1. Introduction
The healthcare of newborns and premature infants is of utmost importance in a hospital, requiring continuous attention and care in neonatal units. These infants are particularly vulnerable, as many of their physiological systems, including those regulating sleep, brain activity, and autonomic functions, are still in the process of development. The neonatal intensive care unit (NICU) must therefore not only ensure the survival of these infants but also support their long-term developmental outcomes.
In addition to monitoring neonates’ vital physiological functions, it is of relevance to systematically observe and analyze specific parameters, including sleep–wake cycles, behavioral states, and the temporal dynamics of circa- and ultradian rhythms. These observations yield significant insights into the multifaceted dimensions of neonatal development. Accurate detection and interpretation of sleep patterns are especially crucial, as sleep plays a vital role in the maturation of the central nervous system. Notably, sleep duration assumes a pivotal role in proper brain development and fostering the optimal maturation of newborns. The intricate regulation of sleep, in conjunction with its cyclical patterns, emerges as a fundamental driver in shaping cerebral development, early neurobehavioral progress, cognitive learning, memory consolidation, and the preservation of neural plasticity [
1,
2].
Sleep deprivation or other problems concerning sleep can inhibit the creation of permanent neural circuits for the primary sensory systems and can have a negative effect on the brain development of neonates [
3]. Sleep disturbance can also occur due to the frequent scheduling of medical procedures (bathing, changing diapers, feeding, treatments, etc.), many of which require waking the infant, thereby interrupting natural sleep cycles and imposing unnecessary stress [
4,
5,
6,
7]. Repeated disturbances in sleep are linked to altered stress regulation, reduced growth, and delayed neurodevelopmental outcomes.
To address these challenges, hospitals that follow developmentally supportive care models have adopted frameworks such as the Newborn Individualized Developmental Care and Assessment Program (NIDCAP). In these settings, medical staff strive to maintain a calm and nurturing environment that respects the infants’ behavioral cues. Caregivers monitor the sleep and activity states of the babies according to the NIDCAP scale, which is further informed by the FINE (Family and Infant Neurodevelopmental Education) tutorial materials developed by NIDCAP-certified experts [
8]. However, manual monitoring of sleep and activity phases is time-consuming, labor-intensive, and prone to subjectivity. This presents a barrier to widespread implementation, especially in resource-constrained settings.
From a clinical perspective, automating the classification of neonatal behavioral states according to the NIDCAP framework is a highly worthwhile objective, offering both practical and developmental benefits. Such systems can provide continuous, objective observation of infants’ sleep and activity states, enabling more timely and individualized care. Real-time classification allows healthcare professionals to better synchronize caregiving tasks—such as feeding, medical procedures, or environmental adjustments—with the infant’s most receptive states, thereby minimizing stress and supporting improved developmental outcomes. Furthermore, automated monitoring facilitates earlier detection of abnormal patterns, reduces the burden on clinical staff, and promotes a more structured and developmentally sensitive workflow. Ultimately, the integration of intelligent monitoring tools has the potential to transform neonatal care from reactive to proactive, optimizing both clinical efficiency and long-term health trajectories.
Advancements in machine learning, especially in the field of deep learning, have opened new possibilities for healthcare applications, including neonatal care. Recent studies have explored the use of neural networks for analyzing video, audio, and physiological data to identify behavioral states or detect early signs of distress. Recurrent neural networks (RNNs), particularly gated recurrent units (GRUs) and long short-term memory (LSTM) architectures, have shown strong capabilities in modeling temporal sequences, making them ideal for understanding sleep–wake patterns and physiological fluctuations over time. In neonatal care, combining computer vision with physiological signal processing offers a promising avenue to reduce the reliance on subjective assessments and to enhance the continuity and precision of monitoring.
In this study, we propose a novel approach to address this challenge using a GRU-stack-based method that determines the activity and sleep phases based on visual features extracted from camera footage. In addition, we demonstrate that integrating physiological signals—specifically pulse rate and pulse rate variability derived from photoplethysmography (PPG) recordings from standard patient monitors—significantly enhances the accuracy and reliability of the classification. Our work represents an important step toward non-invasive, automated, and scalable solutions for developmental care in neonatal settings.
Behavioral monitoring via the NIDCAP protocol is crucial for optimizing newborn and preterm infant development, offering comprehensive insights beyond traditional vital signs to support critical brain development, as sleep duration significantly impacts neural circuit formation. Clinically, this approach enables personalized care, precisely timing medical interventions during an infant’s most receptive states to minimize stress and maximize therapeutic efficacy, with early behavioral patterns serving as valuable predictors for later cognitive and neurological outcomes. The integration of a GRU-stack-based automated system significantly enhances monitoring accuracy and continuity, eliminating labor-intensive manual observations and providing continuous 24-h surveillance unattainable by human observation alone. This technology uses video actigraphy and pulse rate variability data from existing hospital equipment, thus avoiding additional disturbing sensors while supporting better developmental outcomes. Ultimately, this system optimizes care scheduling and improves clinical decision making by providing objective, continuous data on sleep–wake cycles, thereby reducing staff workload and enhancing overall neonatal care quality.
2. Related Work
2.1. Behavioral State Monitoring
The most commonly used method for monitoring infant sleep is polysomnography (PSG), which involves attaching multiple electrodes and sensors to the infant’s body. While PSG remains the clinical gold standard due to its comprehensive nature, its application in neonatal settings is challenging. The multitude of cables can interfere with caregiving procedures, limit infant mobility, and irritate the baby’s sensitive skin—potentially increasing the risk of infection.
In recent years, numerous attempts have been made to develop non-invasive or minimally invasive alternatives for assessing sleep and activity in infants. Notably, Xi Long et al. [
9] developed a system of actigraphy combined with video for monitoring sleep and wake in infants. Similarly Huang et al. [
10] applied video measurements as well for sleep–wake cycle observations. Although these works mark important progress toward non-contact behavioral assessment, they are limited in scope—typically distinguishing only between binary states (sleep vs. wakefulness), without accounting for the nuanced behavioral categories used in developmental care protocols.
To the best of our knowledge, we know of no algorithm that categorizes infants into behavioral phases according to the globally recognized NIDCAP (Newborn Individualized Developmental Care and Assessment Program) protocol while also accounting for situations such as medical interventions—thus providing a truly holistic monitoring approach.
In response to these gaps, we have developed a holistic, non-contact monitoring system designed for use in clinical environments. In our earlier work [
11], we introduced an algorithm capable of identifying quiet periods, estimating pulse and respiration rates, and recognizing disruptive events such as medical procedures—all from a single video stream. In this study, we build on that foundation by presenting a GRU-stack-based classification model that maps visual features to NIDCAP behavioral categories. We further enhance this approach by incorporating physiological data, specifically pulse rate and pulse rate variability, extracted from photoplethysmography (PPG) signals recorded by standard patient monitors.
2.2. Pulse Rate Variability (PRV) for Sleep State Categorization
The motivation for incorporating pulse rate variability (PRV) as an additional feature in behavioral state classification lies in its potential to reflect underlying autonomic nervous system (ANS) activity.
Heart rate variability is regarded as a reliable reflection of the many physiological factors modulating the normal rhythm of the heart; for example, it provides a powerful means of observing the interplay between the sympathetic and parasympathetic nervous systems [
12]. There are three frequency components of HRV associated with the autonomic nervous system: very low frequency (0.04 Hz, VLF), low frequency (0.04–0.15 Hz, LF), and high frequency (0.15–0.4 Hz, HF) components [
13].
Although the physiological interpretation of these components is not fully understood, HF power is frequently used to quantify parasympathetic activity [
14]. LF power is considered as a function of both sympathetic and parasympathetic activity [
15]. The ratio of LF to HF power is used as an indicator of sympathetic–parasympathetic balance [
15]. This ratio has been used for sleep classification [
16,
17,
18,
19], since there is a major difference in the autonomic nervous system’s interplay during different stages of sleep [
14]. During REM sleep, the cardiovascular system is unstable and greatly influenced by surges in sympathetic activity. It is also characterized by more irregular breathing and heart rate [
14]. Whereas during NREM sleep, the cardiovascular system is stable, parasympathetic cardiac modulation is stronger [
20].
It is important to note that in this work, the output classes of the applied model are not entirely the same as the sleep stages (e.g., NREM, REM, etc.), although in the literature light and REM sleep, deep and NREM sleep are used interchangeably in certain contexts. There are also findings showing significantly higher HRV values during active sleep than during quiet sleep specifically [
21].
Since every newborn is constantly monitored with a pulse oximeter, but not with an ECG, we decided to use the pulse signal and elaborate on PRV metrics, even though it is mentionable that there is not a clear settlement on the substitutability of HRV and PRV values [
22]. However, the results speak in favor of sufficient accuracy when subjects are at rest in a resting position [
23]. Thus, we used these metrics as a feature for the sleep stage categorization to investigate its effectiveness.
3. Experimental Setup
The dataset was collected in cooperation with the Department of Obstetrics and Gynecology at Semmelweis Medical University. The data collection system [
11] consisted of a conventional RGB camera (Basler acA2040-55uc) equipped with a near-infrared (NIR) night illuminator, enabling recording during both day and night without disturbing the infants. Videos were recorded at 20 frames per second with a resolution of 500 × 500 pixels and stored using a lossless compression format to preserve all relevant visual details for post-processing.
Physiological signals were acquired in parallel using Philips IntelliVue MP20/MP50 patient monitors. These included photoplethysmographic (PPG) pulse signals, skin resistance, and blood oxygen saturation. All signals were synchronized with the video recordings using NTP-based time alignment. Custom-developed software (written in Python 3.11.13) was used for data acquisition, storage, and annotation.
In total, 402 h of video and physiological data were collected from neonates (primarily preterm at gestational ages 32–38 weeks) in neonatal intensive care units. More details from the population of the recorded babies and their properties can be found in
Table 1. After excluding caregiving periods and empty incubator intervals, 108 h of usable recordings were retained. Behavioral states were annotated manually by three trained reviewers with expertise in neonatal care, following the NIDCAP system [
24,
25,
26]. Each reviewer annotated a distinct, non-overlapping subset of the recordings. All reviewers received standardized training to ensure consistency in applying the annotation criteria. The six behavioral states were the following: (1) deep sleep, (2) light sleep, (3) drowsy, (4) quiet awake, (5) active awake, and (6) highly aroused/crying. For the purposes of this study, we merged the three awake-related classes (4–6), including crying, into a single “awake” category. This decision was motivated by the primary clinical objective: to reliably distinguish between sleep and wakefulness. Our medical collaborators emphasized that the most clinically meaningful information lies in understanding how much time the infant spends in each major state—awake, drowsy, light sleep, or deep sleep—rather than in differentiating between subtle subtypes of wakefulness. In addition, we aimed to incorporate physiological data, particularly pulse rate and derived metrics. However, during episodes of crying or active wakefulness, the accuracy of PPG measurements is often compromised due to sensor displacement or motion artifacts. For this reason, it was methodologically sound to focus on more stable, quieter states—such as drowsiness and sleep phases—where physiological signals are more reliable. Moreover, this study was conducted as a pilot, with a focus on feasibility and clinical alignment. In this context, reducing label granularity within the awake class helped mitigate annotation ambiguity in short observation windows and facilitated more robust model development. Future work will aim to refine and expand the label taxonomy, particularly within the awake state, as data volume and annotation reliability increase.
4. Methodology
Our method comprised four main stages: (1) visual and physiological feature extraction, (2) PRV calculation, (3) feature fusion and dimensionality reduction, and (4) behavioral state classification using a recurrent neural network. This pipeline is illustrated in
Figure 1. Video data were acquired from a camera positioned above the neonate, while pulse signals were recorded using Philips IntelliVue MP20/MP50 patient monitors. Scalar intra- and interframe statistics [
11] were computed independently within semantically segmented regions, on a frame-by-frame basis, to capture temporal dynamics. These features were subsequently concatenated with derived pulse rate variability (PRV) statistics. The resulting one-dimensional descriptors formed a dynamic sequence of numerical features, which served as input to a recurrent neural network (RNN) for behavioral state classification.
4.1. Video Processing
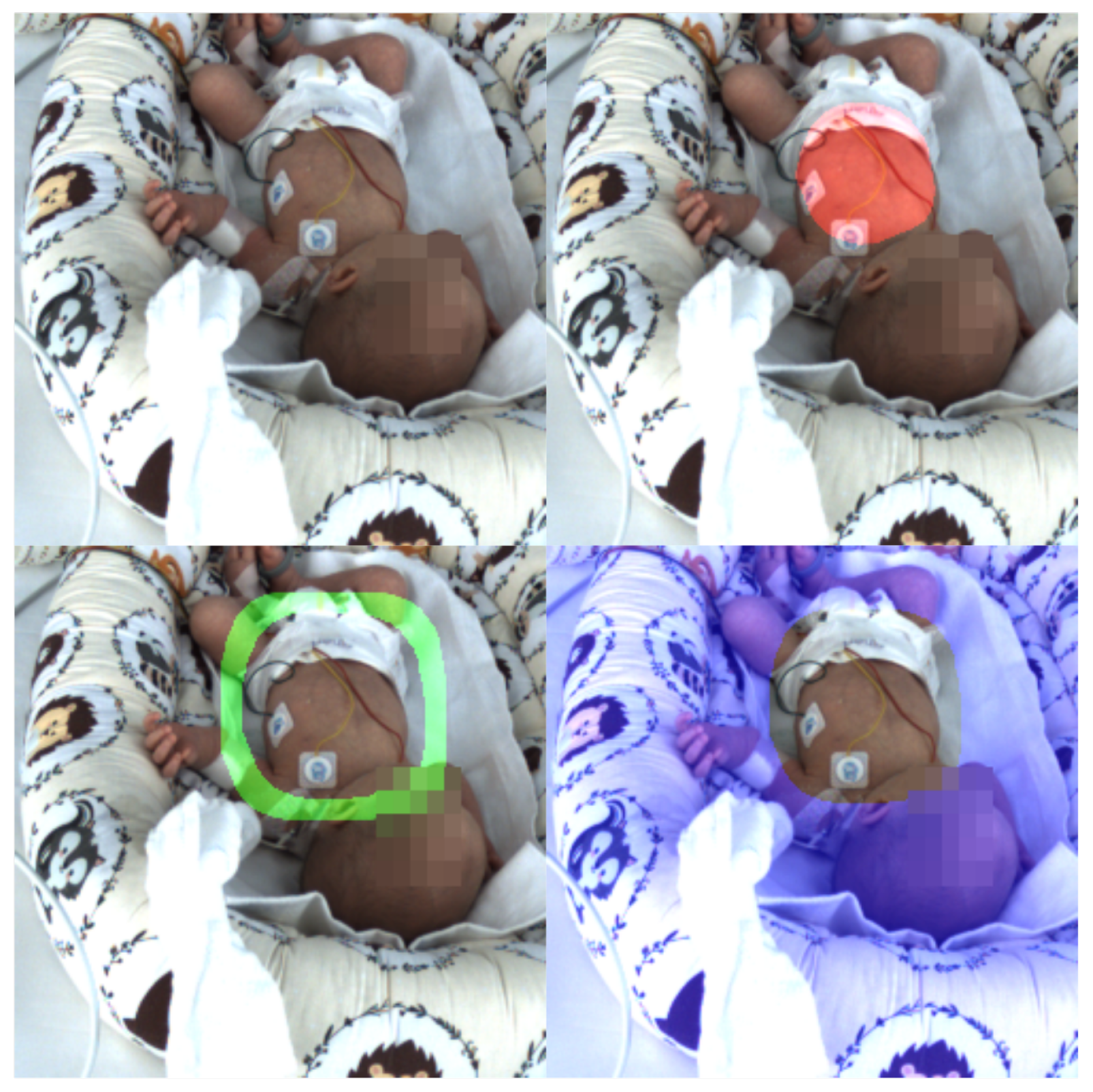
The features of the visual data were gathered from multiple different classical and machine learning (ML)-based methods for real-time classification, enabling the capture of various activities, identification of regions of particular interest, and the extraction of additional information, notably for motion activity monitoring and different nursing activities. Our semantic segmentation methodology relies on the fact that distinct motion patterns and areas gain prominence in different behavioral states of the neonate. For instance, key regions include the infant’s abdominal region, where respiratory activity is observable, and the infant’s face, where open eyes are visible during periods of wakefulness. In this study, four such ROIs were defined and segmented, as visualized in
Figure 2.
To elucidate our rationale for selecting and extracting features from these aforementioned ROIs, first we elaborate on our criteria for ROI selection. Our motivation relies on the observation that abdominal movements often correlate with respiration, a vital physiological signal in discriminating sleep–wakefulness stages. Accordingly, we have employed a UNET architecture for the segmentation of the infant’s abdominal region in images. It is worth noting that this method may encounter limitations when dealing with draped infants. However, this challenge is mitigated by our supplementary motion-based approach for identifying respiratory movement locations. Once the abdominal region is segmented, we can easily identify the surrounding area within a specified distance from the abdomen, a region of particular importance, as it may encompass the infant’s limbs, depending on the ring’s parameter settings. Consequently, we can obtain insights into limb movements independently of respiration. Finally, the outermost area, farther from the neonate’s body, serves to suppress external events.
4.2. PRV Acquisition and Processing
To ensure reliable PRV estimation, signal quality checks were applied to the PPG signals prior to analysis, and segments with excessive artefacts were excluded from further processing. Pulse rate variability (PRV) was then computed from the PPG signals using a peak detector specifically designed for pulse waveforms [
27]. The analysis was performed using a 5-min sliding window, following established conventions in the literature [
28]. Two standard time-domain metrics were calculated, as defined in Equations (
1) and (
2). These metrics were chosen due to their established use in reflecting autonomic nervous system dynamics, with RMSSD being sensitive to parasympathetic activity and SDNN capturing overall heart rate variability [
28].
Here, denotes the time (in seconds) between two consecutive R peaks, is the i-th RR interval, is the mean of all RR intervals, and N is the total number of RR intervals.
To obtain frequency-domain features, we followed the methodology described by Peralta et al. [
29].
The extracted PRV features were subsequently concatenated with vision-based features, resulting in a combined feature matrix of shape , where 31 denotes the number of feature channels and 250 the number of time steps. Principal Component Analysis (PCA) was then applied row-wise for normalization, using parameters computed on the training set.
4.3. Classification
The final step is the classification itself, for which we used a variation of recurrent neural networks, as this architecture has been successfully used in many applications for the classification of time series [
30,
31,
32]. More specifically, we used GRU (gated reccurrent unit) stack because this type of RNN misses the cell state from the traditional GRU structure and only uses the hidden state. This type of RNN performs better on smaller datasets with relatively long sequences [
33].
4.4. Hardware Used, System Architecture, Computational Environment, and Training Details
The computational experiments were primarily conducted on a purpose-built PC (64-bit Ubuntu 22.04.4 LTS PC with Intel® Core™ i7-8700 CPU and NVIDIA GeForce GTX 1060 6 GB GPU) and in the Google Colaboratory (Colab) environment. This platform provides access to cloud-based graphics processing units (GPUs), specifically NVIDIA GPUs, which were used to accelerate the training and inference processes of deep neural networks.
The core of the developed solution is built upon a Python-based deep learning framework, employing several established libraries and tools. Data management and preprocessing were handled by NumPy 2.0.2 and Scikit-learn 1.6.1. Additionally, we used Matplotlib 3.10.0 and torchmetrics for utilities and visualization.
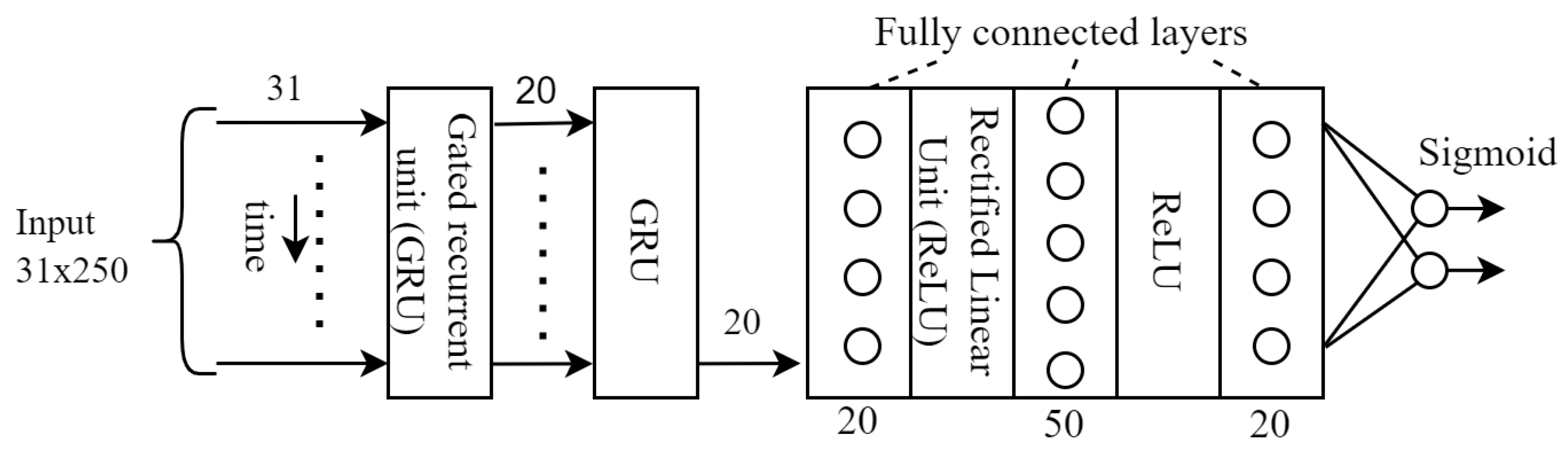
The used loss function was cross-entropy that is usually used in case of classifications. The Adam optimizer was used as optimizer in the training. In the GRU block (see
Figure 3), the GRU itself was followed by 3 fully connected layers. The training and test sets were split as usual in a ratio of 75% and 25%.
Considering the hyperparameters, it is worth noting that the value of lstm_hidden_ dimension was 35, the number of lstm_layers was 2, and the value of learning_rate was 1 × 10−3.
5. Results
The proposed GRU-based model achieved high classification performance across all key evaluation metrics, as summarized in
Table 2. When both visual and physiological features were used, the model attained an accuracy of 92.90%, demonstrating robust performance in classifying neonatal behavioral states.
We further evaluated the contribution of PRV features by comparing the model’s performance with and without physiological data. As shown in
Table 2, incorporating PRV features improved overall accuracy by approximately 8%, confirming their complementary role. The physiological features were particularly helpful in distinguishing between intermediate and deep sleep phases, where motion activity is minimal. In contrast, during more active periods (e.g., awake states), visual and motion-based statistics alone (derived from video actigraphy) provided sufficient discriminative information.
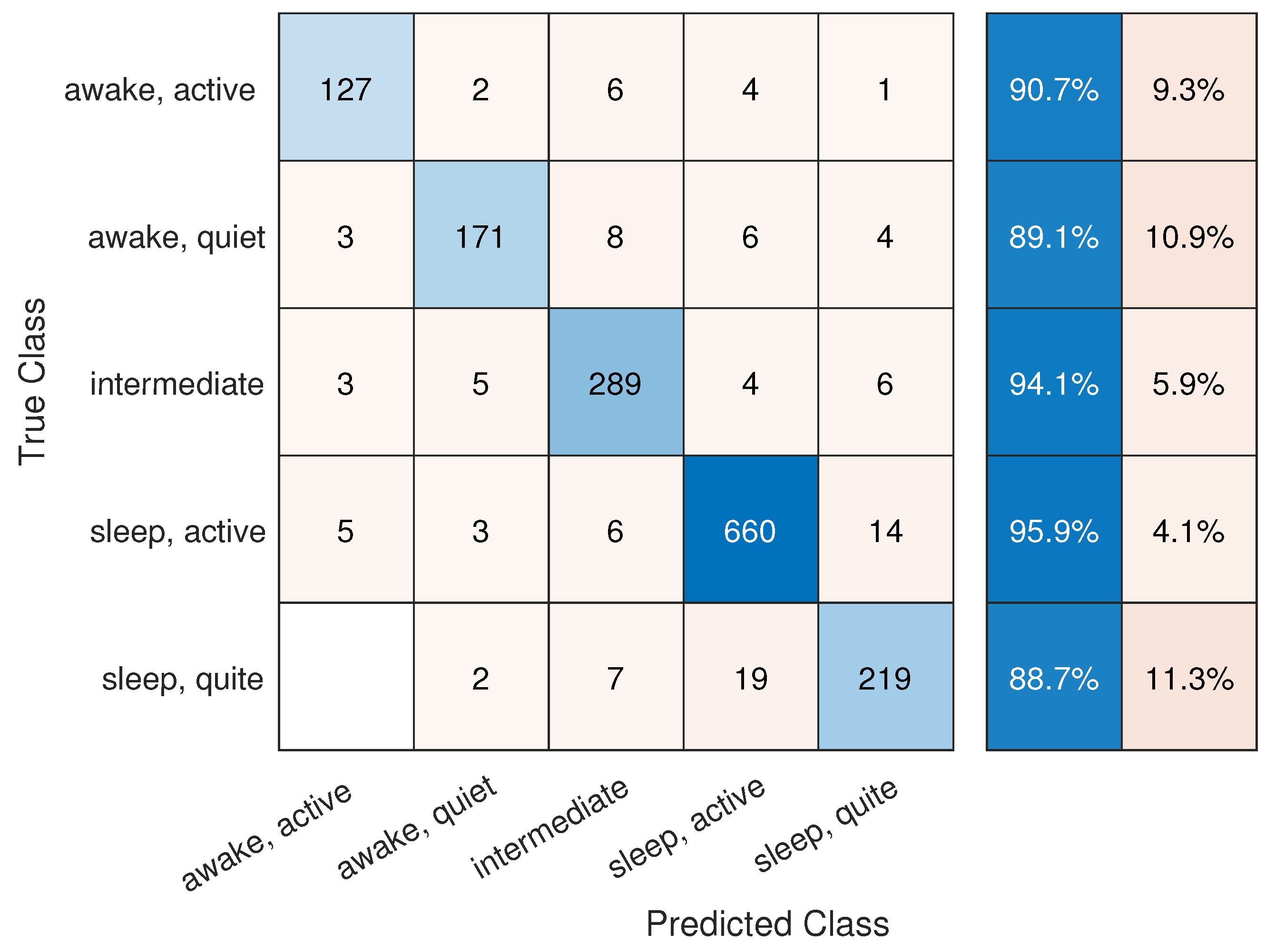
To gain further insight into class-wise performance,
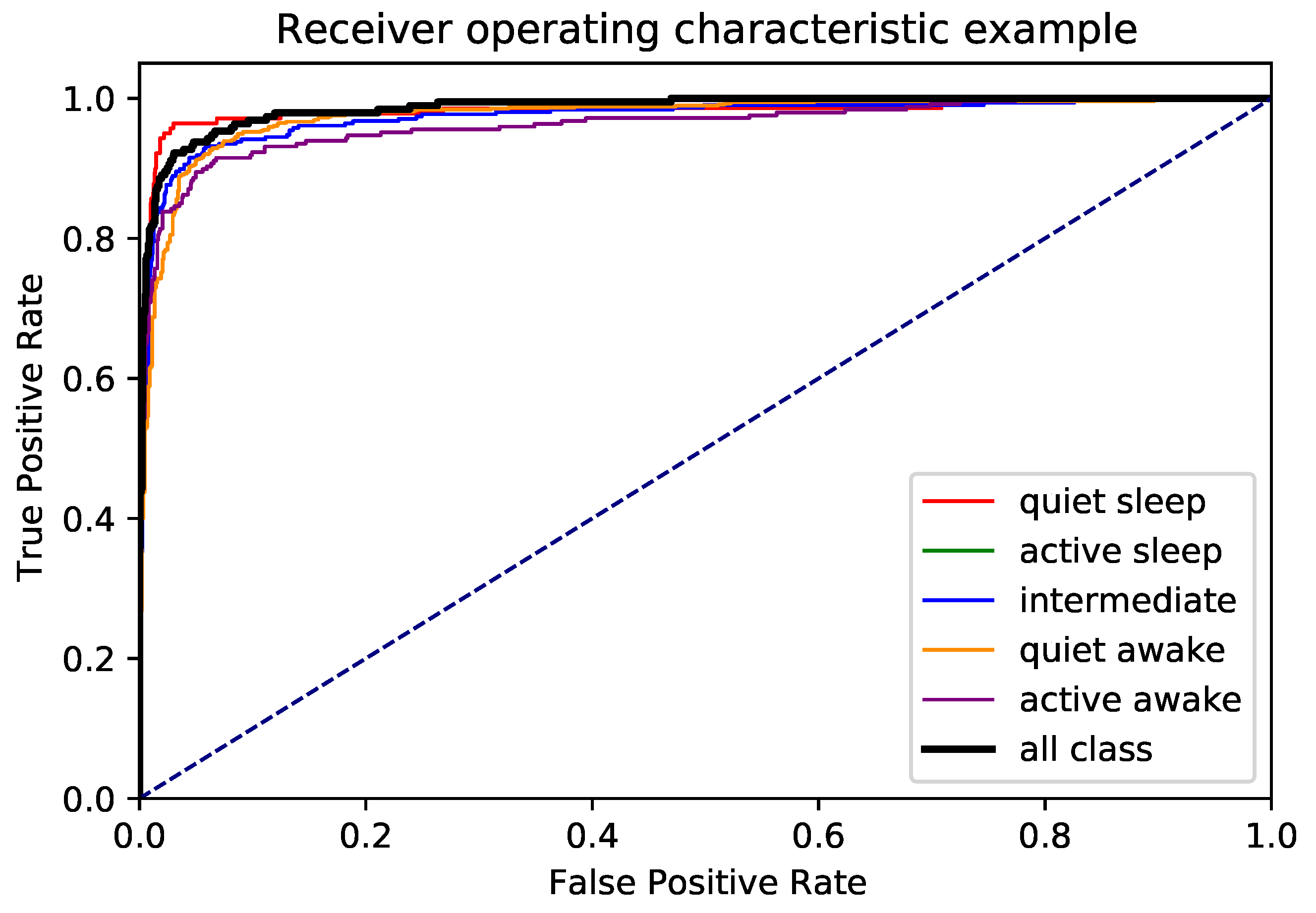
Figure 4 presents the normalized confusion matrix. The model performed well across all categories, with minimal misclassification between adjacent states. Additionally, the ROC curves in
Figure 5 indicate high separability for each class, confirming the robustness of the model.
6. Discussion
Our results demonstrate the feasibility of automatically classifying neonatal behavioral states using a multimodal approach that combines video-derived motion features and pulse rate variability (PRV). The model achieved high overall accuracy and performed well across all behavioral categories, including subtle transitions between sleep phases. These findings align with and extend earlier observations on the clinical value of PRV features in assessing infant sleep and wakefulness.
Importantly, this study highlights the potential for clinical integration. Current behavioral assessments in neonatal intensive care units (NICUs) often rely on manual observation, which is time-consuming, subjective, and limited in temporal resolution. Our automated system, by contrast, enables continuous, real-time monitoring of behavioral states with no disruption to the infant. This aligns well with the goals of individualized care frameworks such as the NIDCAP, which emphasize sensitive, developmentally appropriate interactions based on the infant’s behavioral cues.
The ability to reliably detect whether an infant is asleep, drowsy, or awake can support several clinical applications. For instance, caregivers could time interventions to coincide with optimal states of receptivity or avoid disrupting deep sleep phases that are critical for neurodevelopment. Moreover, quantitative metrics derived from behavioral state distributions over time may serve as early indicators of neurological maturation or dysregulation.
The added value of PRV features was particularly evident in sleep phases, where motion is minimal and visual signals alone may be ambiguous. This supports the inclusion of physiological data in future monitoring systems, especially in contexts where fine-grained state differentiation is clinically relevant. At the same time, the visual modality alone provided strong performance during more active states, which opens the possibility of tailoring sensing strategies to the infant’s condition or care environment.
6.1. Clinical Implementation Considerations
The proposed system is designed with real-world clinical applicability in mind, particularly in neonatal intensive care units of tertiary care or university-affiliated hospitals. Our solution leverages standard patient monitoring (Philips IntelliVue MP20/MP50) and affordable, off-the-shelf RGB cameras mounted above the infant’s bed. Near-infrared (NIR) illumination ensures recording under low-light conditions, enabling 24/7 monitoring without disturbing the infant.
From a computational standpoint, the system is efficient: the GRU-based model operates on precomputed features and can run in real time on standard GPU-equipped clinical workstations. A real-time implementation is currently under development, and preliminary performance benchmarks confirm its feasibility.
Despite this, practical deployment in NICUs presents several challenges. One major issue is camera stability—cameras are sometimes repositioned by clinical staff during care routines, necessitating automated recalibration or realignment. Additionally, night-time recordings must avoid overexposure or signal dropout, especially when bright lights or reflections interfere with the infrared spectrum. These technical factors affect both segmentation and motion feature quality.
Beyond hardware constraints, successful implementation depends heavily on clinical acceptance and usability. A key barrier is the level of trust clinical staff place in automated systems. Throughout this project, we have worked closely with neonatologists and NICU nurses to ensure that the system aligns with practical workflows and clinical expectations. Ongoing efforts focus on developing an intuitive, interpretable user interface and incorporating clinician feedback through iterative usability testing.
In summary, our findings suggest that automated behavioral state classification in neonates is not only technically feasible but also clinically promising. With further development, such systems may enhance individualized care, reduce caregiver workload, and offer novel insights into infant neurodevelopment within real-world clinical settings.
6.2. Limitations and Future Work
As a proof-of-concept study, our work has several limitations that should be addressed in future research. First, the sample size is relatively small and limited to a single clinical site. Although the results are promising, larger-scale studies across multiple NICU environments are required to assess the generalizability and robustness of the approach.
Second, the behavioral annotations used as ground truth were based solely on human observation while adapting the NIDCAP framework for our purposes. While this provides a clinically relevant baseline, it remains inherently subjective. Future work will incorporate additional objective modalities such as EEG or polysomnography to refine and validate state definitions, especially for transitions and ambiguous cases.
Third, although our system is capable of real-time inference, this study did not yet include full real-time deployment or integration into clinical workflows. Implementing the system in live clinical environments will require solving practical challenges such as robust camera stabilization, automatic recalibration, adaptive lighting management, and reliable data handling within hospital IT infrastructures.
Finally, the social and human factors surrounding clinical adoption represent a non-trivial challenge. Successful implementation will depend on building trust among healthcare staff, designing intuitive user interfaces, and demonstrating the system’s reliability in daily clinical operations. We are currently working with clinical stakeholders to address these aspects through usability testing, iterative design, and continued interdisciplinary collaboration.
Future directions will also explore semi-supervised or self-supervised learning techniques to reduce the burden of manual annotation and the incorporation of additional physiological signals to further enhance state classification accuracy.
7. Conclusions
This proof-of-concept study demonstrates the feasibility of automatically classifying infant behavioral states—guided by the NIDCAP approach—using a GRU-based deep learning model that integrates visual and physiological features. By combining video actigraphy with pulse rate variability (PRV) statistics, we achieved high classification performance across key behavioral categories, including various sleep and wake phases.
To our knowledge, this is the first deep learning-based system to perform NIDCAP-inspired behavioral classification with merged alert states using only passive sensing modalities, namely, overhead video and standard hospital pulse oximetry. While no direct benchmarking with existing approaches was conducted due to a lack of comparable datasets and methods aligned with the NIDCAP, our results suggest that this multimodal strategy is both technically effective and clinically meaningful. The inclusion of PRV metrics, which are easily accessible from existing hospital infrastructure, significantly improves classification performance during low-motion sleep states.
In future applications, such systems may help alleviate the burden of continuous behavioral observation from NICU staff while providing richer and more objective 24-h information about infant sleep–wake patterns. Ongoing work focuses on real-time deployment, broader clinical validation, and deeper integration into neonatal care workflows.
Author Contributions
Conceptualization, M.S. (Miklós Szabó); Methodology, Á.N., Z.L.R., P.F. and J.V.; Software, Á.N.; Validation, Á.N. and P.F.; Investigation, Z.L.R. and M.S. (Máté Siket); Data curation, Z.L.R. and I.J.; Writing—original draft, Á.N., Z.L.R., I.J. and M.S. (Máté Siket); Writing—review & editing, Á.Z., Á.N. and Z.L.R.; Supervision, Á.Z.; Funding acquisition, P.F. and Á.Z. All authors have read and agreed to the published version of the manuscript.
Funding
Support by the the European Union project RRF-2.3.1-21-2022-00004 withinthe framework of the Artificial Intelligence National Laboratory.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the National Scientific and Ethical Committee (ETT-TUKEB IV/3383-3/2022/EKU).
Informed Consent Statement
Written informed consent was obtained from parents to use the anonimized visual records for study purposes. Informed consent documents are stored as part of patient documentation.
Data Availability Statement
Restrictions apply to the availability of these data. Data were obtained from the participants given consent and are available from the authors only with the permission of the participants. Should a requirement arise for access to the code, kindly make contact the research team through the provided email address.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dahl, R.E. The regulation of sleep and arousal: Development and psychopathology. Dev. Psychopathol. 1996, 8, 3–27. [Google Scholar] [CrossRef]
- Graven, S. Sleep and brain development. Clin. Perinatol. 2006, 33, 693–706, vii. [Google Scholar] [CrossRef] [PubMed]
- Graven, S.; Browne, J. Sleep and Brain Development: The Critical Role of Sleep in Fetal and Early Neonatal Brain Development. Newborn Infant Nurs. Rev. 2008, 8, 173–179. [Google Scholar] [CrossRef]
- Brandon, D.H.; Holditch-Davis, D.; Beylea, M. Nursing care and the development of sleeping and waking behaviors in preterm infants. Res. Nurs. Health 1999, 22, 217–229. [Google Scholar] [CrossRef]
- Liaw, J.J.; Yang, L.; Lo, C.; Yuh, Y.S.; Fan, H.C.; Chang, Y.C.; Chao, S.C. Caregiving and positioning effects on preterm infant states over 24 hours in a neonatal unit in Taiwan. Res. Nurs. Health 2012, 35, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Griffith, T.; Rankin, K.; White-Traut, R. The Relationship Between Behavioral States and Oral Feeding Efficiency in Preterm Infants. Adv. Neonatal Care 2017, 17, E12–E19. [Google Scholar] [CrossRef]
- Newnham, C.A.; Inder, T.E.; Milgrom, J. Measuring preterm cumulative stressors within the NICU: The Neonatal Infant Stressor Scale. Early Hum. Dev. 2009, 85, 549–555. [Google Scholar] [CrossRef]
- Warren, I.; Mat-Ali, E.; Green, M.; Nyathi, D. Evaluation of the Family and Infant Neurodevelopmental Education (FINE) programme in the UK. J. Neonatal Nurs. 2018, 25, 93–98. [Google Scholar] [CrossRef]
- Long, X.; Otte, R.; Sanden, E.v.d.; Werth, J.; Tan, T. Video-Based Actigraphy for Monitoring Wake and Sleep in Healthy Infants: A Laboratory Study. Sensors 2019, 19, 1075. [Google Scholar] [CrossRef]
- Huang, D.; Yu, D.; Zeng, Y.; Song, X.; Pan, L.; He, J.; Ren, L.; Yang, J.; Lu, H.; Wang, W. Generalized camera-based infant sleep-wake monitoring in nicus: A multi-center clinical trial. IEEE J. Biomed. Health Inform. 2024, 28, 3015–3028. [Google Scholar] [CrossRef]
- Nagy, A.; Földesy, P.; Janoki, I.; Terbe, D.; Siket, M.; Szabó, M.; Varga, J.; Zarándy, A. Continuous Camera-Based Premature-Infant Monitoring Algorithms for NICU. Appl. Sci. 2021, 11, 7215. [Google Scholar] [CrossRef]
- Acharya, U.R.; Joseph, K.P.; Kannathal, N.; Lim, C.M.; Suri, J.S. Heart rate variability: A review. Med. Biol. Eng. Comput. 2006, 44, 1031–1051. [Google Scholar] [CrossRef] [PubMed]
- Task Force of the European Society of Cardiology the North American Society of Pacing Electrophysiology. Heart Rate Variability: Standards of Measurement, Physiological Interpretation, and Clinical Use. Circulation 1996, 93, 1043–1065. [Google Scholar] [CrossRef]
- Dehkordi, P.; Garde, A.; Karlen, W.; Wensley, D.; Ansermino, J.M.; Dumont, G.A. Sleep stage classification in children using photoplethysmogram pulse rate variability. In Proceedings of the Computing in Cardiology 2014, Cambridge, MA, USA, 7–10 September 2014; pp. 297–300. [Google Scholar]
- Malliani, A.; Pagani, M.; Lombardi, F.; Cerutti, S. Cardiovascular neural regulation explored in the frequency domain. Circulation 1991, 84, 482–492. [Google Scholar] [CrossRef]
- Boudreau, P.; Yeh, W.H.; Dumont, G.A.; Boivin, D.B. Circadian Variation of Heart Rate Variability Across Sleep Stages. Sleep 2013, 36, 1919–1928. [Google Scholar] [CrossRef]
- Penzel, T.; Kantelhardt, J.; Grote, L.; Peter, J.; Bunde, A. Comparison of detrended fluctuation analysis and spectral analysis for heart rate variability in sleep and sleep apnea. IEEE Trans. Biomed. Eng. 2003, 50, 1143–1151. [Google Scholar] [CrossRef]
- Cui, J.; Huang, Z.; Wu, J. Automatic Detection of the Cyclic Alternating Pattern of Sleep and Diagnosis of Sleep-Related Pathologies Based on Cardiopulmonary Resonance Indices. Sensors 2022, 22, 2225. [Google Scholar] [CrossRef]
- Karlen, W.; Floreano, D. Adaptive Sleep–Wake Discrimination for Wearable Devices. IEEE Trans. Biomed. Eng. 2011, 58, 920–926. [Google Scholar] [CrossRef]
- Bušek, P.; Vaňková, J.; Opavský, J.; Salinger, J.; Nevšímalová, S. Spectral analysis of heart rate variability in sleep. Physiol. Res. 2005, 369–376. [Google Scholar] [CrossRef]
- DeHaan, R.; Patrick, J.; Chess, G.F.; Jaco, N.T. Definition of sleep state in the newborn infant by heart rate analysis. Am. J. Obstet. Gynecol. 1977, 127, 753–758. [Google Scholar] [CrossRef]
- Mejía-Mejía, E.; May, J.M.; Torres, R.; Kyriacou, P.A. Pulse rate variability in cardiovascular health: A review on its applications and relationship with heart rate variability. Physiol. Meas. 2020, 41, 07TR01. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Vagedes, J. How accurate is pulse rate variability as an estimate of heart rate variability? Int. J. Cardiol. 2013, 166, 15–29. [Google Scholar] [CrossRef]
- Als, H. NIDCAP Program Guide; Children’s Hospital: Boston, MA, USA, 2007. [Google Scholar]
- Als, H. A synactive model of neonatal behavioral organization: Framework for the assessment of neurobehavioral development in the premature infant and for support of infants and parents in the neonatal intensive care environment. Phys. Occup. Ther. Pediatr. 1986, 6, 3–53. [Google Scholar] [CrossRef]
- Als, H.; Butler, S.; Kosta, S.; McAnulty, G. The Assessment of Preterm Infants’ Behavior (APIB): Furthering the understanding and measurement of neurodevelopmental competence in preterm and full-term infants. Ment. Retard. Dev. Disabil. Res. Rev. 2005, 11, 94–102. [Google Scholar] [CrossRef]
- Charlton, P.H.; Kotzen, K.; Mejía-Mejía, E.; Aston, P.J.; Budidha, K.; Mant, J.; Pettit, C.; Behar, J.A.; Kyriacou, P.A. Detecting beats in the photoplethysmogram: Benchmarking open-source algorithms. Physiol. Meas. 2022, 43, 085007. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, F.; Ginsberg, J.P. An Overview of Heart Rate Variability Metrics and Norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef]
- Peralta, E.; Lazaro, J.; Bailon, R.; Marozas, V.; Gil, E. Optimal fiducial points for pulse rate variability analysis from forehead and finger photoplethysmographic signals. Physiol. Meas. 2019, 40, 025007. [Google Scholar] [CrossRef]
- Wang, P.; Jiang, A.; Liu, X.; Shang, J.; Zhang, L. LSTM-based EEG Classification in Motor Imagery Tasks. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 2086–2095. [Google Scholar] [CrossRef]
- Zeng, H.; Yang, C.; Dai, G.; Qin, F.; Zhang, J.; Kong, W. EEG classification of driver mental states by deep learning. Cogn. Neurodynamics 2018, 12, 597–606. [Google Scholar] [CrossRef]
- Hu, X.; Yuan, S.; Xu, F.; Leng, Y.; Yuan, K. Scalp EEG classification using deep Bi-LSTM network for seizure detection. Comput. Biol. Med. 2020, 124, 103919. [Google Scholar] [CrossRef]
- Yang, S.; Yu, X.; Zhou, Y. LSTM and GRU neural network performance comparison study: Taking yelp review dataset as an example. In Proceedings of the 2020 International Workshop on Electronic Communication and Artificial Intelligence (IWECAI), Shanghai, China, 12–14 June 2020; IEEE: New York, NY, USA, 2020; pp. 98–101. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).