The Applications of MALDI-TOF MS in the Diagnosis of Microbiological Food Contamination
Abstract
1. Introduction
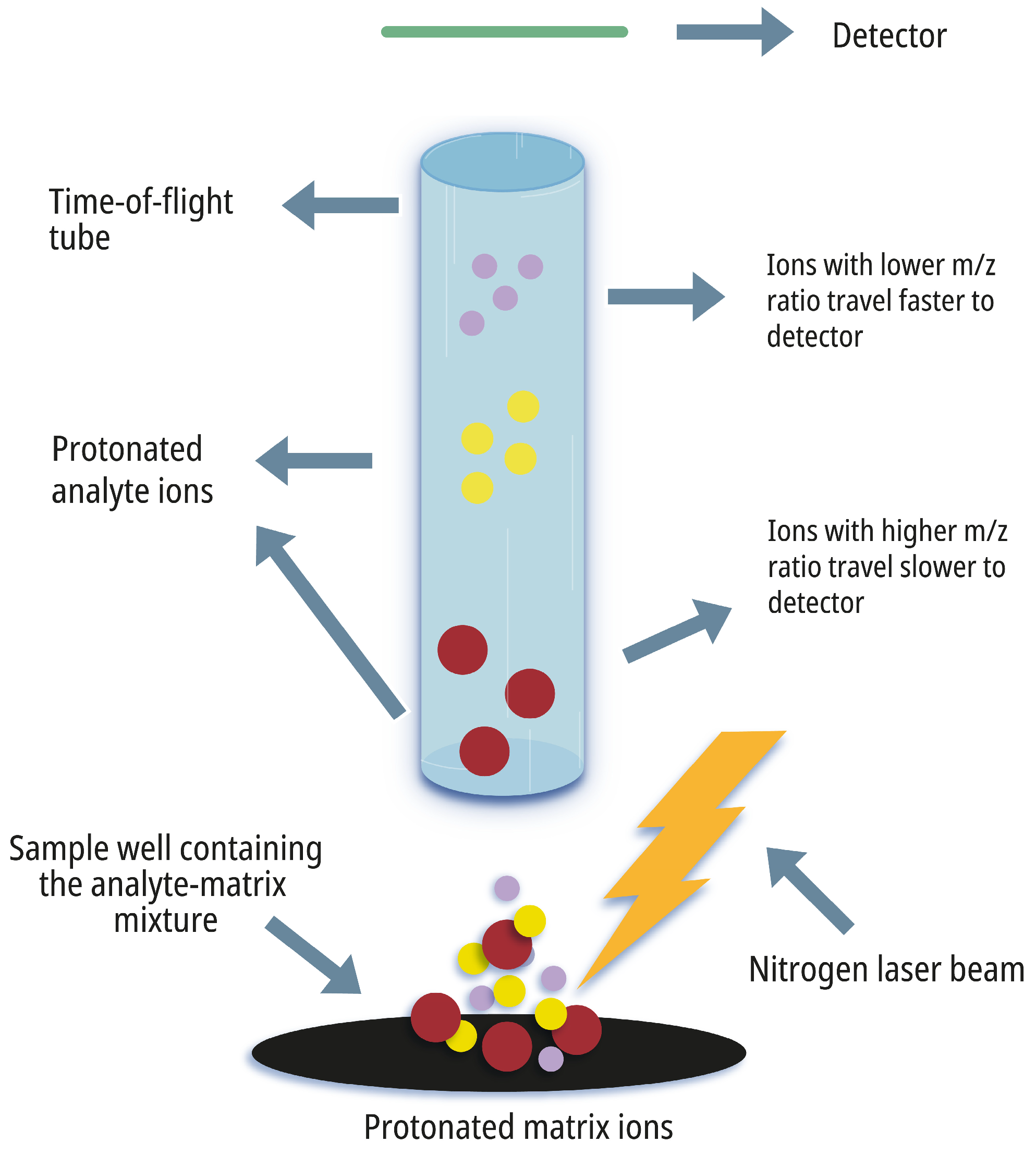
2. The Principles of MALDI-TOF MS
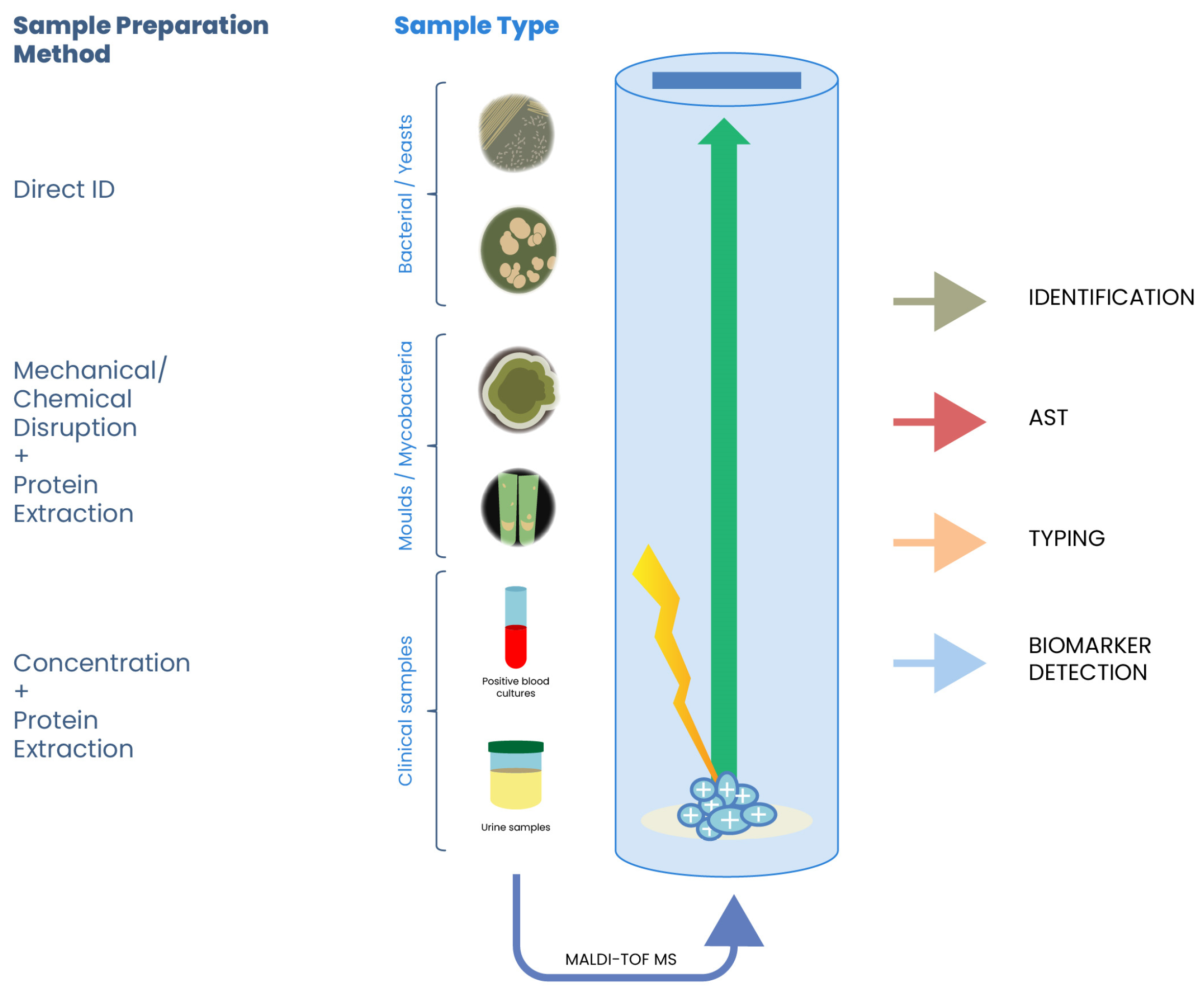
2.1. Sample Preparation
2.2. Laser Desorption/Ionization
2.3. Time-of-Flight Mass Analysis
2.4. Spectral Interpretation and Identification
2.5. Key Analytical Considerations
- (a)
- Resolution and Accuracy:
- (b)
- Reproducibility:
- (c)
- Databases:
2.6. Advantages of MALDI-TOF over Conventional Techniques
- (a)
- Rapid Turnaround Time:
- (b)
- High Accuracy and Specificity:
- (c)
- Cost Efficiency and Low Per-Sample Expense:
- (d)
- Minimal Sample Preparation and High Throughput:
- (e)
- Broad Taxonomic Coverage:
- (f)
- Reduced Dependence on Reagents and Kits:
- (g)
- Compatibility with Emerging Data Analytics
3. Applications in Food Microbiology
3.1. Identification of Foodborne Pathogens
3.2. Detection of Spoilage Organisms
3.3. Authentication and Traceability
3.4. Section Summary
4. Limitations and Challenges
4.1. Dependence on Reference Databases
4.2. Limited Discrimination Between Closely Related Strains
4.3. Inability to Detect Viable but Non-Culturable (VBNC) Organisms
4.4. Sample-Matrix Interference
4.5. Identification of Mixed Cultures
4.6. Instrument and Operational Limitations
4.7. Regulatory and Standardization Gaps
4.8. Section Summary
5. Future Perspectives
5.1. Expansion and Curation of Databases
5.2. Direct-from-Sample Identification
5.3. Strain-Level Discrimination and Typing
5.4. Integration with Food Quality Management Systems
5.5. Applications Beyond Bacteria: Mycotoxins, Viruses, and Metabolomics
- (a)
- Mycotoxin detection: by modifying matrix chemistry and ionization protocols, MALDI-TOF MS could be adapted for the rapid screening of fungal toxins (e.g., aflatoxins, ochratoxins) in grain, nut, and dairy products. However, it is still needed to develop methods for simultaneous detection of multiple mycotoxins [3,151,152].
- (b)
- (c)
5.6. Regulatory Acceptance and Standardization
5.7. Section Summary
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fung, F.; Wang, H.S.; Menon, S. Food safety in the 21st century. Biomed. J. 2018, 41, 88–95. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Lichtveld, M.; Mazet, J.A.; Togami, E.; Miller, S.A. Plant health and its effects on food safety and security in a One Health framework: Four case studies. One Health Outlook 2021, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Ansari, F.; Lee, C.-C.; Rashidimehr, A.; Eskandari, S.; Ashaolu, T.J.; Mirzakhani, E.; Pourjafar, H.; Jafari, S.M. The role of probiotics in improving food safety: Inactivation of pathogens and biological toxins. Curr. Pharm. Biotechnol. 2024, 25, 962–980. [Google Scholar] [CrossRef]
- Sobiczewski, P.; Iakimova, E.T. Plant and human pathogenic bacteria exchanging their primary host environments. J. Hortic. Res. 2022, 30, 11–30. [Google Scholar] [CrossRef]
- Sahoo, M.; Panigrahi, C.; Aradwad, P. Management strategies emphasizing advanced food processing approaches to mitigate food borne zoonotic pathogens in food system. Food Front. 2022, 3, 641–665. [Google Scholar] [CrossRef]
- Abdel-Aziz, S.M.; Asker, M.M.; Keera, A.A.; Mahmoud, M.G. Microbial Food Spoilage: Control Strategies for Shelf Life Extension. In Microbes in Food and Health; Springer: Cham, Switzerland, 2016; pp. 239–264. [Google Scholar]
- Rawat, S. Food Spoilage: Microorganisms and their prevention. Asian J. Plant Sci. Res. 2015, 5, 47–56. [Google Scholar]
- Teshome, E.; Forsido, S.F.; Rupasinghe, H.V.; Olika Keyata, E. Potentials of natural preservatives to enhance food safety and shelf life: A review. Sci. World J. 2022, 2022, 9901018. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.A.; Ayivi, R.D.; Zimmerman, T.; Siddiqui, S.A.; Altemimi, A.B.; Fidan, H.; Esatbeyoglu, T.; Bakhshayesh, R.V. Lactic acid bacteria as antimicrobial agents: Food safety and microbial food spoilage prevention. Foods 2021, 10, 3131. [Google Scholar] [CrossRef]
- Smith, A.; Terry, S.; Detken, D. 10 years of the European Food Safety Authority (EFSA) and the EU food safety system. Eur. Food Feed Law Rev. 2012, 7, 111–116. [Google Scholar]
- European Food Safety Authority (EFSA). The food classification and description system FoodEx 2 (revision 2). EFSA J. 2015, 12, 804E. [Google Scholar]
- European Food Safety Authority (EFSA). Use of the EFSA comprehensive European food consumption database in exposure assessment. EFSA J. 2011, 9, 2097. [Google Scholar]
- Alamri, M.S.; Qasem, A.A.; Mohamed, A.A.; Hussain, S.; Ibraheem, M.A.; Shamlan, G.; Alqah, H.A.; Qasha, A.S. Food packaging’s materials: A food safety perspective. Saudi J. Biol. Sci. 2021, 28, 4490–4499. [Google Scholar] [CrossRef] [PubMed]
- Butz, H.; Patócs, A. Brief Summary of the Most Important Molecular Genetic Methods (PCR, qPCR, Microarray, Next-Generation Sequencing, etc.). In Genetics of Endocrine Diseases and Syndromes; Springer: Cham, Switzerland, 2019; pp. 33–52. [Google Scholar]
- Gabaldón, T. Recent trends in molecular diagnostics of yeast infections: From PCR to NGS. FEMS Microbiol. Rev. 2019, 43, 517–547. [Google Scholar]
- Kaprou, G.D.; Bergšpica, I.; Alexa, E.A.; Alvarez-Ordóñez, A.; Prieto, M. Rapid methods for antimicrobial resistance diagnostics. Antibiotics 2021, 10, 209. [Google Scholar] [CrossRef]
- Schlaberg, R.; Chiu, C.Y.; Miller, S.; Procop, G.W.; Weinstock, G.; Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology; Microbiology Resource Committee of the College of American Pathologists. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab. Med. 2017, 141, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Pham, D.; Sivalingam, V.; Tang, H.M.; Montgomery, J.M.; Chen, S.C.A.; Halliday, C.L. Molecular diagnostics for invasive fungal diseases: Current and future approaches. J. Fungi 2024, 10, 447. [Google Scholar] [CrossRef]
- Aladhadh, M. A review of modern methods for the detection of foodborne pathogens. Microorganisms 2023, 11, 1111. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-generation sequencing technology: Current trends and advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Shah, H.N.; Shah, A.J.; Belgacem, O.; Ward, M.; Dekio, I.; Selami, L.; Duncan, L.; Bruce, K.; Xu, Z.; Mkrtchyan, H.V.; et al. MALDI-TOF MS and currently related proteomic technologies in reconciling bacterial systematics. In Trends in the Systematics of Bacteria and Fungi; CABI: Wallingford, UK, 2021; pp. 93–118. [Google Scholar]
- Yamin, D.; Uskoković, V.; Wakil, A.M.; Goni, M.D.; Shamsuddin, S.H.; Mustafa, F.H.; Alfouzan, W.A.; Alissa, M.; Alshengeti, A.; Almaghrabi, R.H.; et al. Current and future technologies for the detection of antibiotic-resistant bacteria. Diagnostics 2023, 13, 3246. [Google Scholar] [CrossRef]
- Neilson, A.P.; O’Keefe, S.F.; Bolling, B.W. High-molecular-weight proanthocyanidins in foods: Overcoming analytical challenges in pursuit of novel dietary bioactive components. Annu. Rev. Food Sci. Technol. 2016, 7, 43–64. [Google Scholar] [CrossRef]
- Torres-Sangiao, E.; Leal Rodriguez, C.; García-Riestra, C. Application and perspectives of MALDI–TOF mass spectrometry in clinical microbiology laboratories. Microorganisms 2021, 9, 1539. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yi, J.; Han, G.; Qiao, L. MALDI-TOF mass spectrometry in clinical analysis and research. ACS Meas Sci. Au 2022, 2, 385–404. [Google Scholar] [CrossRef] [PubMed]
- Darie-Ion, L.; Whitham, D.; Jayathirtha, M.; Rai, Y.; Neagu, A.N.; Darie, C.C.; Petre, B.A. Applications of MALDI-MS/MS-based proteomics in biomedical research. Molecules 2022, 27, 6196. [Google Scholar] [CrossRef]
- Mohar Lorbeg, P.; Golob, M.; Kramer, M.; Treven, P.; Bogovič Matijašić, B. Evaluation of dietary supplements containing viable bacteria by cultivation/MALDI-TOF mass spectrometry and PCR identification. Front. Microbiol. 2021, 12, 700138. [Google Scholar] [CrossRef]
- Haider, A.; Ringer, M.; Kotroczó, Z.; Mohácsi-Farkas, C.; Kocsis, T. The current level of MALDI-TOF MS applications in the detection of microorganisms: A short review of benefits and limitations. Microbiol. Res. 2023, 14, 80–90. [Google Scholar] [CrossRef]
- Yoon, E.J.; Jeong, S.H. MALDI-TOF mass spectrometry technology as a tool for the rapid diagnosis of antimicrobial resistance in bacteria. Antibiotics 2021, 10, 982. [Google Scholar] [CrossRef]
- Weis, C.; Cuénod, A.; Rieck, B.; Dubuis, O.; Graf, S.; Lang, C.; Oberle, M.; Brackmann, M.; Søgaard, K.K.; Osthoff, M.; et al. Direct antimicrobial resistance prediction from clinical MALDI-TOF mass spectra using machine learning. Nat. Med. 2022, 28, 164–174. [Google Scholar] [CrossRef]
- Rakotonirina, A.; Pol, M.; Raharimalala, F.N.; Ballan, V.; Kainiu, M.; Boyer, S.; Kilama, S.; Marcombe, S.; Russet, S.; Barsac, E.; et al. MALDI-TOF MS: An effective tool for a global surveillance of dengue vector species. PLoS ONE 2022, 17, e0276488. [Google Scholar] [CrossRef]
- Feucherolles, M.; Frache, G. MALDI mass spectrometry imaging: A potential game-changer in a modern microbiology. Cells 2022, 11, 3900. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Han, S.S.; Jeong, Y.S.; Choi, S.K. Current scenario and challenges in the direct identification of microorganisms using MALDI TOF MS. Microorganisms 2021, 9, 1917. [Google Scholar] [CrossRef] [PubMed]
- Solntceva, V.; Kostrzewa, M.; Larrouy-Maumus, G. Detection of species-specific lipids by routine MALDI TOF mass spectrometry to unlock the challenges of microbial identification and antimicrobial susceptibility testing. Front. Cell. Infect. Microbiol. 2021, 10, 621452. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Chezzi, C. MALDI-TOF MS: A reliable tool in the real life of the clinical microbiology laboratory. Microorganisms 2024, 12, 322. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Martinez-Chapa, S.O. Principles and mechanism of MALDI-ToF-MS analysis. In Fundamentals of MALDI-ToF-MS Analysis: Applications in Bio-Diagnosis, Tissue Engineering and Drug Delivery; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–19. [Google Scholar]
- Welker, M.; Fastner, J.; Erhard, M.; von Döhren, H. Applications of MALDI-TOF MS analysis in cyanotoxin research. Environ. Toxicol. 2002, 17, 367–374. [Google Scholar] [CrossRef]
- Welker, M.; Van Belkum, A.; Girard, V.; Charrier, J.P.; Pincus, D. An update on the routine application of MALDI-TOF MS in clinical microbiology. Expert Rev. Proteomics 2019, 16, 695–710. [Google Scholar] [CrossRef]
- Beale, D.J.; Karpe, A.V.; Jadhav, S.; Muster, T.H.; Palombo, E.A. Omics-based approaches and their use in the assessment of microbial-influenced corrosion of metals. Corros. Rev. 2016, 34, 1–15. [Google Scholar] [CrossRef]
- Topić Popović, N.; Kazazić, S.P.; Bojanić, K.; Strunjak-Perović, I.; Čož-Rakovac, R. Sample preparation and culture condition effects on MALDI-TOF MS identification of bacteria: A review. Mass Spectrom. Rev. 2023, 42, 1589–1603. [Google Scholar] [CrossRef]
- Drevinek, M.; Dresler, J.; Klimentova, J.; Pisa, L.; Hubalek, M. Evaluation of sample preparation methods for MALDI-TOF MS identification of highly dangerous bacteria. Lett. Appl. Microbiol. 2012, 55, 40–46. [Google Scholar] [CrossRef]
- Przybilla, L.; Brand, J.D.; Yoshimura, K.; Räder, H.J.; Müllen, K. MALDI-TOF mass spectrometry of insoluble giant polycyclic aromatic hydrocarbons by a new method of sample preparation. Anal. Chem. 2000, 72, 4591–4597. [Google Scholar] [CrossRef]
- Pan, C.; Xu, S.; Zhou, H.; Fu, Y.; Ye, M.; Zou, H. Recent developments in methods and technology for analysis of biological samples by MALDI-TOF-MS. Anal. Bioanal. Chem. 2007, 387, 193–204. [Google Scholar] [CrossRef]
- Schubert, S.; Weinert, K.; Wagner, C.; Gunzl, B.; Wieser, A.; Maier, T.; Kostrzewa, M. Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J. Mol. Diagn. 2011, 13, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Schulthess, B.; Bloemberg, G.V.; Zbinden, R.; Böttger, E.C.; Hombach, M. Evaluation of the Bruker MALDI Biotyper for identification of Gram-positive rods: Development of a diagnostic algorithm for the clinical laboratory. J. Clin. Microbiol. 2014, 52, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Chen, Z.Y.; Wu, H.F. Surface tuning laser desorption/ionization mass spectrometry (STLDI-MS) for the analysis of small molecules using quantum dots. Anal. Bioanal. Chem. 2017, 409, 4943–4950. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.; Walzer, K.; Leo, K. Analysis of complete organic semiconductor devices by laser desorption/ionization time-of-flight mass spectrometry. Adv. Funct. Mater. 2008, 18, 2541–2547. [Google Scholar] [CrossRef]
- Hrabák, J.; Chudáčková, E.; Walková, R. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: From research to routine diagnosis. Clin. Microbiol. Rev. 2013, 26, 103–114. [Google Scholar] [CrossRef]
- Chen, X.F.; Hou, X.; Xiao, M.; Zhang, L.; Cheng, J.W.; Zhou, M.L.; Huang, J.J.; Zhang, J.J.; Xu, Y.C.; Hsueh, P.R. Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) analysis for the identification of pathogenic microorganisms: A review. Microorganisms 2021, 9, 1536. [Google Scholar] [CrossRef]
- Angeletti, S. Matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology. J. Microbiol. Methods 2017, 138, 20–29. [Google Scholar] [CrossRef]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current status of matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) in clinical diagnostic microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef]
- Harris, L.G.; El-Bouri, K.; Johnston, S.; Rees, E.; Frommelt, L.; Siemssen, N.; Christner, M.; Davies, A.P.; Rohde, H.; Mack, D. Rapid identification of staphylococci from prosthetic joint infections using MALDI-TOF mass-spectrometry. Int. J. Artif. Organs 2010, 33, 568–574. [Google Scholar] [CrossRef]
- Moothoo-Padayachie, A.; Kandappa, H.R.; Krishna, S.B.N.; Maier, T.; Govender, P. Biotyping Saccharomyces cerevisiae strains using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Eur. Food Res. Technol. 2013, 236, 351–364. [Google Scholar] [CrossRef]
- Cassagne, C.; Normand, A.C.; L’Ollivier, C.; Ranque, S.; Piarroux, R. Performance of MALDI-TOF MS platforms for fungal identification. Mycoses 2016, 59, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Lay, J.O., Jr. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 2001, 20, 172–194. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, S.; Dufresne, P.J.; Soualhine, H.; Domingo, M.C.; Bekal, S.; Lefebvre, B.; Tremblay, C. A side by side comparison of Bruker Biotyper and VITEK MS: Utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS ONE 2015, 10, e0144878. [Google Scholar] [CrossRef]
- Wieser, A.; Schneider, L.; Jung, J.; Schubert, S. MALDI-TOF MS in microbiological diagnostics—Identification of microorganisms and beyond (mini review). Appl. Microbiol. Biotechnol. 2012, 93, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Do, T.; Guran, R.; Adam, V.; Zitka, O. Use of MALDI-TOF mass spectrometry for virus identification: A review. Analyst 2022, 147, 3131–3154. [Google Scholar] [CrossRef]
- Takach, E.J.; Hines, W.M.; Patterson, D.H.; Juhasz, P.; Falick, A.M.; Vestal, M.L.; Martin, S.A. Accurate mass measurements using MALDI-TOF with delayed extraction. J. Protein Chem. 1997, 16, 363–369. [Google Scholar] [CrossRef]
- Hillenkamp, F.; Peter-Katalinic, J. (Eds.) MALDI MS: A practical Guide to Instrumentation, Methods and Applications; John Wiley & Sons: New York, NY, USA, 2013. [Google Scholar]
- Hortin, G.L. The MALDI-TOF mass spectrometric view of the plasma proteome and peptidome. Clin. Chem. 2006, 52, 1223–1237. [Google Scholar] [CrossRef]
- Veloo, A.C.M.; Elgersma, P.E.; Friedrich, A.W.; Nagy, E.; Van Winkelhoff, A.J. The influence of incubation time, sample preparation and exposure to oxygen on the quality of the MALDI-TOF MS spectrum of anaerobic bacteria. Clin. Microbiol. Infect. 2014, 20, O1091–O1097. [Google Scholar] [CrossRef]
- Albrethsen, J. Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin. Chem. 2007, 53, 852–858. [Google Scholar] [CrossRef]
- Rodriguez-Temporal, D.; Alcaide, F.; Mareković, I.; O’cOnnor, J.A.; Gorton, R.; van Ingen, J.; Bossche, A.V.D.; Héry-Arnaud, G.; Beauruelle, C.; Orth-Höller, D.; et al. Multicentre study on the reproducibility of MALDI-TOF MS for nontuberculous mycobacteria identification. Sci. Rep. 2022, 12, 1237. [Google Scholar] [CrossRef]
- Christner, M.; Trusch, M.; Rohde, H.; Kwiatkowski, M.; Schlüter, H.; Wolters, M.; Aepfelbacher, M.; Hentschke, M.; Greub, G. Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-toxigenic Escherichia coli. PLoS ONE 2014, 9, e101924. [Google Scholar] [CrossRef] [PubMed]
- Jurinke, C.; Oeth, P.; van den Boom, D. MALDI-TOF mass spectrometry: A versatile tool for high-performance DNA analysis. Mol. Biotechnol. 2004, 26, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.L.; Andrzejewski, D.; Lay, J.O.; Musser, S.M. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 2003, 14, 342–351. [Google Scholar] [CrossRef]
- Spielmann, G.; Huber, I.; Maggipinto, M.; Haszprunar, G.; Busch, U.; Pavlovic, M. Comparison of five preparatory protocols for fish species identification using MALDI-TOF MS. Eur. Food Res. Technol. 2018, 244, 685–694. [Google Scholar] [CrossRef]
- Cuénod, A.; Aerni, M.; Bagutti, C.; Bayraktar, B.; Boz, E.S.; Carneiro, C.B.; Casanova, C.; Coste, A.T.; Damborg, P.; van Dam, D.W.; et al. Quality of MALDI-TOF mass spectra in routine diagnostics: Results from an international external quality assessment including 36 laboratories from 12 countries using 47 challenging bacterial strains. Clin. Microbiol. Infect. 2023, 29, 190–199. [Google Scholar] [CrossRef]
- Sandrin, T.R.; Goldstein, J.E.; Schumaker, S. MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom. Rev. 2013, 32, 188–217. [Google Scholar] [CrossRef]
- Patel, R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef]
- Barker, K.R.; Kus, J.V.; Normand, A.-C.; Gharabaghi, F.; McTaggart, L.; Rotstein, C.; Richardson, S.E.; Campigotto, A.; Tadros, M.; Hanson, K.E. A practical workflow for the identification of Aspergillus, Fusarium, Mucorales by MALDI-TOF MS: Database, medium, and incubation optimization. J. Clin. Microbiol. 2022, 60, e01032-22. [Google Scholar] [CrossRef]
- Lasch, P.; Beyer, W.; Bosch, A.; Borriss, R.; Drevinek, M.; Dupke, S.; Ehling-Schulz, M.; Gao, X.; Grunow, R.; Jacob, D.; et al. A MALDI-ToF mass spectrometry database for identification and classification of highly pathogenic bacteria. Sci. Data 2025, 12, 187. [Google Scholar] [CrossRef]
- Pinar-Méndez, A.; Fernández, S.; Baquero, D.; Vilaró, C.; Galofré, B.; González, S.; Rodrigo-Torres, L.; Arahal, D.R.; Macián, M.C.; Ruvira, M.A.; et al. Rapid and improved identification of drinking water bacteria using the Drinking Water Library, a dedicated MALDI-TOF MS database. Water Res. 2021, 203, 117543. [Google Scholar] [CrossRef]
- Teke, L.; Barış, A.; Bayraktar, B. Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) systems for non-albicans Candida and uncommon yeast isolates. J. Microbiol. Methods 2021, 185, 106232. [Google Scholar] [CrossRef] [PubMed]
- Thelen, P.; Graeber, S.; Schmidt, E.; Hamprecht, A. A side-by-side comparison of the new VITEK MS PRIME and the MALDI Biotyper sirius in the clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Hankins, J.D.; Amerson-Brown, M.H.; Brown, C.A.; Riegler, A.N.; Muldrew, K.L.; Dunn, J.J. Comparison of Bruker Biotyper® and Vitek® MS matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry platforms for the identification of filamentous fungi. Future Microbiol. 2023, 18, 553–561. [Google Scholar] [CrossRef]
- Quintela-Baluja, M.; Böhme, K.; Fernández-No, I.C.; Alnakip, M.E.; Caamano, S.; Barros-Velázques, J.; Calo-Mata, P. MALDI-TOF mass spectrometry, a rapid and reliable method for the identification of bacterial species in food-microbiology laboratories. In Novel Food Preservation and Microbial Assessment Technique; CRC Press: Boca Raton, FL, USA, 2014; pp. 353–385. [Google Scholar]
- Engel, K.M.; Prabutzki, P.; Leopold, J.; Nimptsch, A.; Lemmnitzer, K.; Vos, D.N.; Hopf, C.; Schiller, J. A new update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 2022, 86, 101145. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Temporal, D.; Adrados, D.; Alastruey-Izquierdo, A.; Alkorta, M.; Candela, A.; Canut, A.; Castro, C.; Cilla, C.G.; Caballero, J.D.D.; Ercibengoa, M.; et al. Current Performance of MALDI–TOF Mass Spectrometry Databases for the Identification of Dermatophyte Species. J. Fungi 2025, 11, 356. [Google Scholar] [CrossRef]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for identification of environmental bacteria: A review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef]
- Zengin Canalp, H.; Bayraktar, B. Direct rapid identification from positive blood cultures by MALDI-TOF MS: Specific focus on turnaround times. Microbiol. Spectr. 2021, 9, e01103-21. [Google Scholar] [CrossRef]
- Kim, S.H.; Chon, J.W.; Jeong, H.W.; Song, K.Y.; Kim, D.H.; Bae, D.; Kim, H.; Seo, K.H. Identification and phylogenetic analysis of Enterococcus isolates using MALDI-TOF MS and VITEK 2. AMB Express 2023, 13, 21. [Google Scholar] [CrossRef]
- Colaninno, P.M. Identification of gram-positive organisms. In Practical Handbook of Microbiology; CRC Press: Boca Raton, FL, USA, 2014; pp. 51–58. [Google Scholar]
- Dichtl, K.; Klugherz, I.; Greimel, H.; Luxner, J.; Köberl, J.; Friedl, S.; Steinmetz, I.; Leitner, E.; McElvania, E. A head-to-head comparison of three MALDI-TOF mass spectrometry systems with 16S rRNA gene sequencing. J. Clin. Microbiol. 2023, 61, e01913-22. [Google Scholar] [CrossRef]
- Cobo, F.; Pérez-Carrasco, V.; Martín-Hita, L.; García-Salcedo, J.A.; Navarro-Marí, J.M. Comparative evaluation of MALDI-TOF MS and 16S rRNA gene sequencing for the identification of clinically relevant anaerobic bacteria: Critical evaluation of discrepant results. Anaerobe 2023, 82, 102754. [Google Scholar] [CrossRef]
- Bielen, A.; Babić, I.; Vuk Surjan, M.; Kazazić, S.; Šimatović, A.; Lajtner, J.; Udiković-Kolić, N.; Mesić, Z.; Hudina, S. Comparison of MALDI-TOF mass spectrometry and 16S rDNA sequencing for identification of environmental bacteria: A case study of cave mussel-associated culturable microorganisms. Environ. Sci. Pollut. Res. 2024, 31, 21752–21764. [Google Scholar] [CrossRef]
- Honsig, C.; Selitsch, B.; Hollenstein, M.; Vossen, M.G.; Spettel, K.; Willinger, B. Identification of filamentous fungi by MALDI-TOF mass spectrometry: Evaluation of three different sample preparation methods and validation of an in-house species cutoff. J. Fungi 2022, 8, 383. [Google Scholar] [CrossRef] [PubMed]
- Barnini, S.; Ghelardi, E.; Brucculeri, V.; Morici, P.; Lupetti, A. Rapid and reliable identification of Gram-negative bacteria and Gram-positive cocci by deposition of bacteria harvested from blood cultures onto the MALDI-TOF plate. BMC Microbiol. 2015, 15, 124. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa, M.; Sparbier, K.; Maier, T.; Schubert, S. MALDI-TOF MS: An upcoming tool for rapid detection of antibiotic resistance in microorganisms. Proteomic Clin. Appl. 2013, 7, 767–778. [Google Scholar] [CrossRef]
- Alexandrov, T. MALDI imaging mass spectrometry: Statistical data analysis and current computational challenges. BMC Bioinform. 2012, 13 (Suppl. S16), S11. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Sanchez, B.; Oviaño, M. MALDI-TOF mass spectrometry in the 21st century clinical microbiology laboratory. J. Clin. Microbiol. 2020, 58, e01512-19. [Google Scholar]
- Pavlovic, M.; Huber, I.; Konrad, R.; Busch, U. Application of MALDI-TOF MS for the identification of food borne bacteria. Open Microbiol. J. 2013, 7, 135. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Hamada, M.; Al-Dubaib, M.; Alyamani, E.; Moussa, I.M.; AlRowaidhan, A.; Hemeg, H.A. Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiol. 2017, 40, 269–278. [Google Scholar]
- De Oliveira Mota, J.; Boué, G.; Prévost, H.; Maillet, A.; Jaffres, E.; Maignien, T.; Arnich, N.; Sanaa, M.; Federighi, M. Environmental monitoring program to support food microbiological safety and quality in food industries: A scoping review of the research and guidelines. Food Control 2021, 130, 108283. [Google Scholar] [CrossRef]
- Schalli, M.; Inwinkl, S.M.; Platzer, S.; Baumert, R.; Reinthaler, F.F.; Ofner-Kopeinig, P.; Haas, D. Cefsulodin and vancomycin: A supplement for chromogenic coliform agar for detection of Escherichia coli and coliform bacteria from different water sources. Microorganisms 2022, 10, 2499. [Google Scholar] [CrossRef]
- Jadhav, S.; Gulati, V.; Fox, E.M.; Karpe, A.; Beale, D.J.; Sevior, D.; Bhave, M.; Palombo, E.A. Rapid identification and source-tracking of Listeria monocytogenes using MALDI-TOF mass spectrometry. Int. J. Food Microbiol. 2015, 202, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ojima-Kato, T.; Yamamoto, N.; Takahashi, H.; Tamura, H. Matrix-assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) can precisely discriminate the lineages of Listeria monocytogenes and species of Listeria. PLoS ONE 2016, 11, e0159730. [Google Scholar] [CrossRef]
- Paauw, A.; Jonker, D.; Roeselers, G.; Heng, J.M.; Mars-Groenendijk, R.H.; Trip, H.; Molhoek, E.M.; Jansen, H.-J.; van der Plas, J.; de Jong, A.L.; et al. Rapid and reliable discrimination between Shigella species and Escherichia coli using MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 2015, 305, 446–452. [Google Scholar] [CrossRef] [PubMed]
- van den Beld, M.J.; Rossen, J.W.; Evers, N.; Kooistra-Smid, M.A.; Reubsaet, F.A. MALDI-TOF MS using a custom-made database, biomarker assignment, or mathematical classifiers does not differentiate Shigella spp. and Escherichia coli. Microorganisms 2022, 10, 435. [Google Scholar] [CrossRef]
- Santos, I.C.; Hildenbrand, Z.L.; Schug, K.A. Applications of MALDI-TOF MS in environmental microbiology. Analyst 2016, 141, 2827–2837. [Google Scholar] [CrossRef]
- Asadi, A.; Angerjas, A.; Paalme, V.; Huseynli, L.; Sarand, I. Assessment of spoilage microbial communities in modified atmosphere-packed ready-to-eat salad during cold storage: A comparative study using MALDI-TOF MS identification and PacBio full-length 16S rRNA and ITS sequencing. Int. J. Food Microbiol. 2025, 440, 111268. [Google Scholar] [CrossRef]
- Bächli, P.; Baars, S.; Simmler, A.; Zbinden, R.; Schulthess, B. Impact of MALDI-TOF MS identification on anaerobic species and genus diversity in routine diagnostics. Anaerobe 2022, 75, 102554. [Google Scholar] [CrossRef]
- Song, D.; Dong, K.; Liu, S.; Fu, S.; Zhao, F.; Man, C.; Jiang, Y.; Zhao, K.; Qu, B.; Yang, X. Research advances in detection of food adulteration and application of MALDI-TOF MS: A review. Food Chem. 2024, 456, 140070. [Google Scholar] [CrossRef] [PubMed]
- Chalupová, J.; Raus, M.; Sedlářová, M.; Šebela, M. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnol. Adv. 2014, 32, 230–241. [Google Scholar] [CrossRef]
- Zeller-Péronnet, V.; Brockmann, E.; Pavlovic, M.; Timke, M.; Busch, U.; Huber, I. Potential and limitations of MALDI-TOF MS for discrimination within the species Leuconostoc mesenteroides and Leuconostoc pseudomesenteroides. J. Verbraucherschutz Lebensmittelsicherh. 2013, 8, 205–214. [Google Scholar] [CrossRef]
- Chen, L.; Gao, W.; Tan, X.; Han, Y.; Jiao, F.; Feng, B.; Xie, J.; Li, B.; Zhao, H.; Tu, H.; et al. MALDI-TOF MS is an effective technique to classify specific microbiota. Microbiol. Spectr. 2023, 11, e00307-23. [Google Scholar] [CrossRef]
- Boyaci Gunduz, C.P.; Agirman, B.; Erten, H. Identification of yeasts in fermented foods and beverages using MALDI-TOF MS. FEMS Yeast Res. 2022, 22, foac056. [Google Scholar] [CrossRef]
- Šedo, O.; Roblíčková, A.; Ježek, F.; Gintar, P.; Kameník, J.; Zdráhal, Z. Discriminatory power of MALDI-TOF MS protein profiling analysis of pork meat and meat products. Food Chem. 2024, 449, 139155. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, E.; Million, M.; Henry, M.; Raoult, D. Rapid and accurate bacterial identification in probiotics and yoghurts by MALDI-TOF mass spectrometry. J. Food Sci. 2011, 76, M568–M572. [Google Scholar] [CrossRef]
- Rodhouse, L.; Carbonero, F. Overview of craft brewing specificities and potentially associated microbiota. Crit. Rev. Food Sci. Nutr. 2019, 59, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Zhang, L.; Yang, R.; Wang, X.; Yu, L.; Yue, X.; Ma, F.; Mao, J.; Wang, X.; Zhang, W.; et al. Mass spectrometry in food authentication and origin traceability. Mass Spectrom. Rev. 2023, 42, 1772–1807. [Google Scholar] [CrossRef]
- Arena, S.; Salzano, A.M.; Scaloni, A. Identification of protein markers for the occurrence of defrosted material in milk through a MALDI-TOF-MS profiling approach. J. Proteom. 2016, 147, 56–65. [Google Scholar] [CrossRef]
- Akimowicz, M.; Bucka-Kolendo, J. MALDI-TOF MS–application in food microbiology. Acta Biochim. Pol. 2020, 67, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Jussiaux, F.; Miot-Sertier, C.; Nguyen-Lopez, D.; Badet, C.; Samot, J. Reliability of MALDI-TOF mass spectrometry to identify oral isolates of Streptococcus salivarius and Lactobacillus spp. Arch. Oral Biol. 2021, 121, 104983. [Google Scholar]
- de Koster, C.G.; Brul, S. MALDI-TOF MS identification and tracking of food spoilers and food-borne pathogens. Curr. Opin. Food Sci. 2016, 10, 76–84. [Google Scholar] [CrossRef]
- Feucherolles, M.; Cauchie, H.M.; Penny, C. MALDI-TOF mass spectrometry and specific biomarkers: Potential new key for swift identification of antimicrobial resistance in foodborne pathogens. Microorganisms 2019, 7, 593. [Google Scholar] [CrossRef]
- Neoh, H.M.; Tan, X.E.; Sapri, H.F.; Tan, T.L. Pulsed-field gel electrophoresis (PFGE): A review of the “gold standard” for bacteria typing and current alternatives. Infect. Genet. Evol. 2019, 74, 103935. [Google Scholar] [CrossRef] [PubMed]
- Weis, C.V.; Jutzeler, C.R.; Borgwardt, K. Machine learning for microbial identification and antimicrobial susceptibility testing on MALDI-TOF mass spectra: A systematic review. Clin. Microbiol. Infect. 2020, 26, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, I.S.; Adesiyun, A.; Seepersadsingh, N.; Rahaman, S. Investigation for possible source (s) of contamination of ready-to-eat meat products with Listeria spp. and other pathogens in a meat processing plant in Trinidad. Food Microbiol. 2006, 23, 359–366. [Google Scholar] [CrossRef]
- Coeuret, V.; Dubernet, S.; Bernardeau, M.; Gueguen, M.; Vernoux, J.P. Isolation, characterisation and identification of lactobacilli focusing mainly on cheeses and other dairy products. Le Lait 2003, 83, 269–306. [Google Scholar] [CrossRef]
- Jamali, H.; Chai, L.C.; Thong, K.L. Detection and isolation of Listeria spp. and Listeria monocytogenes in ready-to-eat foods with various selective culture media. Food Control 2013, 32, 19–24. [Google Scholar] [CrossRef]
- Gribble, A.; Brightwell, G. Spoilage characteristics of Brochothrix thermosphacta and campestris in chilled vacuum packaged lamb, and their detection and identification by real time PCR. Meat Sci. 2013, 94, 361–368. [Google Scholar] [CrossRef]
- Nguyen, D.T.L.; Van Hoorde, K.; Cnockaert, M.; De Brandt, E.; Aerts, M.; Thanh, L.B.; Vandamme, P. A description of the lactic acid bacteria microbiota associated with the production of traditional fermented vegetables in Vietnam. Int. J. Food Microbiol. 2013, 163, 19–27. [Google Scholar] [CrossRef]
- Henry, M.; Fouladkhah, A. Outbreak history, biofilm formation, and preventive measures for control of Cronobacter sakazakii in infant formula and infant care settings. Microorganisms 2019, 7, 77. [Google Scholar] [CrossRef]
- Fukuda, A.; Tsunashima, R.; Usui, M. Antimicrobial Resistant Bacteria Monitoring in Raw Seafood Retailed: A Pilot Study Focused on Vibrio and Aeromonas. Food Safety 2023, 11, 65–77. [Google Scholar] [CrossRef]
- Moore, G.; Griffith, C. A comparison of surface sampling methods for detecting coliforms on food contact surfaces. Food Microbiol. 2002, 19, 65–73. [Google Scholar] [CrossRef]
- Junaid, K.; Ejaz, H.; Younas, S.; Alanazi, A.; Yasmeen, H.; Rehman, A. Detection of Klebsiella pneumoniae antibiotic-resistant genes: An impending source of multidrug resistance dissemination through raw food. Saudi J. Biol. Sci. 2022, 29, 3347–3353. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Oh, S.W. Rapid detection of E. coli O157: H7 by a novel access with combination of improved sample preparation and real-time PCR. Food Sci. Biotechnol. 2020, 29, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Välimaa, A.L.; Tilsala-Timisjärvi, A.; Virtanen, E. Rapid detection and identification methods for Listeria monocytogenes in the food chain—A review. Food Control 2015, 55, 103–114. [Google Scholar] [CrossRef]
- England, P.; Tang, W.; Kostrzewa, M.; Shahrezaei, V.; Larrouy-Maumus, G. Discrimination of bovine milk from non-dairy milk by lipids fingerprinting using routine matrix-assisted laser desorption ionization mass spectrometry. Sci. Rep. 2020, 10, 5160. [Google Scholar] [CrossRef]
- Singh, N.; Goel, G.; Raghav, M. Prevalence and characterization of Cronobacter spp. from various foods, medicinal plants, and environmental samples. Curr. Microbiol. 2015, 71, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Yehia, H.M.; Ibraheim, S.M.; Hassanein, W.A. Prevalence of Listeria species in some foods and their rapid identification. Trop. J. Pharm. Res. 2016, 15, 1047–1052. [Google Scholar] [CrossRef]
- Mezal, E.H.; Sabol, A.; Khan, M.A.; Ali, N.; Stefanova, R.; Khan, A.A. Isolation and molecular characterization of Salmonella enterica serovar Enteritidis from poultry house and clinical samples during 2010. Food Microbiol. 2014, 38, 67–74. [Google Scholar] [CrossRef]
- Normand, A.C.; Cassagne, C.; Gautier, M.; Becker, P.; Ranque, S.; Hendrickx, M.; Piarroux, R. Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiol. 2017, 17, 25. [Google Scholar] [CrossRef]
- Zhu, B.; Xiao, D.; Zhang, H.; Zhang, Y.; Gao, Y.; Xu, L.; Lv, J.; Wang, Y.; Zhang, J.; Shao, Z.; et al. MALDI-TOF MS distinctly differentiates nontypable Haemophilus influenzae from Haemophilus haemolyticus. PLoS ONE 2013, 8, e56139. [Google Scholar] [CrossRef]
- Hettick, J.M.; Kashon, M.L.; Slaven, J.E.; Ma, Y.; Simpson, J.P.; Siegel, P.D.; Mazurek, G.N.; Weissman, D.N. Discrimination of intact mycobacteria at the strain level: A combined MALDI-TOF MS and biostatistical analysis. Proteomics 2006, 6, 6416–6425. [Google Scholar] [CrossRef]
- Ramamurthy, T.; Ghosh, A.; Pazhani, G.P.; Shinoda, S. Current perspectives on viable but non-culturable (VBNC) pathogenic bacteria. Front. Public Health 2014, 2, 103. [Google Scholar] [CrossRef] [PubMed]
- Mantini, D.; Petrucci, F.; Pieragostino, D.; Del Boccio, P.; Di Nicola, M.; Di Ilio, C.; Federici, G.; Sacchetta, P.; Comani, S.; Urbani, A. LIMPIC: A computational method for the separation of protein MALDI-TOF-MS signals from noise. BMC Bioinform. 2007, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Wijetunge, C.D.; Saeed, I.; Boughton, B.A.; Roessner, U.; Halgamuge, S.K. A new peak detection algorithm for MALDI mass spectrometry data based on a modified Asymmetric Pseudo-Voigt model. BMC Genom. 2015, 16, S12. [Google Scholar] [CrossRef]
- Tabb, D.L.; Jeong, K.; Druart, K.; Gant, M.S.; Brown, K.A.; Nicora, C.; Zhou, M.; Couvillion, S.; Nakayasu, E.; Williams, J.E.; et al. Comparing top-down proteoform identification: Deconvolution, PrSM overlap, and PTM detection. J. Proteome Res. 2023, 22, 2199–2217. [Google Scholar] [CrossRef]
- Bailey, D.; Diamandis, E.P.; Greub, G.; Poutanen, S.M.; Christensen, J.J.; Kostrzew, M. Use of MALDI-TOF for diagnosis of microbial infections. Clin. Chem. 2013, 59, 1435–1441. [Google Scholar] [CrossRef]
- Hajduk, J.; Matysiak, J.; Kokot, Z.J. Challenges in biomarker discovery with MALDI-TOF MS. Clin. Chim. Acta 2016, 458, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa, M.; Maier, T. Criteria for development of MALDI-TOF mass spectral database. In MALDI-TOF and Tandem MS for Clinical Microbiolog; Wiley: Hoboken, NJ, USA, 2017; pp. 39–54. [Google Scholar]
- Chatterjee, D.; Ytterberg, A.J.; Son, S.U.; Loo, J.A.; Garrell, R.L. Integration of protein processing steps on a droplet microfluidics platform for MALDI-MS analysis. Anal. Chem. 2010, 82, 2095–2101. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Chen, P.; Huang, X.; Li, S.; Liu, B.F. Microfluidic chip electrophoresis for biochemical analysis. J. Sep. Sci. 2020, 43, 258–270. [Google Scholar] [CrossRef]
- Beaulieu, L.; Thibodeau, J.; Desbiens, M.; Saint-Louis, R.; Zatylny-Gaudin, C.; Thibault, S. Evidence of antibacterial activities in peptide fractions originating from snow crab (Chionoecetes opilio) by-products. Probiotics Antimicrob. Proteins 2010, 2, 197–209. [Google Scholar] [CrossRef]
- Dick, L.W.; McGown, L.B. Aptamer-enhanced laser desorption/ionization for affinity mass spectrometry. Anal. Chem. 2004, 76, 3037–3041. [Google Scholar] [CrossRef]
- Armengaud, J.; Trapp, J.; Pible, O.; Geffard, O.; Chaumot, A.; Hartmann, E.M. Non-model organisms, a species endangered by proteogenomics. J. Proteomics 2014, 105, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Elosta, S.; Gajdošová, D.; Hégrová, B.; Havel, J. MALDI TOF mass spectrometry of selected mycotoxins in barley. J. Appl. Biomed. 2007, 5, 39–47. [Google Scholar] [CrossRef]
- Jeyakumar, J.M.J.; Zhang, M.; Thiruvengadam, M. Determination of mycotoxins by HPLC, LC-ESI-MS/MS, and MALDI-TOF MS in Fusarium species-infected sugarcane. Microb. Pathog. 2018, 123, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Cobo, F. Application of maldi-tof mass spectrometry in clinical virology: A review. Open Virol. J. 2013, 7, 84. [Google Scholar] [CrossRef]
- Calderaro, A.; Arcangeletti, M.-C.; Rodighiero, I.; Buttrini, M.; Gorrini, C.; Motta, F.; Germini, D.; Medici, M.-C.; Chezzi, C.; De Conto, F. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry applied to virus identification. Sci. Rep. 2014, 4, 6803. [Google Scholar] [CrossRef] [PubMed]
- Lathrop, J.T.; Jeffery, D.A.; Shea, Y.R.; Scholl, P.F.; Chan, M.M. US Food and Drug Administration perspectives on clinical mass spectrometry. Clin. Chem. 2016, 62, 41–47. [Google Scholar] [CrossRef]
- Apostol, I.; Bondarenko, P.V.; Ren, D.; Semin, D.J.; Wu, C.H.; Zhang, Z.; Goudar, C.T. Enabling development, manufacturing, and regulatory approval of biotherapeutics through advances in mass spectrometry. Curr. Opin. Biotechnol. 2021, 71, 206–215. [Google Scholar] [CrossRef]
| Software and Database | Microbe Lynx System (Waters Corporation and Manchester Metropolitan University) | Maldi Biotyper (Bruker Daltonics) (Billerica, MA, USA) | SARAMIS (AnagnosTec GmbH) (Potsdam, Germany) | MS-ID (BioM Rieux) (Craponne, France) |
|---|---|---|---|---|
| Mass Spectrometer | Micro MX (Waters Corporation, Milford, MA, USA) | Microflex (Bruker, Billerica, MA, USA) | Axima (Shimadzu, Tokyo, Japan) | Vitek MS (BioM Rieux, Craponne, France) |
| Identification | Areobic/anaerobic bacteria | Bacteria, yeast and filamentous | Bacteria, yeast and filamentous | Bacteria, yeast and filamentous |
| Range | 500–15,000 Da | 2000–20,000 Da | 2000–20,000 Da | 2000–20,000 Da |
| Requires Sample Preparation | Yes (depends on the culture medium) | No | No | No |
| Reproducibility | Low | High | High | High |
| Application Type | Food Matrix | Target Organism(s) | MALDI-TOF MS Role | Reference |
|---|---|---|---|---|
| Routine pathogen identification | Meat | Salmonella spp., Listeria spp. | Rapid identification from colony isolates | [121] |
| Quality control during production | Cheese and milk | E. coli | Strain-level identification | [122] |
| Post-processing contamination detection | Ready-to-eat foods | Listeria monocytogenes | Confirmation of isolates | [123] |
| Detection of spoilage bacteria | Vacuum-packed meat | Brochothrix thermosphacta | Differentiation of spoilage flora | [124] |
| Microbiota profiling | Fermented vegetables | Lactobacillus spp. | Typing of beneficial microorganisms | [125] |
| Outbreak-source tracing | Infant formula | Cronobacter sakazakii | Strain tracking in epidemiology | [126] |
| Seafood pathogen monitoring | Raw seafood | Vibrio spp., Aeromonas spp. | Identification of marine bacteria | [127] |
| Hygiene monitoring | Food-contact surfaces | Various Gram-negative bacteria | Surface swab screening | [128] |
| Detection of resistant strains | Mixed food products | Klebsiella pneumoniae, A. baumannii | Species ID with resistance context | [129] |
| Rapid detection of outbreaks | Mixed retail samples | E. coli O157:H7 | Fast confirmation of pathogen | [130] |
| Rapid detection of Listeria monocytogenes | Various food products | Listeria monocytogenes | Reduced detection time | [131] |
| Detection of food adulterations | Milk | Bovine and non-dairy milk | Lipid fingerprinting | [132] |
| Detection of Cronobacter spp. | Environmental samples | Cronobacter spp. | Identification in environmental surveillance | [133] |
| Identification of atypical Listeria spp. | Various food products | Listeria spp. | Identification of atypical strains | [134] |
| Discrimination of Salmonella Enteritidis | Poultry | Salmonella enteritidis | Rapid serovar discrimination | [135] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kluz, M.I.; Waszkiewicz-Robak, B.; Kačániová, M. The Applications of MALDI-TOF MS in the Diagnosis of Microbiological Food Contamination. Appl. Sci. 2025, 15, 7863. https://doi.org/10.3390/app15147863
Kluz MI, Waszkiewicz-Robak B, Kačániová M. The Applications of MALDI-TOF MS in the Diagnosis of Microbiological Food Contamination. Applied Sciences. 2025; 15(14):7863. https://doi.org/10.3390/app15147863
Chicago/Turabian StyleKluz, Maciej Ireneusz, Bożena Waszkiewicz-Robak, and Miroslava Kačániová. 2025. "The Applications of MALDI-TOF MS in the Diagnosis of Microbiological Food Contamination" Applied Sciences 15, no. 14: 7863. https://doi.org/10.3390/app15147863
APA StyleKluz, M. I., Waszkiewicz-Robak, B., & Kačániová, M. (2025). The Applications of MALDI-TOF MS in the Diagnosis of Microbiological Food Contamination. Applied Sciences, 15(14), 7863. https://doi.org/10.3390/app15147863







