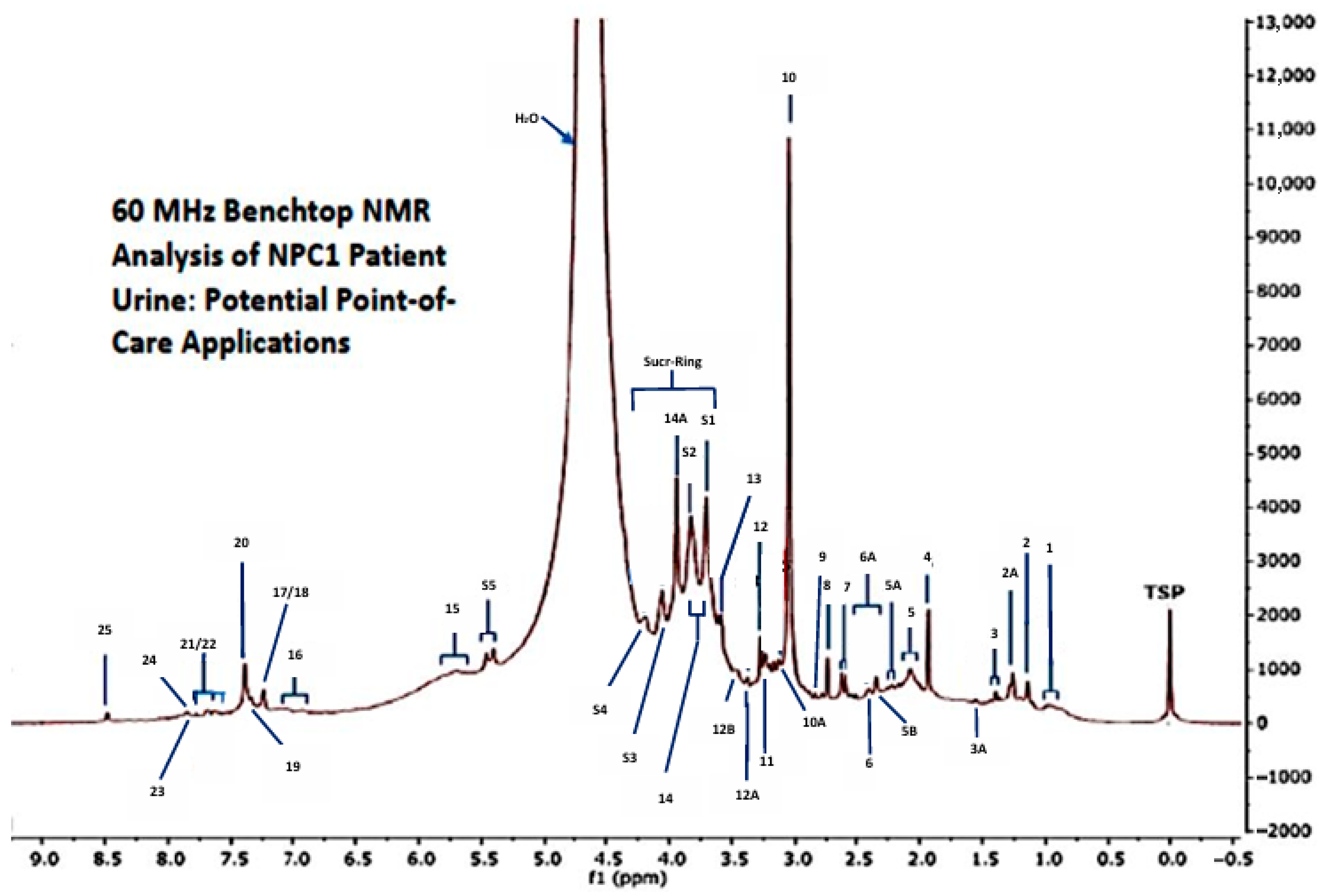

| Assignment | Assignment Code | Chemical Shift (δ) Value (ppm) | Corresponding Coupling Pattern(s) | Superimposition Status |
|---|---|---|---|---|
| BCAA-CH3s (isoleucine/leucine/valine) | 1 | 0.86–1.05 | d/t/dd | +++ |
| 3-Aminoisobutyrate-CH3 | 2 | 1.15 | d | + |
| 3-D-Hydroxybutyrate-CH3 | 2A | 1.24 | d | + |
| Lactate-CH3 | 3 | 1.33 | d | + |
| Alanine-CH3 | 3A | 1.49 | d | ni |
| Acetate-CH3 | 4 | 1.92 | s | ni |
| GlycA/N-Acetylsugar- and N-Acetylamino acid-NH-CO-CH3 | 5 | 2.04 | s (broad) | ++ |
| ¶ Acetone-CH3/* 5-Aminovalerate-α-CH2 | 5A | 2.24 | s/2 × m | +++ |
| ¶ Acetoacetate-CH3/Aminoadipate-γ-CH2 | 5B | 2.28/2.32–2.35 | s/2 × dt | ++ |
| Pyruvate-CH3 and Succinate-CH2s | 6 | 2.38–2.41 | s and s | +++ |
| Glutamate-/Glutamine-γ-CH2 | 6A | 2.34–2.43 | m/m | +++ |
| Citrate-CH2-CO2− | 7 | 2.60 | Apparent d | ni |
| Dimethylamine-N(CH3)2 | 8 | 2.75 | s | ni |
| Dimethylglycine-N(CH3)2 | 9 | 2.83 | s | + |
| Creatine-/Creatinine-N-CH3 | 10 | 3.03 | 2 × s | +++ |
| * Malonate-CH2 | 10A | 3.11 | s | ++ |
| * Dimethylsulphone | 10B | 3.17 | s | +++ |
| Choline/Betaine-N+(CH3)3 | 11 | 3.20 | s/s | +++ |
| Trimethylamine N-oxide-N(CH3)3 | 12 | 3.25 | s | ++ |
| Methanol-CH3 | 12A | 3.38 | s | + |
| * Taurine-CH2SO3− | 12B | 3.46 | t | ++ |
| Glycine-CH2/Sucrose sugar ring-Gluc-C2H | 13 | 3.56/3.55 | s/dd | ++ |
| Amino acid-α-CH protons | 14 | 3.7–3.8 | All m | +++ |
| Creatine-CH2/* Glycolate-CH2/Hippurate-CH2 | 14A | 3.95 | s/s/d | +++ |
| Sucrose sugar ring-Fruc-C1′(-CH2OH) and Gluc-C3H/* Phenylacetylglycine-CH2 | S1 | 3.67 and 3.75/3.66 | s/t/s | +++ |
| Sucrose sugar ring-Fruc-C5′H, Gluc-C5H, Fruc-C6′(-CH2OH) and Gluc-C6(-CH2OH)/* Ethanolamine-CH2OH/* Mannitol-CH2OH | S2 | 3.89/3.87/3.82/3.82/3.84 | dd/dd/m/m/t/dd | +++ |
| Sucrose sugar ring-Fruc-C4′H/Creatinine-CH2 | S3 | 4.04/4.05 | t/s | ++ |
| Sucrose sugar ring-Fruc-C3′H | S4 | 4.21 | d | + |
| Sucrose: D-glucopyranoside-C1αH | S5 | 5.40 | d | ni |
| Urea-CONH2 | 15 | 5.75 | Broad s | + |
| * Tyrosine aromatic ring-C2H,C6H/* Histidine imidazole ring- C4H/Phenol-C2H,C4H/* 4-Hydroxypropionate-C2H,C6H and C3H,C5H/* 3-(3-Hydroxyphenyl) propionate-C2H,C4H | 16 | 6.80–7.12 | d/s/2 × d/2 × m | +++ |
| Indoxylsulphate-C5H, C6H/* Imidazole ring protons-C2H,C4H,C5H/* Tyrosine-aromatic ring-C3H,C5H | 17 | 7.21–7.28 | 2 × dd/s/d | +++ |
| Indoxylsulphate-C2H/* Phenylalanine- aromatic ring-C3H,C4H,C5H | 18 | 7.32–7.44 | s/m | +++ |
| Phenylalanine aromatic ring-C2H,C6H | 19 | 7.30–7.33 | m | ++ |
| Indoxylsulphate-C2H | 20 | 7.40 | s | ++ |
| Indoxylsulphate-C7H/Hippurate-aromatic ring-C3H/C5H | 21 | 7.51–7.54 | d/m | +++ |
| Hippurate-aromatic ring-C4H | 22 | 7.63 | tt | +++ |
| Indoxylsulphate-indole ring-C4H | 23 | 7.77 | d | ++ |
| Hippurate-aromatic ring-C2H,C6H | 24 | 7.81 | dd | + |
| Formate-H | 25 | 8.46 | s | ni |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grootveld, M.; Ruiz-Rodado, V.; Gerdova, A.; Edgar, M. Very First Application of Compact Benchtop NMR Spectrometers to Complex Biofluid Analysis and Metabolite Tracking for Future Metabolomics Studies: A Retrospective Decennial Report from November 2014. Appl. Sci. 2025, 15, 9675. https://doi.org/10.3390/app15179675
Grootveld M, Ruiz-Rodado V, Gerdova A, Edgar M. Very First Application of Compact Benchtop NMR Spectrometers to Complex Biofluid Analysis and Metabolite Tracking for Future Metabolomics Studies: A Retrospective Decennial Report from November 2014. Applied Sciences. 2025; 15(17):9675. https://doi.org/10.3390/app15179675
Chicago/Turabian StyleGrootveld, Martin, Victor Ruiz-Rodado, Anna Gerdova, and Mark Edgar. 2025. "Very First Application of Compact Benchtop NMR Spectrometers to Complex Biofluid Analysis and Metabolite Tracking for Future Metabolomics Studies: A Retrospective Decennial Report from November 2014" Applied Sciences 15, no. 17: 9675. https://doi.org/10.3390/app15179675
APA StyleGrootveld, M., Ruiz-Rodado, V., Gerdova, A., & Edgar, M. (2025). Very First Application of Compact Benchtop NMR Spectrometers to Complex Biofluid Analysis and Metabolite Tracking for Future Metabolomics Studies: A Retrospective Decennial Report from November 2014. Applied Sciences, 15(17), 9675. https://doi.org/10.3390/app15179675







