1. Introduction
The liver is one of the most vital organs in the human body. Liver diseases are on the rise [
1]; statistics show that more than 360 million people worldwide suffer from abnormal liver dysfunction. Each year, approximately 1.1 million people are diagnosed with new hepatocellular carcinoma [
2]. The incidence of liver cancer is ranked sixth among all cancers, while it is fourth in terms of mortality [
3]. The liver tumor is characterized by easy relapse, rapid deterioration, and easy metastasis. The success rate for liver surgery is relatively low; therefore, there is an urgent demand to improve it.
Accurate liver segmentation and volume assessment are critical in clinical settings, particularly for surgical planning, tumor monitoring, and treatment response evaluation. However, these tasks remain challenging due to the complex and variable anatomy of the liver, the low contrast between adjacent organs in computed tomography (CT) images, and the frequent presence of imaging artifacts. Moreover, manual segmentation is time-consuming and subject to inter-operator variability, underscoring the need for automated, reliable, and interpretable solutions. It remains challenging due to anatomical variations and pathological conditions that affect its morphology. Recent advances in artificial intelligence (AI), particularly in medical image analysis, have enabled substantial improvements in segmentation performance [
4]. Nonetheless, many AI-based approaches still suffer from poor interpretability and high computational requirements.
Deep learning (DL) techniques, such as convolutional neural networks and U-Net architectures, have shown high accuracy in liver segmentation tasks. However, they often function as black boxes, lacking interpretability, and demand extensive labeled datasets and powerful hardware. Traditional methods like Chan-Vese offer transparent segmentation logic, but suffer from slow execution, especially with high-resolution medical volumes. These limitations underscore the need for a hybrid approach that offers both speed and explainability while maintaining competitive accuracy. Beyond liver imaging, recent deep learning research has shown significant advances in dermatology and breast cancer screening. For example, CNN-based systems have been successfully tailored to enhance recognition of skin lesions [
5], while augmentation strategies have improved deep learning robustness in mammography [
6].
This study addresses these limitations by introducing an explainable approach that significantly reduces computation time and enhances accuracy using a ’Parallel Cropping’ technique, overcoming the current constraints identified in recent literature.
Liver segmentation and volume estimation are challenging tasks in medical imaging [
7]. The low contrast between the adjacent organs and the liver is the main problem with liver segmentation [
8]. Liver images contain a lot of unnecessary data that affects the overall process’s speed and the liver segmentation’s accuracy. Many algorithms have recently been developed for liver segmentation. We classify these approaches into two categories: traditional and learning approaches. Seeded region growth stands out as a traditional approach for segmentation and volume assessment [
9,
10,
11,
12]. Xuesong et al. proposed automatic liver segmentation using a multi-dimensional graph cut with shape information [
13]. U-Net is a convolutional network approach used for biomedical image segmentation [
14,
15]. U-Net applied to liver segmentation sometimes segments the part outside the liver boundary [
14]. In contrast, the liver segmentation based on Chan-Vese is slow due to the unwanted data in the medical image [
16,
17].
Learning-based algorithms require extensive datasets for training and validation, utilizing complete liver images obtained by CT [
18]. DL solutions require high computational resources and labeled data availability, which is not always available; it is currently scarce. Numerous studies have advanced the field of liver segmentation using deep learning models. Transformer-based architectures such as Swin-Transformer and hybrid attention-based networks like DRDA-Net have recently demonstrated superior performance on liver and tumor segmentation tasks [
19]. These models excel in accuracy but are computationally intensive and require extensive labeled datasets, which limit their real-time deployment and interpretability. Our work takes a complementary path by focusing on pre-processing techniques that can be integrated into such pipelines, reducing input complexity and accelerating inference.
So, the most common liver dataset is LiTS [
20], which provides only 131 livers. Recent DL-based proposals in prestigious journals have used LiTS. RMAU-Net [
21] consists of a segmentation network incorporating residual and multi-scale attention mechanisms to enhance liver and tumor segmentation from CT scans. Ref. [
22] explores a hybrid approach combining deep residual networks with a biplane segmentation method to optimize liver boundary detection and volume estimation. Both articles present high-performing segmentation methods. Their reliance on computational power and large datasets limits their applicability in real-time, resource-limited environments.
DL models operate as black boxes. The decision-making process behind the segmentation results is not easily interpretable, making it difficult for clinicians to understand why the model produces certain outputs, especially in edge cases or when anomalies are present. In the medical field, transparency and the ability to explain outcomes are crucial for gaining trust and acceptance from healthcare professionals. The lack of interpretability in these DL models poses a barrier to clinical adoption, as practitioners might be hesitant to rely on a system that cannot provide clear justifications for its results. When a model performs poorly or produces incorrect segmentations, it is difficult to diagnose and address the cause of the error without a clear understanding of how the model processes the data. This lack of insight makes it challenging to fine-tune or adapt the model for different datasets or clinical environments, limiting its flexibility and robustness.
In contrast, traditional methods provide transparency and robustness but suffer from slow execution, particularly when handling large volumetric medical images containing extensive irrelevant regions outside the organ of interest (RoI) [
23]. These unwanted elements within liver images pose challenges for real-time segmentation implementation, detrimentally affecting segmentation accuracy [
9]. Processing medical images entails managing vast amounts of data, often taxing memory resources [
24]. When executed on a graphics processing unit (GPU), insufficient GPU memory hampers the segmentation process [
25]. GPUs are pivotal in accelerating processes within image-guided surgery (IGS) [
9,
10]. Employing cropping techniques aids in eliminating unwanted elements from medical images, enhancing overall process efficiency.
We aim to achieve a rapid parallel cropped image for liver segmentation and volume assessment while minimizing intermediate memory transfers between the central processing unit (CPU) and GPU. This strategy reduces processing overhead on unnecessary image elements, obtaining significant speed improvements. Our proposal is based on the persistent thread block (PT) approach [
26]. The PT approach relies on the number of computing blocks available. In cases where there are insufficient computing blocks to process image elements independently, the remaining elements’ workload is distributed among existing blocks. These blocks iterate over all image elements until complete processing, utilizing a grid-stride loop described by Gupta et al. [
26]. Persistence and grid-stride loop-based cropping offer the flexibility to harness parallelism while minimizing the processing of unwanted voxel elements. This technique consists of two parts: first, calculating diagonally opposite cropping points in a single GPU kernel call, followed by efficient data transfer from the original to the cropped image.
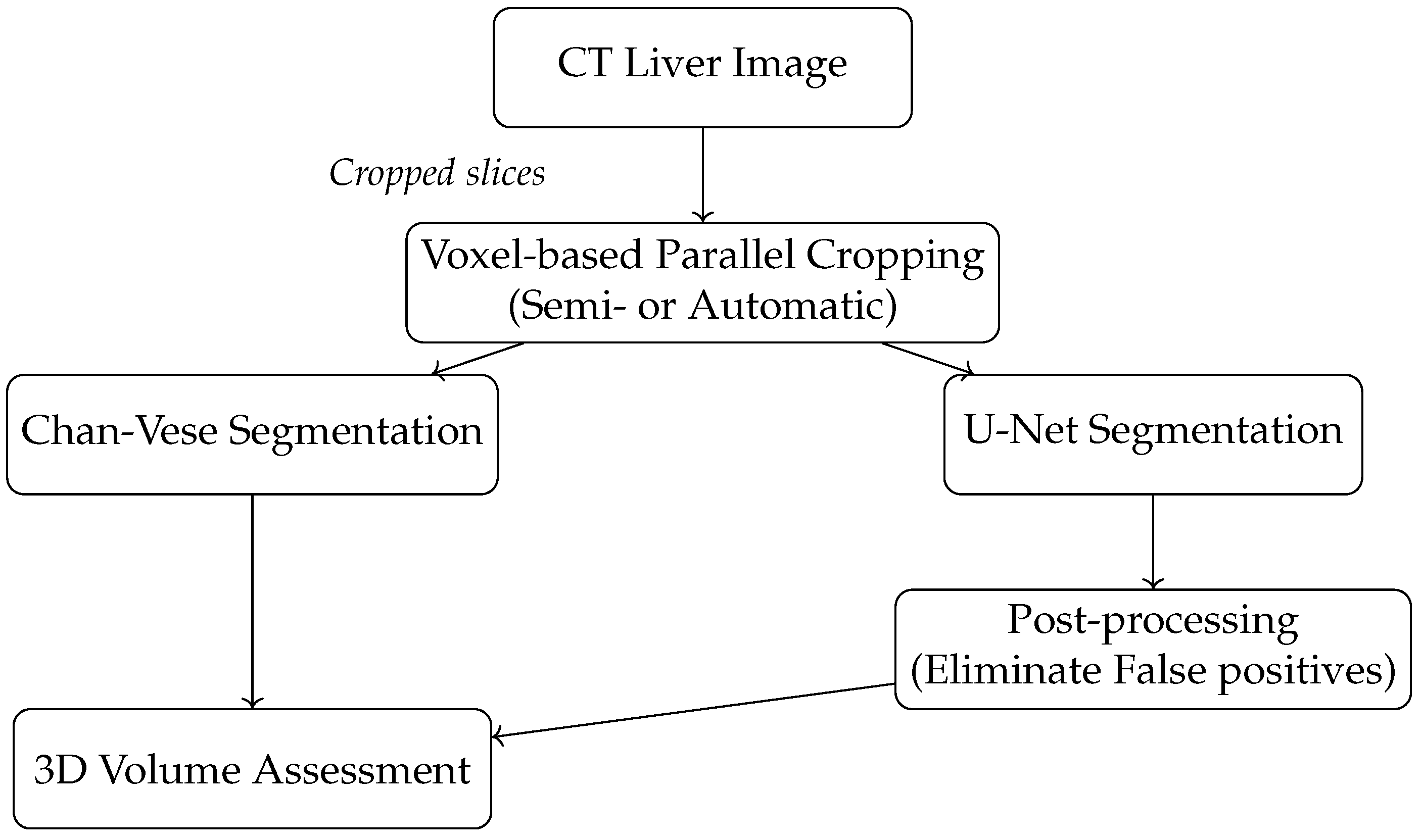
The main contributions of this work are as follows: (1) A novel voxel-based parallel cropping algorithm is used that significantly reduces unnecessary image content before segmentation. (2) We demonstrate that this approach improves segmentation accuracy and computational efficiency when integrated with traditional (Chan-Vese) and deep learning (U-Net) models, but other models could be integrated, such as U-net++ [
27], Swin-UNet [
28] or TransUNet [
29]. (3) Our voxel-based approach computes cropping boundaries in a single pass across all dimensions, unlike previous slice-based methods in 3D volume assessment, minimizing memory transfers and redundant kernel launches, resulting in a 2.29× speedup without sacrificing accuracy. (4) A clinically interpretable, resource-efficient alternative to DL-based segmentation pipelines is provided, offering practical deployment advantages for real-time medical imaging applications. A new pipeline diagram (
Figure 1) illustrates our method’s integration into existing segmentation workflows.
The rest of the paper is structured as follows.
Section 2 briefly reviews the relevant background and motivation for crop images and liver segmentation.
Section 3 explains the parallel GPU approaches for automatic and semiautomatic cropping.
Section 4 and
Section 5 discuss the application of parallel cropping to liver segmentation and volume assessment.
Section 6 analyzes performance results and the persistent and grid-stride loop-based parallel cropping comparison for liver segmentation and volume assessment.
Section 7 summarizes the main conclusions.
2. Background and Motivation
In medical image analysis, particularly for liver segmentation, input data often contains substantial irrelevant information that does not contribute to the target RoI. Processing these unnecessary voxels increases memory usage, causes longer computation times, and reduces overall system responsiveness. Traditional slice-based cropping approaches involve multiple kernel invocations and memory transfers between CPU and GPU. Removing unnecessary data reduces memory usage, avoiding redundant memory transfers and unnecessary computations and, thus, reducing execution time [
9,
30]. Cropping helps select the desired RoI of the targeted application in the analysis of medical imaging data. Due to poor image quality, the object’s boundary in RoI is sometimes diffused in a CT liver image, making it challenging to find the RoI automatically [
31].
Previous works such as [
9,
30] have illustrated the computational drawbacks of these multi-pass approaches. We propose a GPU-based voxel-level cropping technique integrated into the segmentation pipeline to discard irrelevant data in a single pass, dramatically reducing kernel calls and intermediate memory operations.
From a clinical standpoint, accelerating liver segmentation pipelines is essential for time-sensitive workflows such as preoperative planning or intraoperative navigation [
7]. In hospitals with constrained computational resources, the proposed cropping method enables the deployment of segmentation models with smaller memory footprints and faster execution, facilitating broader accessibility and practical integration into daily radiological operations [
23,
31].
Yu et al. [
32] proposed the creation of a robust segmentation model that maintains high accuracy even when incomplete overlap between liver lesions in different imaging phases exists. Ao et al. [
33] proposed a novel Transformer–CNN combined network using multi-scale feature learning for three-dimensional (3D) medical image segmentation. Smistad et al. [
30,
34] have discussed parallel cropping for image segmentation. Tran et al. explored U-Net derived from a traditional U-Net for the liver and liver–tumor segmentation. Their method utilized dilated convolution and dense structure [
35]. In [
36], Liu et al. used artificial intelligence based on the k-means clustering method for liver segmentation. Their model shows improved performance when compared with the region-growing method. Li et al. [
37] proposed Attention U-Net++ (a nested attention-aware segmentation network) with deep supervised encoder–decoder architecture and a redesigned dense skip connection. Fan et al. [
38] presented a multi-scale attention net combing self-attention algorithm that adaptively integrates local features with global dependencies. Nawandhar et al. [
39] developed a stratified squamous epithelium biopsy image classifier-based sequential computing environment for image segmentation. Finally, Gul et al. [
23] provided an overview of the available deep learning approaches for segmenting the liver and detecting liver tumors and their evaluation metrics, including accuracy, volume overlap error, dice coefficient, and mean square distance.
Most recently, some DL implementations have been developed for liver and tumor segmentation. Ref. [
40] proposes a deep learning architecture based on the Swin-Transformer structure, which is employed to capture the spatial relationships and global information for liver and tumor segmentation. The DRDA-Net framework [
41] analyzes CT liver images, integrating deep residual learning and dual-attention mechanisms to extract local and global contextual features. Finally, ref. [
42] presents a hybrid ResUNet model combining the ResNet and U-Net models.
They crop the medical image dataset before processing. The CPU allocates the memory equivalent to the cropped size to copy data to the cropped image on the GPU. Further seeded region growing is performed for image segmentation. This is the simplistic representation of the work by Smistad et al. [
30]. They have proposed a non-PT (non-persistent thread) approach for cropping and seeded region growth. Persistence implies the number of active computing threads processing a complete image. If the number of active computing threads is lesser than the image elements, then the task of the remaining image elements is distributed amongst the threads. The total number of active persistent threads implies the grid size.
Smistad et al. [
30] implemented a slice-based parallel cropping algorithm. Slice-based parallel cropping needs three kernel invocations to estimate cropping points and one kernel invocation to generate cropped data within cropping points. There are four kernel calls for 3D parallel cropping, which impacts the performance. Once the boundary is detected using cropping points, it is necessary to crop the image properly. Over-cropping of the liver crops part of the liver and destroys the segmentation quality. Under-cropping crops a small amount of data from the image, resulting in a limited speedup. It is necessary to balance the speedup and accuracy using cropping and parallelization to avoid the above-mentioned problems. Hence, we propose persistence and voxel-based parallel cropping to improve the performance of liver segmentation and volume assessment by reducing unwanted computations and avoiding intermediate memory transfers between CPU and GPU. We discuss parallel cropping in the following section in more detail.
3. Methodology
Our hypothesis proposes a voxel-based approach to compute all cropping boundaries in a single pass across all dimensions. This should minimize memory transfers and redundant kernel launches, increasing performance without sacrificing accuracy. We propose a GPU-optimized parallel cropping method that replaces traditional multi-pass, slice-based approaches with a single, volume-level processing step to achieve this. The technique leverages persistent threads and grid-stride loops to efficiently localize and extract the RoI, improving speed and memory utilization.
We describe two novel cropping techniques: semiautomatic and automatic. At first, semiautomatic cropping is proper for low-quality images with unclear boundaries, requiring minimal user interaction to indicate two diagonally opposite points that surround the liver. Automatic cropping, on the other hand, is fully data-driven and relies on voxel-level thresholding to identify the RoI without manual input.
Both cropping variants are implemented as GPU kernels that compute the RoI’s bounding coordinates and then transfer only the relevant voxel data to a new memory buffer. This compact representation is then passed to the segmentation module, significantly reducing input size and enabling faster downstream processing.
We integrate the proposed cropping step into two segmentation pipelines: traditional Chan-Vese and deep learning-based models. The Chan-Vese segmentation method is particularly well-suited for cropped images due to its robustness and mathematical transparency. It iteratively evolves a contour to fit the liver boundary and its performance benefits significantly from the reduced input size achieved via cropping.
In parallel, we apply the cropped images to a U-Net model trained on the LiTS dataset. The reduction in input complexity accelerates the model’s inference time. However, resizing steps introduced by the fixed-size input requirement may reduce raw segmentation accuracy. To address this, a post-processing filter is applied to retain only the largest connected component, effectively eliminating false positives. This combination enables high-precision segmentation with reduced computational demands.
Both segmentation outputs—whether from Chan-Vese or post-processed U-Net—are inputs to the final volume assessment stage. We compute the total liver volume by applying a 3D-seeded region growing on the segmented volume. The accuracy and efficiency of this stage are directly enhanced by the compactness of the cropped data, which reduces voxel traversal and memory overhead.
3.1. Semiautomatic Cropping
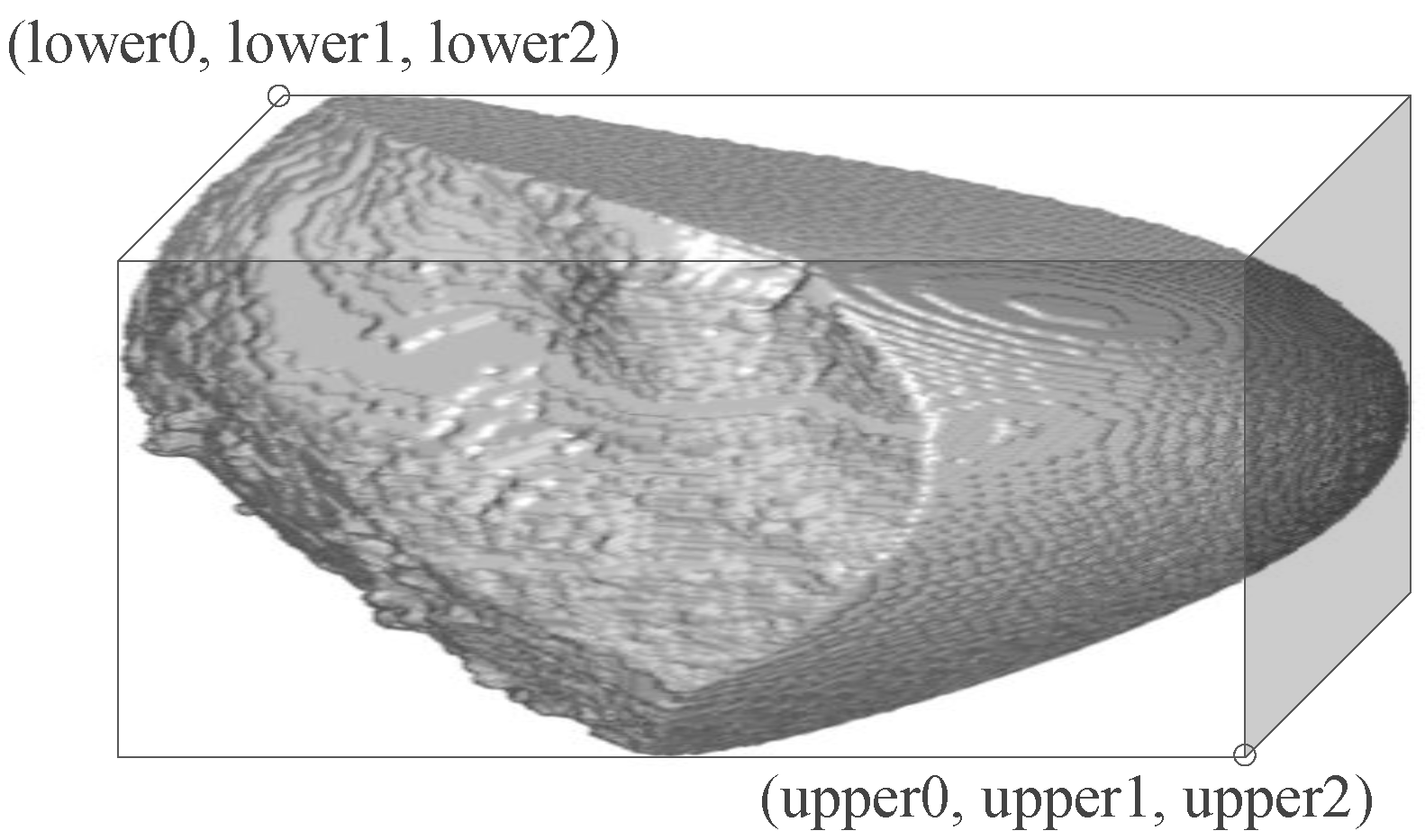
CT liver images are sometimes poor in image quality. The boundary of the liver in poor-quality images is diffused sometimes, requiring manual intervention during the processing of the liver images. In semiautomatic cropping, at first, two diagonally opposite points are provided by the clinician expert. Then, we calculate the number of voxels inside the cuboid (in 3D volume, as shown in
Figure 2) within the cropping points to allocate the required GPU memory for the cropped image in the GPU.
The CPU calls a kernel on the GPU to copy cropped data from the original image. The advantage is computing units process tiles within the cropped image rather than processing the whole image, as done in the non-PT approach. The process is shown in Algorithm 1.
For each voxel in parallel, the GPU calculates the position of the voxel on the original and cropped image given by “index” and “index_crop”, respectively. It checks if this voxel is within the diagonally opposite cropping points. The data within the cropping points is copied on the GPU to create a cropped image. The semiautomatic cropping is essential where the boundaries are diffused enough that it is difficult to calculate the RoI automatically. This study explores liver segmentation as one of the applications for semiautomatic cropping. In the next section, we discuss automatic cropping for liver volume assessment.
| Algorithm 1: Copy Cropped Data |
- 1:
for each voxel from crop size in parallel - 2:
index=g(voxel.x, voxel.y, voxel.z); - 3:
index_crop=f(voxel_crop.x, voxel_crop.y, voxel_crop.z); - 4:
if && then - 5:
if && then - 6:
if && then - 7:
- 8:
end if - 9:
end if - 10:
end for
|
3.2. Automatic Cropping
We aim to automate the process of cropping in medical image analysis. Automatic cropping does not involve any user interaction. This approach finds diagonally opposite points automatically to generate the cropped image. For each voxel, the method determines if it belongs to the RoI. The process is shown in Algorithm 2. There are two diagonal opposite points, lower (i.e., lower0, lower1, lower2) and upper (i.e., upper0, upper1, upper2), as shown in
Figure 2. We initialize lower points by the size of the image and upper points to 0. For each voxel in parallel, if it takes a value of 1, the lower values (lower0, lower1, lower2) are assigned to the minimum values of the indices, and the upper values (upper0, upper1, upper2) are assigned to the maximum values of the indices from the voxels in x, y and z directions.
| Algorithm 2: Voxel-based Cropping |
- 1:
initialization of global variables; - 2:
lower0=width; lower1=height; lower2=depth; - 3:
upper0=upper1=upper2=0; - 4:
for each voxel in parallel perform - 5:
if threshold0<value(voxel)<threshold1 then - 6:
lower0=atomicMin(&lower0,voxel.x); - 7:
lower1=atomicMin(&lower1,voxel.y); - 8:
lower2=atomicMin(&lower2,voxel.z); - 9:
upper0=atomicMax(&upper0,voxel.x); - 10:
upper1=atomicMax(&upper1,voxel.y) - 11:
upper2=atomicMax(&upper2,voxel.z) - 12:
end if
|
The voxels are processed by threads in parallel and the threads may overlap inaccurate values of the diagonally opposite points. Hence, these operations of finding maximum and minimum values need to be atomic so that valid data is assigned to each of the diagonally opposite points, “lower” and “upper”. When all the voxels are processed, the final RoI is represented by a cuboid shape, as shown in
Figure 2. Once the cropping points are obtained, the task is to create a cropped image applying the same approach described in Algorithm 1 for semiautomatic cropping. We use automatic cropping for 3D volume assessment and discuss the applications of fast parallel cropping to liver segmentation and 3D volume assessment in the following sections.
3.3. Post-Processing
We apply a post-processing step after the initial prediction to improve the quality of U-Net-based liver segmentation. This step filters out small, spurious regions by retaining only the largest connected component in the segmentation mask. The rationale is that the liver typically forms the most voluminous continuous structure in the field of view.
The post-processing procedure is presented in Algorithm 3.
| Algorithm 3: Post-processing after U-Net Segmentation |
- 1:
Label all connected components in SegmentationMask - 2:
for each component do - 3:
Compute voxel count (volume) - 4:
end for - 5:
Identify the largest component - 6:
CleanedMask← binary mask containing only - 7:
return CleanedMask
|
This step is particularly effective in removing false positives that result from U-Net’s tendency to segment outside the target region when encountering low-contrast structures or noise. We observed that applying this step leads to a marked improvement in Dice scores and contour accuracy.
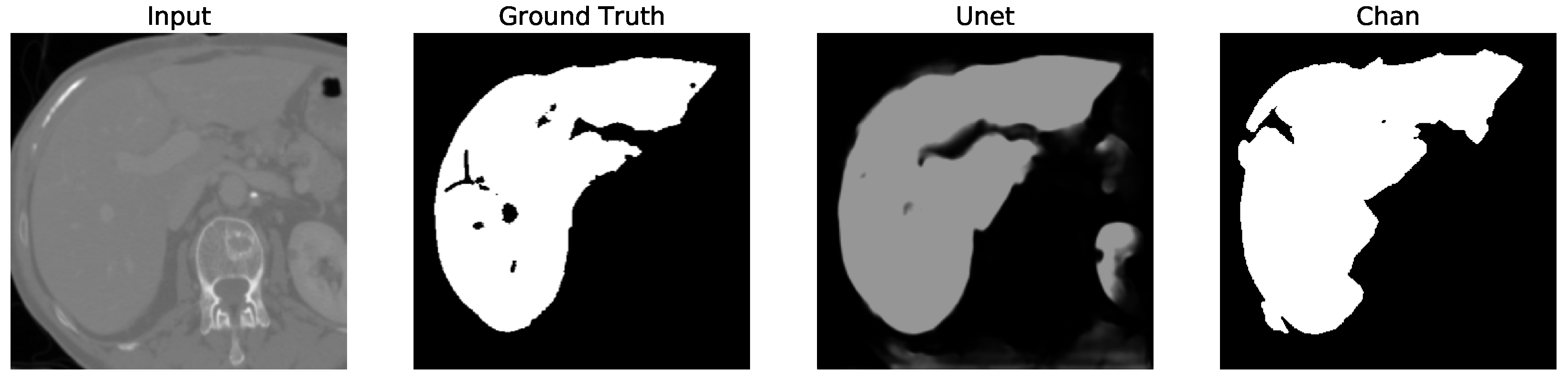
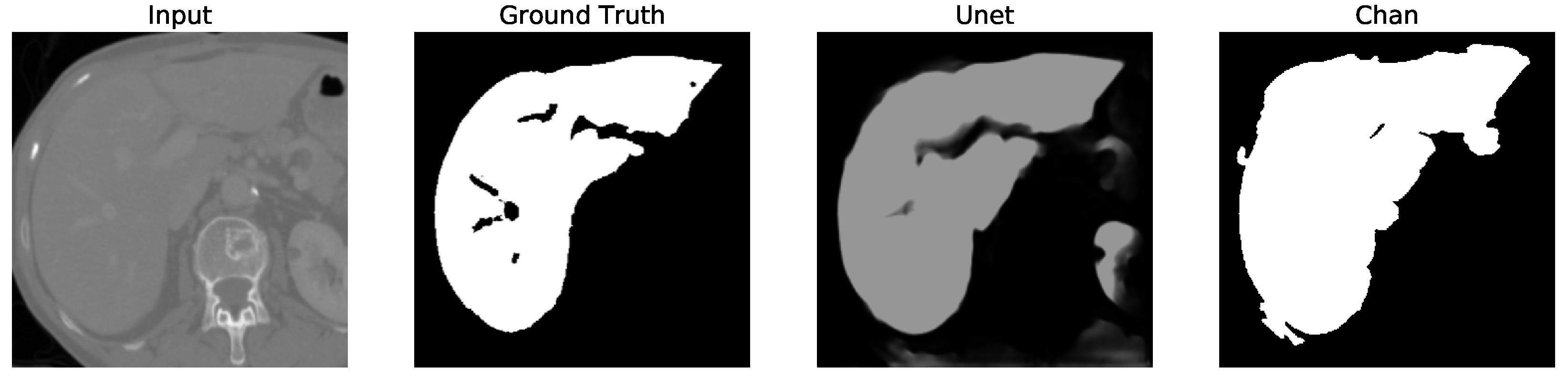
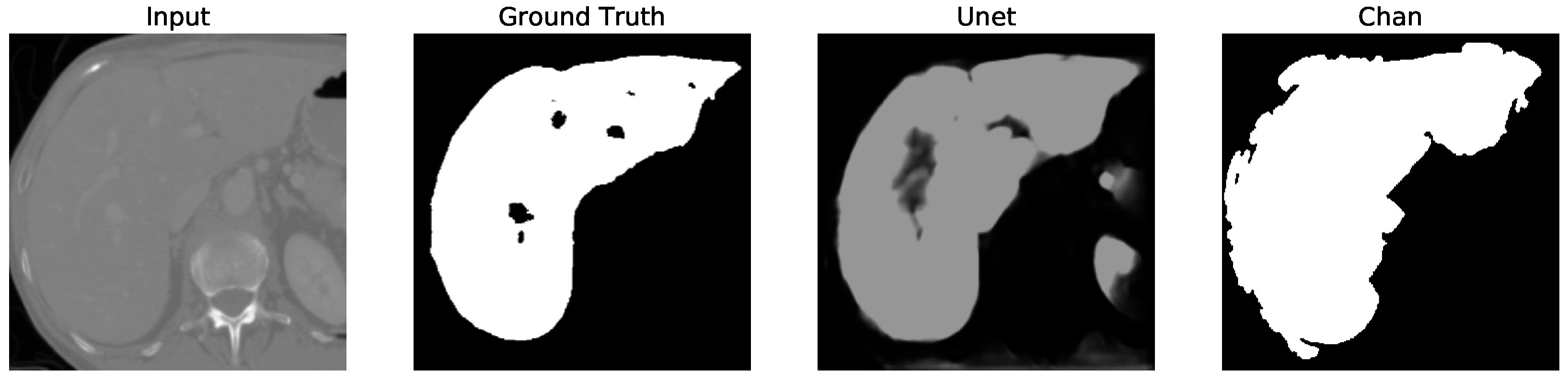
Despite U-Net’s high accuracy on the full dataset, it occasionally segments structures outside the liver boundary, particularly in low-contrast regions. This often results in unwanted elements (false positives), as shown in U-Net, Figures 5–12 where the model erroneously includes adjacent organs. To correct this, a post-processing step was introduced to retain only the largest connected component in the output mask (U-Net Figures 13–16, effectively isolating the liver and improving segmentation fidelity.
Finally, it is necessary to resize cropped inputs back to the fixed input shape required by U-Net.
4. Application to Liver Segmentation
This study examines the impact of the cropping on the liver segmentation and the application of semiautomatic cropping to segment the liver. The Chan-Vese implementation by Siri and Latte [
16] and the U-Net framework by Ronneberger et al. [
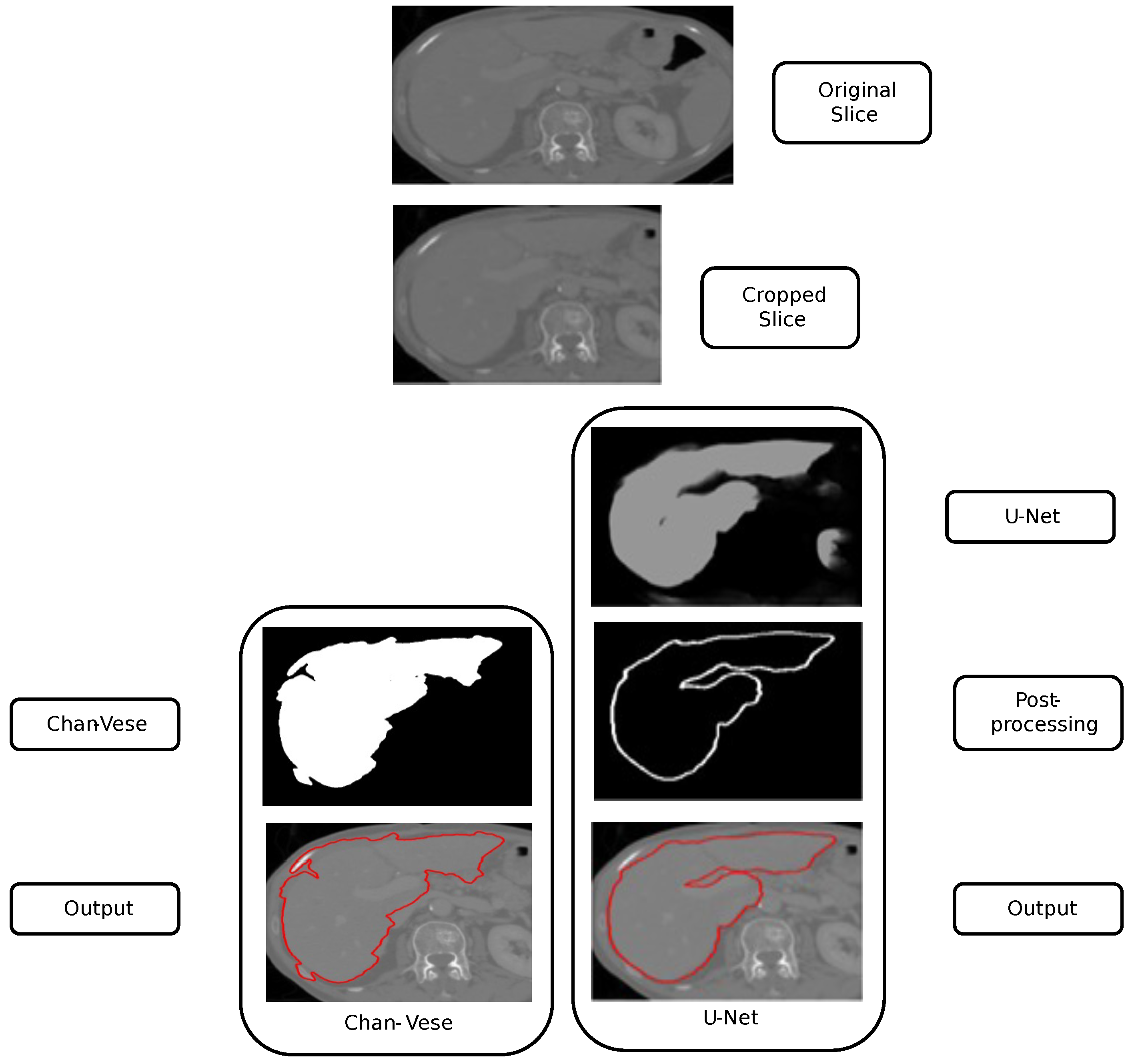
14] are discussed in this section for liver segmentation. We crop the original liver slice using the proposed voxel-based parallel cropping approach. The cropped liver slice is provided as an input to both models, i.e., traditional liver segmentation using Chan-Vese and learning-based liver segmentation using U-Net. The study analyzes segmented liver output from both models and the impact of cropping on liver segmentation.
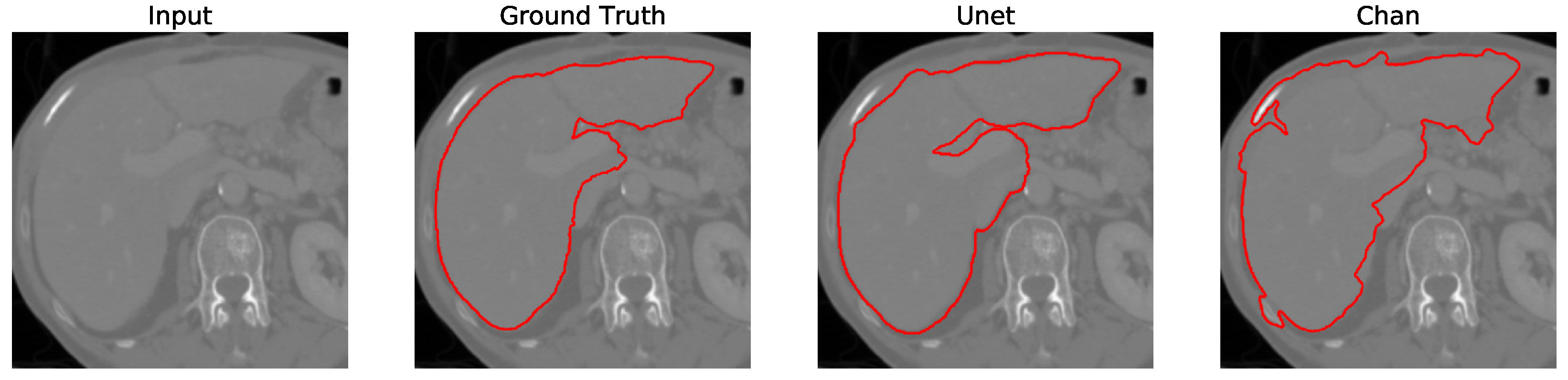
Liver segmentation using the Chan-Vese and U-Net approaches is shown in
Figure 3. We apply parallel cropping on the input liver slice to obtain the cropped liver slice. On the left, the Chan-Vese algorithm is applied, obtaining the final segmentation. Similarly, on the right, liver segmentation using U-Net is computed. The U-Net result presents an unwanted segmentation outside the liver, on the right. This unwanted segmentation outside the liver is removed in the post-processing step, obtaining the image’s maximum component and filtering out the other unwanted segmentation. U-Net has been trained using the (LiTS) dataset [
20], which is composed of 131 CT liver volumes (58638 CT liver slices). We used 100 volumes for training, 10 for validation, and 21 for testing.
The U-Net model was trained for liver segmentation using 58,638 axial slices derived from 131 annotated CT volumes in the LiTS dataset. We applied the Adam optimizer with a learning rate of , values , and a weight decay of for regularization. Training was performed for 250–300 epochs with early stopping and learning rate scheduling, which allowed fine segmentation convergence without overfitting. To improve generalization, we employed data augmentation including random horizontal and vertical flips as well as random rotations.
The loss function for binary segmentation combined binary cross-entropy (BCE) and Dice loss in an equal proportion:
where
represents binary cross-entropy loss, and
denotes the Dice loss computed between the predicted and ground truth masks. This formulation balances pixel-wise classification accuracy with region-level overlap, which is particularly important for liver boundary precision and accurate volume estimation.
We assess the quality of liver segmentation on original and cropped images and discuss the results and improvement in segmentation quality in the performance evaluation section. In the next section, the study analyzes liver volume assessment as an application for 3D cropping.
5. Application to 3D Volume Assessment
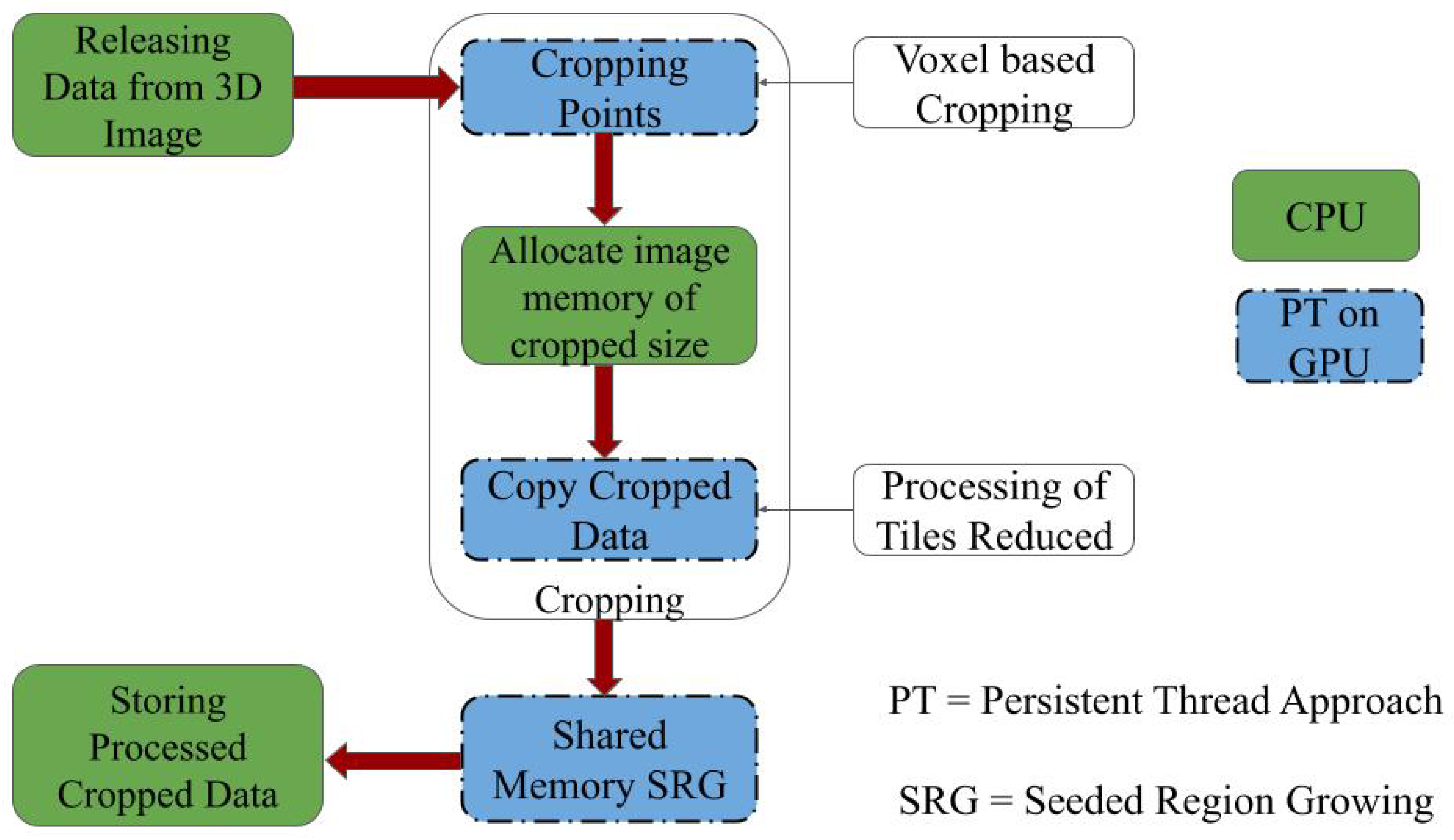
The study aims to assess liver volume as efficiently as possible. We propose fast parallel voxel-based 3D cropping for volume assessment. Parallel automatic cropping is implemented as the initial stage, and 3D seeded region growing is applied. The complete process is described below.
By applying parallel voxel-based cropping before seeded region growing (SRG), we significantly reduce the number of voxels involved in the segmentation process. This reduction lowers memory consumption on the GPU and accelerates the segmentation computation. The cropping ensures that seeded region growing operates within a tightly bounded region of interest (RoI), enhancing efficiency without sacrificing accuracy. This is particularly beneficial in high-resolution CT volumes, where traditional full-volume SRG methods are prohibitively expensive.
The SRG algorithm is implemented using GPU-based persistent threads with kernel relaunching (KTRL) to enable effective exploration of 3D neighborhoods. After cropping, the seed point is automatically determined within the binary mask, and parallel region growing is used to aggregate connected liver tissue voxels. The volume is computed as the product of labeled voxel count and voxel spacing, yielding precise liver volume estimates directly from CT data. The proposed parallel approach for 3D volume assessment is shown in
Figure 4.
The CPU stores the 3D images on the GPU. We apply PT and voxel-based parallel cropping to obtain the necessary RoI on the CT liver image. The 3D volume of the desired organ inside this RoI is assessed using PT and shared memory-based SRG. The parallel implementation of SRG is carried out using kernel termination and relaunch (KTRL) [
34]. The SRG algorithm is implemented using GPU-based persistent threads with kernel relaunching (KTRL) to enable effective exploration of 3D neighborhoods. After cropping, the seed point is automatically determined within the binary mask, and parallel region growing is used to aggregate connected liver tissue voxels. Volume is computed as the product of labeled voxel count and voxel spacing, yielding precise liver volume estimates directly from CT data. The proposed parallel approach for 3D volume assessment is shown in
Figure 4.
The CPU transfers the CT liver image to the GPU. The liver image is cropped to obtain the necessary RoI. We apply KTRL-based SRG to the cropped image. The volume of the desired organ is assessed using KTRL-based SRG on the static RoI of the image tiles, and the processed image is stored back on the CPU.
To demonstrate the generalizability of the proposed cropping method, we evaluated its performance on multiple datasets from the 3D Image Reconstruction for Comparison of Algorithm Database (3DIRACDb [
43]), including vessel, skin, and lung CT volumes. In all cases, the voxel-based cropping consistently accelerated the preprocessing and volume estimation stages when compared to conventional slice-based methods. This shows that our approach is not limited to liver imaging but is broadly applicable to other anatomical structures.
In the next section, we evaluate the impact of cropping on liver segmentation and volume assessment, discuss the speedup obtained by cropping, and analyze the segmentation accuracy.
6. Experimentation and Discussion
In this section, we analyze the performance improvements achieved through the proposed parallel cropping techniques, focusing on liver segmentation and volume assessment. Our evaluation emphasizes the gains in both accuracy and processing speed. The comparison between voxel- and slice-based cropping is discussed for the liver volume assessment. We used Intel(R) Core(TM) i7-7700HQ CPU @ 2.80GHz RAM 24 GB, NVIDIA GPU GeForce GTX 1050 (RAM 4GB), OpenCL 1.2 and CUDA Toolkit 10.1 for the implementation.
6.1. Liver Segmentation
This study aims to analyze the impact of cropping on liver segmentation. The segmented results using Chan-Vese [
16] and U-Net [
14] are shown for four slices. We show the liver segmentation on the original CT liver slices in
Figure 5,
Figure 6,
Figure 7 and
Figure 8. The liver segmentation on cropped liver slices is shown in
Figure 9,
Figure 10,
Figure 11 and
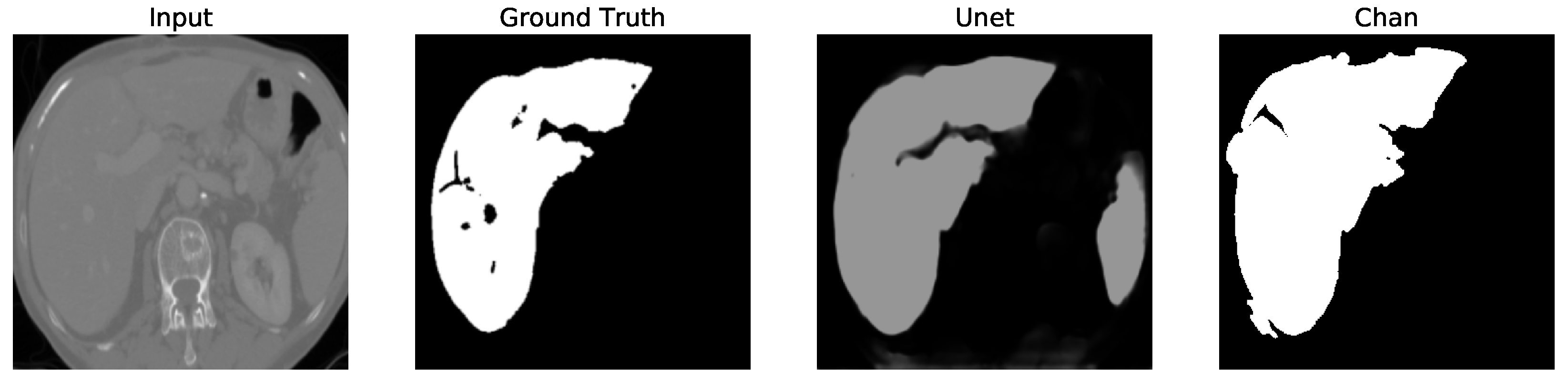
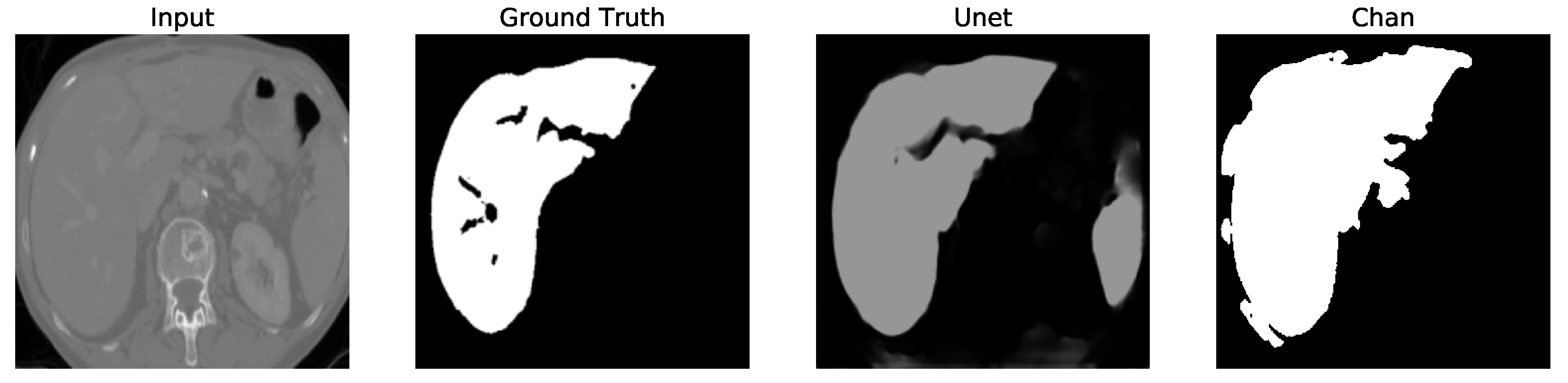
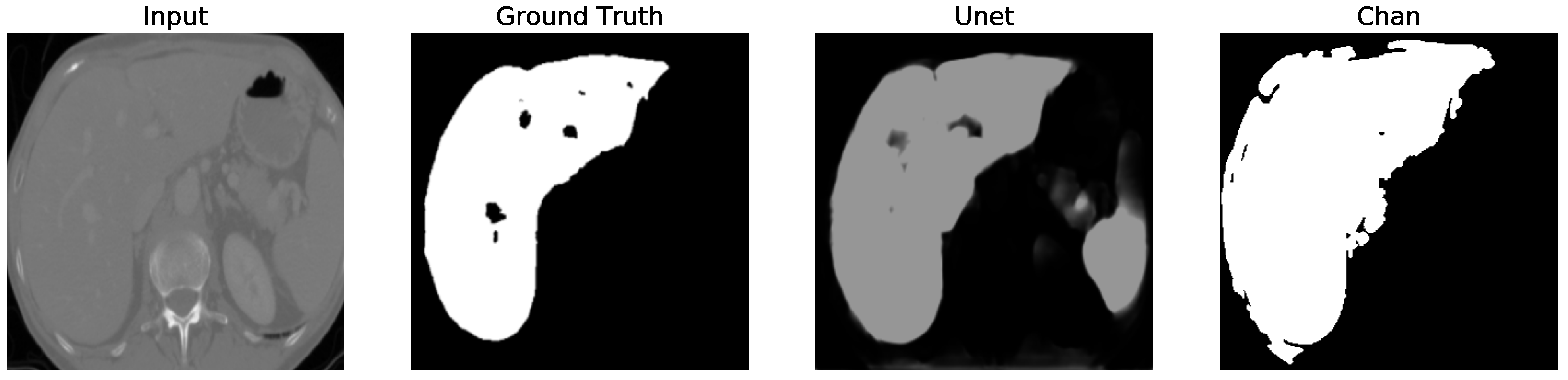
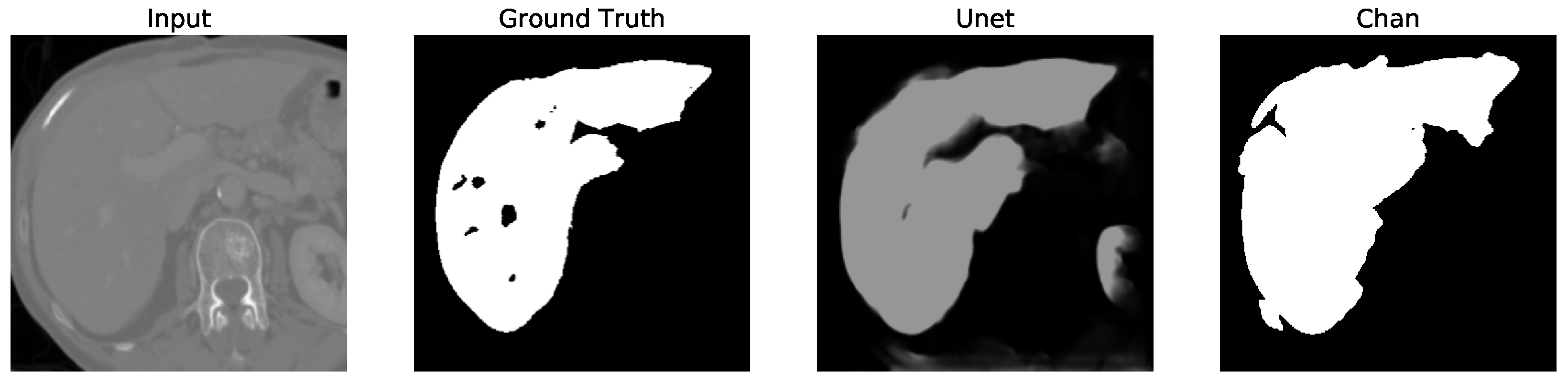
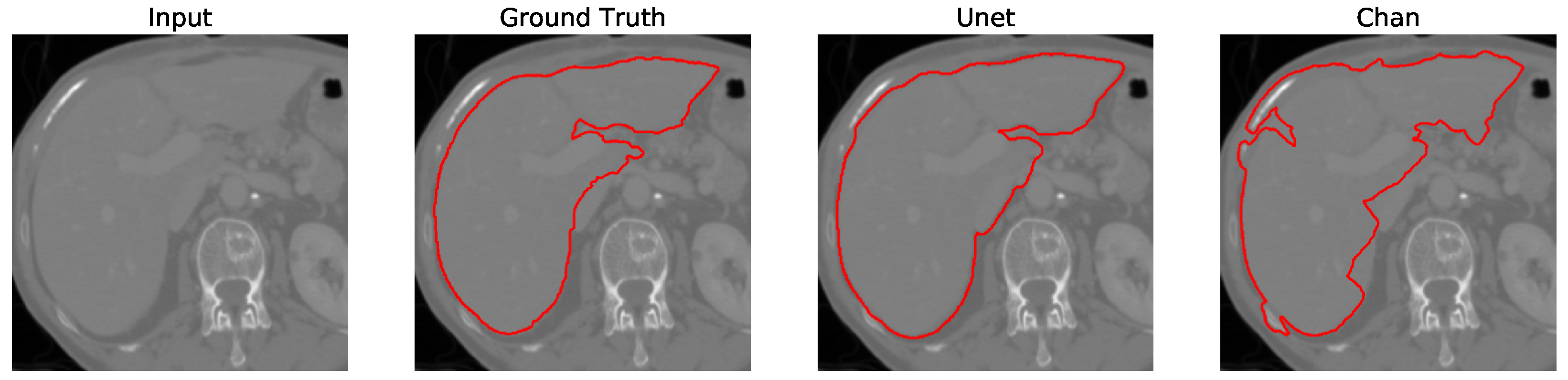
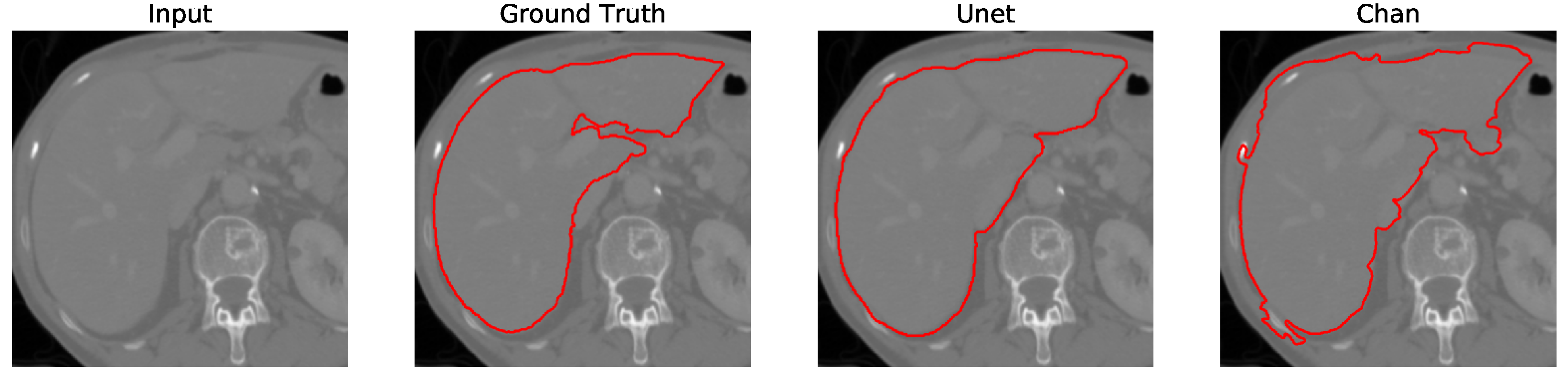
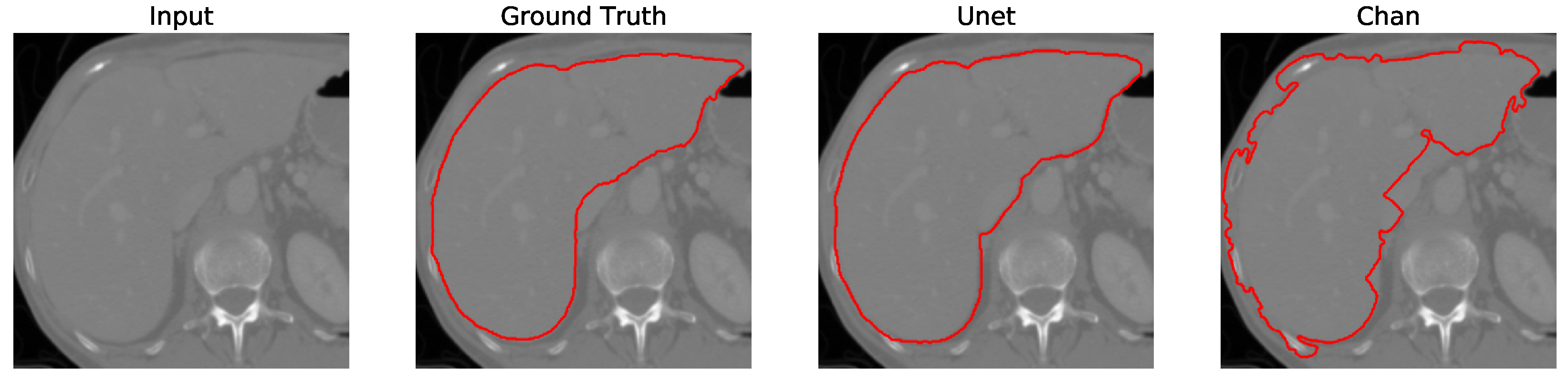
Figure 12. We apply the U-Net and Chan-Vese methods to the original and cropped liver slices. In previous figures the Chan-Vese model does not show any unwanted elements in the segmentation; however, the U-Net model presents unwanted elements.
Figure 13,
Figure 14,
Figure 15 and
Figure 16 present a segmentation comparison between the ground truth, the Chan-Vese approach, and the U-Net algorithm obtained from cropped slices 1–4.
The impact of cropping on liver segmentation was assessed using both the Chan-Vese and U-Net models.
When applied to original CT liver slices, the Chan-Vese model achieved an average dice score of 0.938, with an average processing time of 0.791 s per slice. After applying the cropping technique, the dice score improved to 0.960, and the processing time was reduced to 0.571 s per slice, resulting in an average speedup of 1.387 times. These results affirm that cropping helps to eliminate irrelevant data, focusing computations on the liver, which improves both accuracy and efficiency.
The U-Net model, however, initially showed a decrease in performance when tested on cropped slices, with the dice score dropping from 0.521 to 0.431. This decline was attributed to the resizing of cropped slices to match the input size required by U-Net, which impacted accuracy. Nonetheless, after applying post-processing to remove unwanted segmented areas, the dice score increased to 0.956. Although U-Net’s dice score is comparable to that of Chan-Vese after post-processing, the Chan-Vese model consistently outperformed U-Net in speed and segmentation accuracy on cropped images.
When the slices are cropped, the dice score (defined by Zhao et. al. [
44]) (0.96 ± 0.005) and speedup are improved in the Chan-Vese model, as shown in
Table 1. But the dice score is reduced in U-Net-based liver segmentation before post-processing (0.431 ± 0.04), as shown in
Table 2. Dice score reduction due to cropping in U-Net in
Table 2 is due to the fact that the slices are resized again to map the input of the U-Net network. U-Net is trained on original images and tested on original slices. The study examines U-Net trained on cropped images for testing cropped slices. When we perform post-processing after U-Net-based liver segmentation, this takes the maximum component in the image and filter out the other segmented regions. This results in a better dice score (0.956 ± 0.006) after post-processing, as shown in
Table 2, and the boundaries are more confined to the liver using U-Net-based liver segmentation after post-processing, as shown in the figures dedicated to the cropped CT slice comparison. We do discuss speedups on U-Net, as it takes the fixed-size image as an input to the network.
The dice score is reduced when the input CT slice is cropped using U-Net-based liver segmentation, as shown in
Table 2. But it is improved when we apply the post-processing to remove the unwanted segmentation. The Chan-Vese model for liver segmentation removes the unwanted segmentation completely, giving a better average dice score (0.96 ± 0.005) and speedup (1.387 ± 0.1), as shown in
Table 1. The average time taken by Chan-Vese-based liver segmentation is 0.571 ± 0.038 s after cropping, providing an average speedup of 1.387 (±0.1) times in comparison to the original liver slice (0.791 ± 0.058 s on a liver image of size 512 × 512) on an NVIDIA GPU GeForce GTX 1050 with RAM 4 GB. The average dice score obtained by Chan-Vese (0.96 ± 0.005 in
Table 1) is better compared to U-Net (0.956 ± 0.006 in
Table 2) for liver segmentation in cropped images.
Finally, in all four cases, the U-Net model erroneously segmented adjacent soft tissues with similar intensity values, leading to significant false positives. However, post-processing was able to isolate the true liver region effectively. This highlights both the limitations of slice-wise DL inference and the value of morphology-aware corrections.
6.2. Liver Volume Assessment
In this section, we performed liver volume estimation using an SRG algorithm on the 3D binary masks. The algorithm operates within the cropped region and computes volume as the total number of labeled voxels multiplied by voxel size.
Table 3 gives the analysis of semiautomatic 3D cropping showing the estimated liver volumes compared to ground truth. Results show strong agreement (mean absolute error below 2.5%) across both Chan-Vese and U-Net pipelines. The exact cropped size (ECS) is obtained by counting the number of voxels inside the cuboid, which is defined automatically from ground truths validated by clinicians. The average time to crop the liver slice volume is 8.841 ± 1.505 s. The average semiautomatic cropping time per slice is 11.603 ± 1.913 ms. The average percentage of maximum unnecessary data to be reduced is 92.031 ± 2.731% and semiautomatic cropping helps reduce 90.007 ± 3.406% (average) from the volume of liver slices.
We compare the performance on LiTS liver data [
20] testing on an NVIDIA GPU GeForce GTX 1050. The voxel-based parallel approach improves cropping by 2.289 times compared to slice-based cropping proposed by Smistad et al. [
30]. The cropped volume of liver (with voxels 3097775) is shown in
Figure 17. The proposed parallel cropping algorithm crops liver volume of 512 × 512 × 816 to 285 × 229 × 190 in 44.6 ms.
The time required for cropping is reduced, as four kernel calls are involved (three for finding the cropping points in each dimension and one for copying cropped data) in the state of the art. The average time taken by the non-PT and slice-based parallel cropping is 103.529 ± 8.31 ms, whereas the proposed voxel-based parallel cropping takes 45.227 ± 4.9 ms, providing an average speedup of 2.289 times compared to the state of the art. The average time for the volume assessment of the liver is 0.781 ± 0.271 s.
6.3. Volume Assessment Evaluation of Other Datasets
To assess the generalizability of our cropping and volume assessment pipeline, we applied it to several cases from the 3DIRACDb dataset, including CT scans of skin, lung, and vascular structures. These datasets lack ground truth liver labels but serve as a testbed for efficiency and applicability across anatomical domains.
We assess the volume of the liver dataset from the LiTS challenge (Christ [
20]). We analyze the proposal in vessel, liver, skin, and lung datasets from 3D Image Reconstruction for Comparison of Algorithm Database (3DIRACDb by Li et al. [
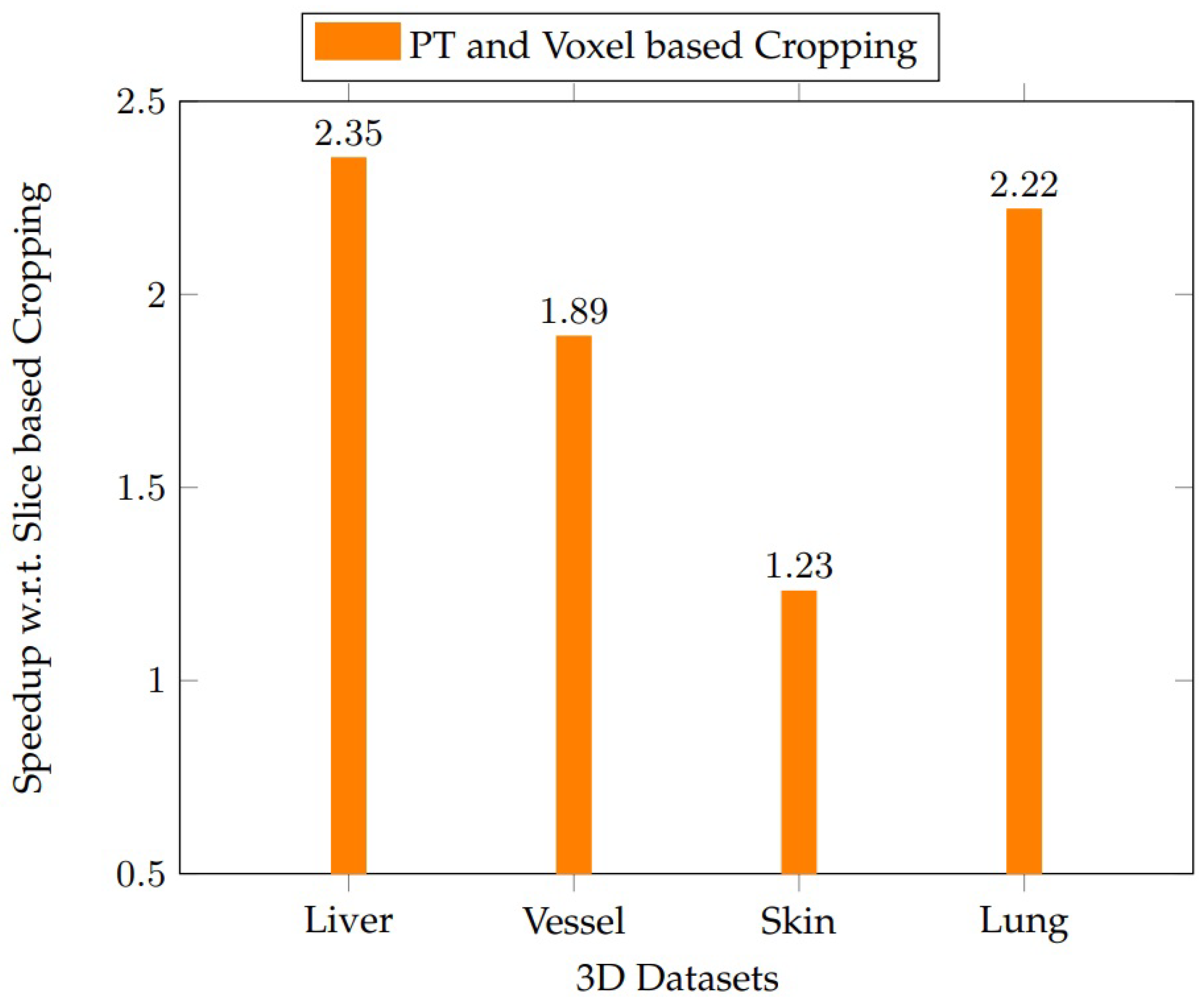
43]). A total of 28 persistent blocks are used for SRG on an NVIDIA GPU RTX 2070. The proposed voxel-based cropping approach outperforms slice-based cropping for all the datasets, as shown in
Figure 18. The speedups obtained by the voxel-based parallel cropping with respect to (w.r.t.) liver, vessel, skin, and lung datasets are 2.35, 1.89, 1.23, and 2.22 times, respectively, compared to the slice-based cropping.
In all cases, voxel-based cropping successfully localized target regions and enabled rapid volume estimation via SRG. As shown in
Table 4, the runtime performance remained consistent, with average speedups of 2.1–2.5× over slice-based methods. Results from non-liver datasets indicate that our pipeline is adaptable to other organs, supporting its deployment in more diverse clinical imaging workflows.
The generalizability of the proposed cropping method was further supported by experiments on 3DIRACDb, which includes non-liver CT data such as vessels, lungs, and skin. Despite anatomical and intensity differences, our voxel-based cropping maintained consistent speedups and localization precision across all evaluated domains. In [
46], the compatibility of preprocessing approaches with variable contrast regimes suggests that voxel-based cropping can be extended to other modalities, such as MRI or PET, especially when combined with contrast normalization techniques.
7. Conclusions
This study analyzes automatic and semiautomatic parallel cropping techniques used to segment and assess the volume of the liver in medical imaging. U-Net produces unwanted segmentation outside the liver’s RoI. However, this was effectively filtered, which increased the dice scores. The average percentage of unwanted data using semiautomatic cropping is 90 ± 3.4%. This implies that only 10% of data are useful from CT volumes. Semi-automatic cropping successfully eliminated irrelevant data. Our proposal improves the accuracy and significantly reduces unnecessary computations.
Applying our proposed method to U-Net for liver segmentation, the dice score impressively rises from 0.52 to 0.94, demonstrating a significant leap in accuracy and the potential to drastically improve clinical outcomes. The semi-automatic cropping technique demonstrated higher robustness when dealing with low-quality images with unclear liver boundaries. This approach ensures that cropping remains precise and efficient even when fully automated techniques fail to identify the correct RoI.
The proposed persistence-based approach was 2.29× faster than traditional slice-based cropping methods, because our voxel-based approach computes cropping boundaries in a single pass across all dimensions. Although the focus was on liver segmentation, the results indicate that this method can also be applied to other areas of medical imaging, such as the volumetric assessment of blood vessels, skin, and lungs.
DL models are popular but operate as black boxes. The decision-making process behind the segmentation results is not interpretable, making it difficult to understand why the model produces certain outputs, especially in edge cases or when anomalies are present. Clinical decisions require transparency and confidence. The lack of interpretability in DL models poses a barrier to clinical adoption, as practitioners might be hesitant to rely on a system that cannot provide clear justifications for its results. Furthermore, incorrect segmentations are difficult to diagnose, and it is difficult to address the cause of the error without a clear understanding of how the model processes the data. This lack of insight makes it challenging to fine-tune or adapt the model for different datasets or clinical environments, limiting its flexibility and robustness.
Our parallel cropping proposal is a robust and explainable alternative to deep learning solutions in scenarios where computational resources are limited, labeled data availability is scarce, and rapid integration and greater interpretability are essential in the clinical setting. This approach allows for efficient and accurate results without the high costs and complexity associated with DL models, making it a practical solution for real-time clinical applications.
Future work will explore adaptive architectures such as UNet++ [
27], Swin-UNet [
28], TransUNet [
29], and other new models with dynamic input resolution. We also aim to further quantify uncertainty and integrate explainability metrics into the segmentation pipeline. We are also interested in implementing these algorithms in the emerging and promising Processing-In-Memory (PIM) paradigm [
47].