1. Introduction
The increasing volume of the scientific literature presents a challenge for knowledge discovery. The manual synthesis of information cannot keep up with the rapid growth of publications across diverse fields, leading to missed connections. Literature-based discovery (LBD) is a data mining approach capable of connecting disparate literature studies [
1] and uncovering new connections within specialized fields, addressing situations where the volume of publications cannot be processed by a human (e.g., [
2,
3]). The original LBD technique, the ABC model [
4]—due to the connection between
A and
C it proposes based on known relations between
A and
B and
B and
C—hinges on the accuracy and quality of the relations extracted from texts, as well as the ability to filter the resulting candidate hidden knowledge pairs (CHKPs) to remove uninformative candidates [
5]. This work introduces a hybrid data analysis approach, which explores the use of large language models (LLMs) to address both of these points.
Early LBD works utilised simple text mining techniques, such as word co-occurrence in titles, to define relations [
6]. This method was effective for investigating suspected (closed) connections where the literature could be reduced to titles containing
A or
C and only the linking term
B was sought. However, even when the relations were refined, the approach had a tendency to generate an overwhelming number of CHKPs when used in open mode, where all connections from every term are followed for two steps [
7].
The need to reduce the quantity of CHKPs generated by open LBD has fuelled research into enhancements to existing methods for effective data mining. Prior approaches have included the following: (a) the removal of terms based on frequency (e.g., [
8,
9]), (b) the mapping of terms to specialized lexicons such as the Unified Medical Language System (UMLS) [
10] to prevent connections via ambiguous terms [
11], (c) the removal of terms based on semantic type (e.g., [
12,
13]), (d) the restriction of the type of discovery (e.g., cancer biology [
14] or protein interaction [
15]), (e) the restriction of relations to be connected [
16], and (f) the re-ranking of generated CHKPs (e.g., [
17,
18,
19,
20,
21]). However, the exponential growth of academic publications [
22] requires a constant refinement of these approaches.
Recent advances in deep learning have improved the performance of many natural language processing (NLP) tasks in the biomedical field [
23], including LBD (for an overview, see [
24]). While pre-trained language models have been used for LBD [
25], many successful applications of data mining, particularly in areas such as drug repurposing, have predominantly relied on knowledge graph-based deep learning, with the knowledge provided by existing tools, such as SemMedDB [
26] (e.g., [
3]), the Global Network of Biomedical Relationships [
27] (e.g., [
28]), or named entity extraction (e.g., [
29]). The use of large, generative, language models (LLMs) has, however, been relatively low for the purpose of LBD since they have a propensity for adhering to information they were trained on rather than proposing new connections [
30].
Through the vast quantity of training data, LLMs have access to an enormous amount of knowledge. While this appears to be a limitation when proposing new connections, it suggests their suitability for use within CHKP filtering. Since a large quantity of proposed CHKPs represent background, well-known knowledge (which, due to being widely familiar, does not appear explicitly in publications [
25]), we propose that LLMs could be used to filter these CHKPs. By instructing them to act as judges—a modification of the LLM-as-a-judge paradigm—they can assess each CHKP against their training data to determine whether it represents generally known, background knowledge. This theoretically distinct approach goes beyond conventional filtering methods that rely on frequency or semantic constraints.
In this work, we therefore present a novel data mining approach that extensively explores the integration of machine learning and AI through LLMs for LBD, involving them in every crucial step except the initial CHKP generation:
Using few-shot learning to extract factual subject–predicate–object relations from publication abstracts, achieving greater coverage than established tools such as SemRep.
Investigating the impact of different training examples within few-shot learning in Step 1, comparing manually annotated instances with examples derived from cited facts.
Using zero-shot learning for filtering background knowledge CHKPs, in an LLM-as-a-judge setup, with hallucinations ruled out using retrieval augmented generation.
The replication of existing discoveries is used to demonstrate the usefulness of the LLM-generated relations. A small-scale timeslicing evaluation indicates their superior suitability for LBD, producing less background knowledge and achieving higher precision against the gold standard than LBD based on SemRep relations.
The remainder of this paper is structured as follows:
Section 2 describes related work, while
Section 3 outlines the experiments carried out in this work. Discussions are presented in
Section 4 with conclusions and future work appearing in
Section 5.
2. Related Work
The growing quantity of the scientific literature indicates that effective knowledge discovery techniques, such as literature-based discovery (LBD), are required. The original LBD approach, the ABC model [
6], uses simple inference to propose a connection between previously unconnected terms
A and
C if there are known, published, connections from
A to
B and from
B to
C, which appear in separate publications. Its functionality was initially demonstrated on a suspected connection between
Raynaud’s syndrome and
fish oil—both
Raynaud’s syndrome and
fish oil were found to have already published links to
blood viscosity, thereby allowing a connection to be confirmed through this (
B) term.
The number of practical applications of LBD is growing: within the biomedical field, this includes, for example, adverse drug event prediction [
31], drug development [
32] and drug repurposing [
33]. Particularly in the latter case, the economic benefits are substantial: a drug that has already been safety tested can bypass 6–7 years of preclinical and early-stage research when being investigated for a new use [
34]. While the novel data analysis approach presented in this paper is applicable to all uses of LBD, our evaluations specifically focus on its utility in drug repurposing.
2.1. Resources
Several resources are frequently employed in large-scale text and data mining for LBD. These will be introduced first, as they are required for relation extraction (
Section 2.2) and subsequently LBD (
Section 2.3).
2.1.1. MEDLINE and PubMed
PubMed, and its biomedical subset MEDLINE, are the National Library of Medicine’s (NLM’s) databases of publications, on which many LBD investigations are based. MEDLINE mainly contains paper titles and abstracts from the biomedical domain, while PubMed offers broader domain coverage and more recently also contains articles’ full texts. Although some works utilise full texts (e.g., [
35]), the majority of LBD systems exploit titles and/or abstracts only.
2.1.2. UMLS
The Unified Medical Language System (UMLS) [
10] is another widely used resource. It comprises the metathesaurus, the semantic network and the SPECIALIST lexicon and tools.
2.1.3. UMLS Metathesaurus
The UMLS metathesaurus unifies a number of source vocabularies into a single database of biomedical and health-related concepts. The concepts, identified using a concept unique identifier (CUI), link together different ways to refer to the same concept in a natural text. A number of manually identified relationships between CUIs (related concepts) are also included within the metathesaurus. The main use of the metathesaurus in LBD is for disambiguation: if terms are accurately mapped to their CUIs, connections made through terms with identical spelling but different meaning can be avoided [
36]. In addition, performing this mapping can link abbreviations to their long form.
2.1.4. The Semantic Network
The Semantic Network groups CUIs together into broad categories known as semantic types with each UMLS concept assigned at least one semantic type. This semantic typing is frequently used to constrain the hidden connections proposed by LBD, for example, to drug (
Chemicals & drugs)–treatment (
Disease or Syndrome) pairs (e.g., [
37]).
2.1.5. MetaMap
MetaMap [
38], a tool within the UMLS suite, is widely used for mapping biomedical text to UMLS concepts. Its availability for local execution and regular releases of pre-processed versions of MEDLINE by the NLM significantly aid large-scale biomedical text mining. In this work, we utilize version 24 of MetaMapped MEDLINE, and MetaMap 2020 with the 2020AA USAbase strict data model is used for local processing.
2.2. Relation Extraction
The extraction of reliable semantic relations is an important component of the ABC model (e.g., [
39] or [
40]). High-quality relations enable further refinements, such as focusing on specific types of interactions (for example, relations between chemicals, genes and diseases as explored by [
27]). Also, the availability of relations allows the construction of large-scale knowledge graphs, which can be exploited by LBD (e.g., [
41]).
2.2.1. SemRep and Refinements
SemRep is a widely used rule-based approach to semantic relation extraction tuned to the biomedical domain. Built upon MetaMap, it incorporates a step for mapping concepts to UMLS CUIs. For example, for the sentence in list 1, SemRep extracts the semantic relations in list 2 (CUIs have been mapped to their term representation).
Raynaud’s phenomenon (RP) is commonly observed in fingers and toes of patients with connective tissue diseases (CTDs).
Connective Tissue Diseases process_of Patients.
Toes part_of Patients.
Fingers location_of Raynaud Phenomenon.
Toes location_of Raynaud Phenomenon.
The tool extracts approximately 70 different predications, with roughly half being negative (such as
neg_treats). A database of SemRep processed abstracts from MEDLINE, SemMedDB [
26], is publicly available and widely used for biomedical data analysis, including LBD (e.g., [
33] or [
42]). However, a recent evaluation of SemRep (version 1.8) on the SemRep test collection [
43] revealed limitations in its performance, with 0.55 precision, 0.34 recall, and 0.41
[
44]. Even after accounting for test collection and evaluation setup issues, the recall remained low at 0.42 (with an
of 0.52), suggesting that the technique requires enhancement for effective data mining.
To address the recall limitations of SemRep, machine learning approaches, such as classification, have been explored [
45]. This involved fine-tuning language models such as PubMedBERT (now BiomedBERT) [
46] on entities extracted by SemRep following a pre-training step using contrastive learning. This approach was found to be complementary to SemRep’s annotations, with 0.81 recall, 0.62 precision and 0.70
.
Beyond rule-based systems such as Public Knowledge Discovery Engine for Java (PKDE4J) [
47] and its transformer-based refinement BioPREP [
48], other applications of transformer techniques to relation extraction, often focusing on specific subsets of relations, have also emerged (e.g., [
49,
50]). However, fine-tuning pre-trained transformer models typically requires a substantial annotated datasets (e.g., [
3]), and the resulting relations are not always optimally suited for LBD.
Recently, there has been a shift towards the use of large language models (LLMs) for relation extraction. Given the enormous quantity of data used to pre-train LLMs, relations can be extracted in a zero-shot setting—with no examples provided in the prompt [
51]—or in a few-shot setting, with a small number of examples included in the prompt [
52]. However, the specific focus of each work (e.g., clinical trials only) and varied different evaluation datasets make direct performance comparisons difficult across different LLM-based approaches.
2.2.2. Factuality Dataset
The publicly available
Factuality dataset allows the confidence level of SemRep-generated relations to be assessed. The distinction between genuine facts, conjectures, or doubtful statements is valuable for high-quality input to LBD (e.g., [
53]). Kilicoglu et al. [
54] manually annotated SemRep relations extracted from 500 PubMed abstracts with one of seven factuality values (
fact,
probable,
possible,
doubtful,
counterfact,
uncommitted, and
conditional), providing a valuable resource for refining relation extraction (available from
https://lhncbc.nlm.nih.gov/ii/tools/SemRep_SemMedDB_SKR.html, accessed 1 August 2024).
2.3. Literature-Based Discovery
The original ABC approach to LBD was integrated into a number of larger-scale systems, such as BITOLA [
55], Arrowsmith [
56] and FACTA+ [
57]. In their open form, these systems often generated an excessively large number of candidate hidden knowledge pairs (CHKPs), making the manual identification of promising avenues an extremely laborious task. To reduce the quantity of CHKPs, co-occurrence was replaced with semantic relations, and the semantic types of the terms involved in connections were restricted [
58]. Further refinements involved the use of graph-based approaches for inference, based on knowledge graphs build from semantic relations extracted from publications (e.g., [
19]). Despite these refinements, even targeted LBD systems can still yield a large quantity of links; for example, Syafiandini et al. [
59] found 2,740,314 paths between 35 Xanthium compounds and three types of diabetes.
Recent advances in deep learning have significantly improved the state of the art across many biomedical domain tasks (as noted in
Section 2.2.1). Many applications of deep learning to LBD exploit the graph structure of extracted relations, whether by using language models to enhance similarity in graph construction [
60] or for filtering relations within the resulting graph [
3]. Other neural network approaches, such as CNNs [
61] and autoencoders [
62], have also been investigated for their utility in LBD.
There is also a clear utility of LLMs within the biomedical domain, for example, by exploiting their ability to answer scientific questions [
63] or aiding clinical decision-making [
64]. LLMs boast a number of advantages over traditional approaches: most importantly, being based on a massive dataset, they do not require structured inputs and are less susceptible to the scalability issues experienced by simple inference. However, so far, works exploiting LLMs for direct hidden knowledge suggestion in LBD have indicated that the proposed hidden knowledge is of low technical depth and novelty [
65], frequently reverting to well-established information [
66]. This observation suggests that it is important to use LLMs where they excel: we therefore propose a hybrid approach, using LLMs to extract knowledge from texts and to identify background knowledge among the proposed CHKPs, while employing the established ABC model for CHKP generation.
2.3.1. Filtering
The effective filtering or re-reranking of the CHKPs resulting from LBD determines its usefulness. We can broadly categorize valid relation-based CHKPs into three groups: (1) connections depicting background knowledge—information that is so widely known that it is unlikely to appear in knowledge bases; (2) minor variations of existing knowledge, e.g., involving synonyms or already closely linked studies such as
fish oil—
Raynaud phenomenon deduced from a known link between
fish oil and
Raynaud disease; and (3) genuinely valuable CHKPs, such as connections between disjointed studies [
67].
Many filtering approaches do not target CHKPs by type. For example, the authors of [
3] reduce the set of predicate types within their LBD system to 15 based on their expected utility. Similarly, restricting the semantic types of subjects (source) and objects (target) of semantic relations (e.g., [
12,
13,
37]) focuses on the application domain rather than eliminating specific CHKP groups. While these methods reduce the overall quantity of CHKPs, type 1 and 2 CHKPs are still prevalent in the remaining lists.
2.3.2. Filtering Using Generative AI
The large quantity of data that LLMs are pre-trained with enables them to be used for content evaluation and filtering, including tasks such as removing noisy data from datasets (e.g., [
68]). However, the application of LLMs for CHKP filtering in LBD has not yet been explored. Within the LLM-as-a-judge paradigm, an LLM is used to evaluate the output of other LLMs, checking their agreement with human annotation, yielding agreement levels of human experts [
69]. We propose modifying this paradigm to utilise an LLM to evaluate the proposed, automatically created CHKPs—which were designed to represent novel hypotheses—to determine whether they represent valuable hidden knowledge or already known background information.
2.3.3. Evaluation of LBD
The evaluation of CHKPs is difficult as there is, by definition, no gold standard available ([
70,
71]). Generally, studies therefore perform one of three evaluations [
5]:
Expert and clinical evaluation, where an expert examines the CHKPs proposed by the LBD system. Such connections are usually proposed by running LBD in a heavily restricted mode to account for experts’ specializations.
The replication of existing discoveries, where a new system is judged on its ability to replicate discoveries made by past LBD systems.
Timeslicing, in which a chronological cut-off date is selected with publications prior to the cut-off point used by the LBD model to generate CHKPs, which are then evaluated against relations extracted from publications after the cut-off point [
72].
It should be noted that the number of past discoveries is limited, restricting this evaluation type [
73] and potentially introducing biases [
74]. However, the timeslicing approach, which—unlike expert evaluation—allows for large-scale evaluation, also suffers from a number of limitations: (1) not all valid CHKPs will have been discovered (yet), and (2) the evaluation is heavily reliant on the accuracy of relations extracted from the literature post cut-off.
5. Conclusions
This work explores the use of LLMs for LBD in line with their strengths: (1) to extract factual semantic relations from biomedical text and (2) to identify knowledge pairs that represent background knowledge so they can be removed from CHKPs generated by simple inference LBD.
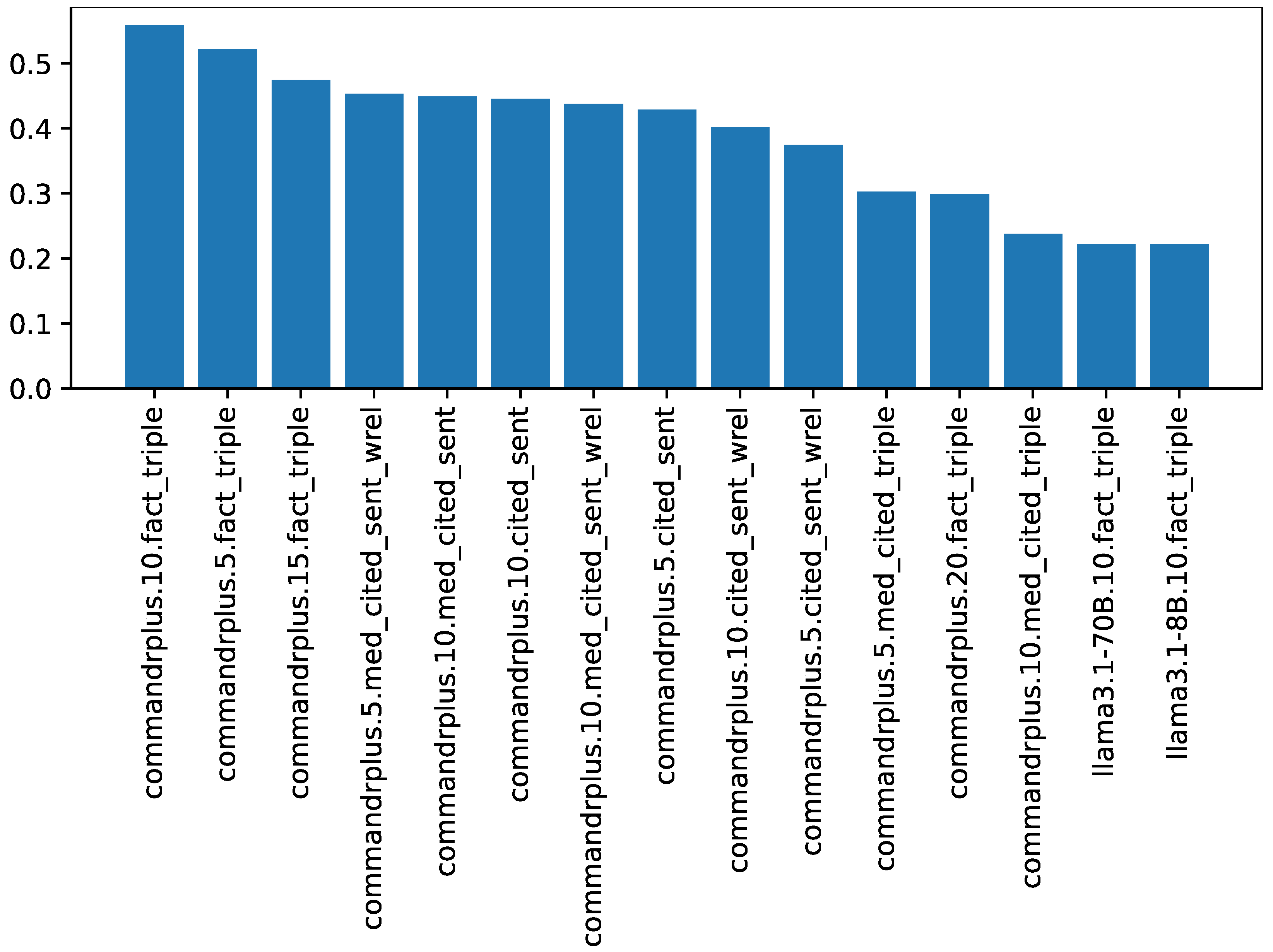
Using a small number of semantic relations representing facts targeted to the LBD task (by restricting the predicate) for the few-shot learning of a large language model is shown to yield a different set of semantic relations to the highly utilised tool, SemRep. This approach exhibits higher coverage than SemRep relations, with more specific terms mentioned. When used for LBD, a higher proportion of the resulting CHKPs are shown to be valid.
This work avoids the shortfall of LLMs mainly suggesting already known or closely related facts [
60] by continuing to employ simple inference for LBD, followed by using LLMs to identify already known CHKPs from the resulting set. The importance of the LLM’s knowledge cut-off for this task is demonstrated and it is shown to successfully filter existing knowledge with hallucination not found to pose a problem.
The next step would involve an expert evaluation of the resulting discoveries, including clinical trials as appropriate. To this end, the discoveries yielded by the timeslicing evaluation presented in the paper are being made publicly available. In addition, expert involvement could evaluate the inverse application of LLMs—when an LLM is prompted to determine whether a possible candidate hidden knowledge pair through its linking term ‘makes sense’. Further re-ranking could also be offered by combining the proposed approach with other methods, such as, e.g., distance within a knowledge graph [
27].