Untargeted Salivary Metabolomics and Proteomics: Paving the Way for Early Detection of Periodontitis
Abstract
1. Introduction
2. Materials and Methods
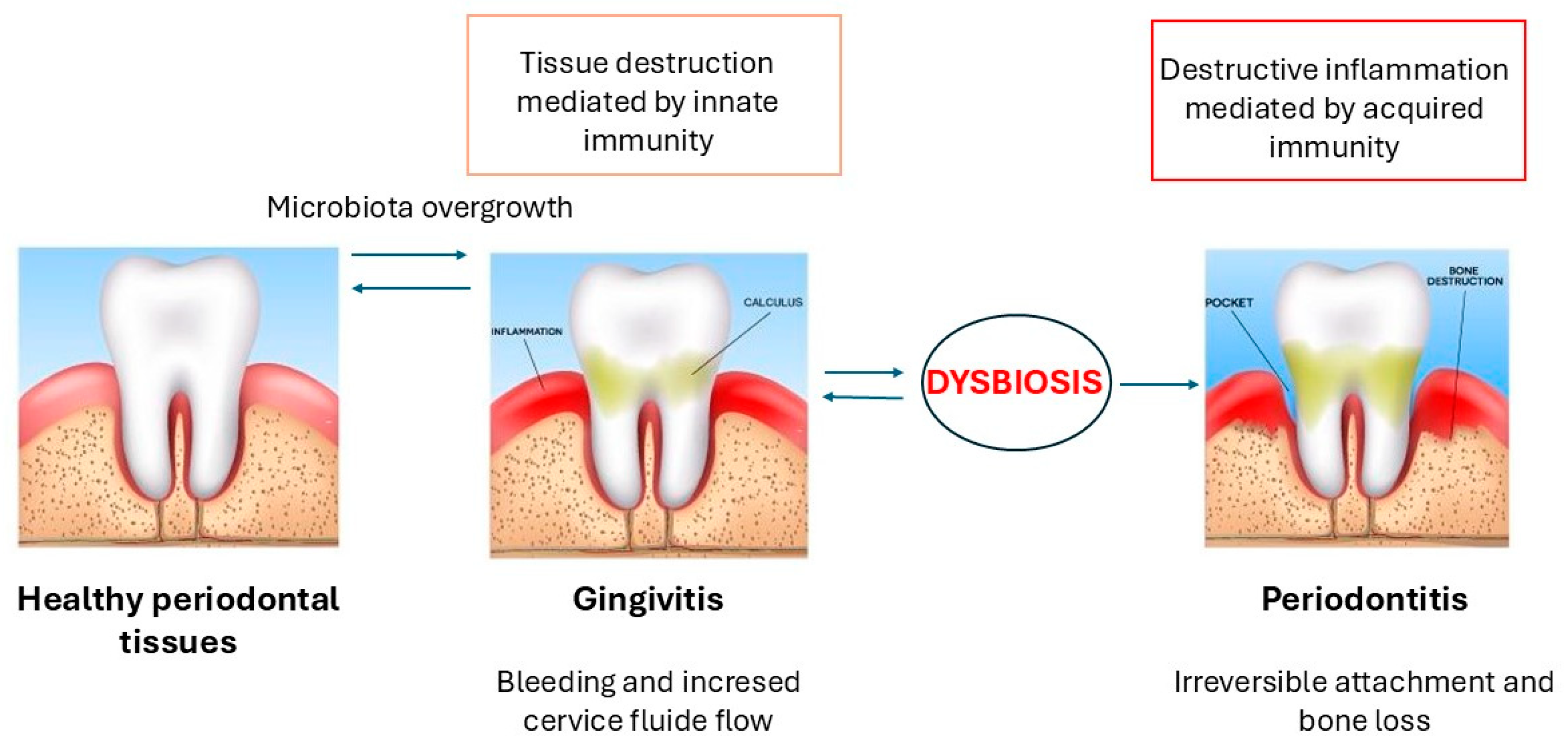
3. Periodontitis: Pathogenesis and Early Diagnosis
3.1. Early Detection and Salivary Diagnosis of Periodontitis
3.2. Saliva Compared with Gingival Crevicular Fluid
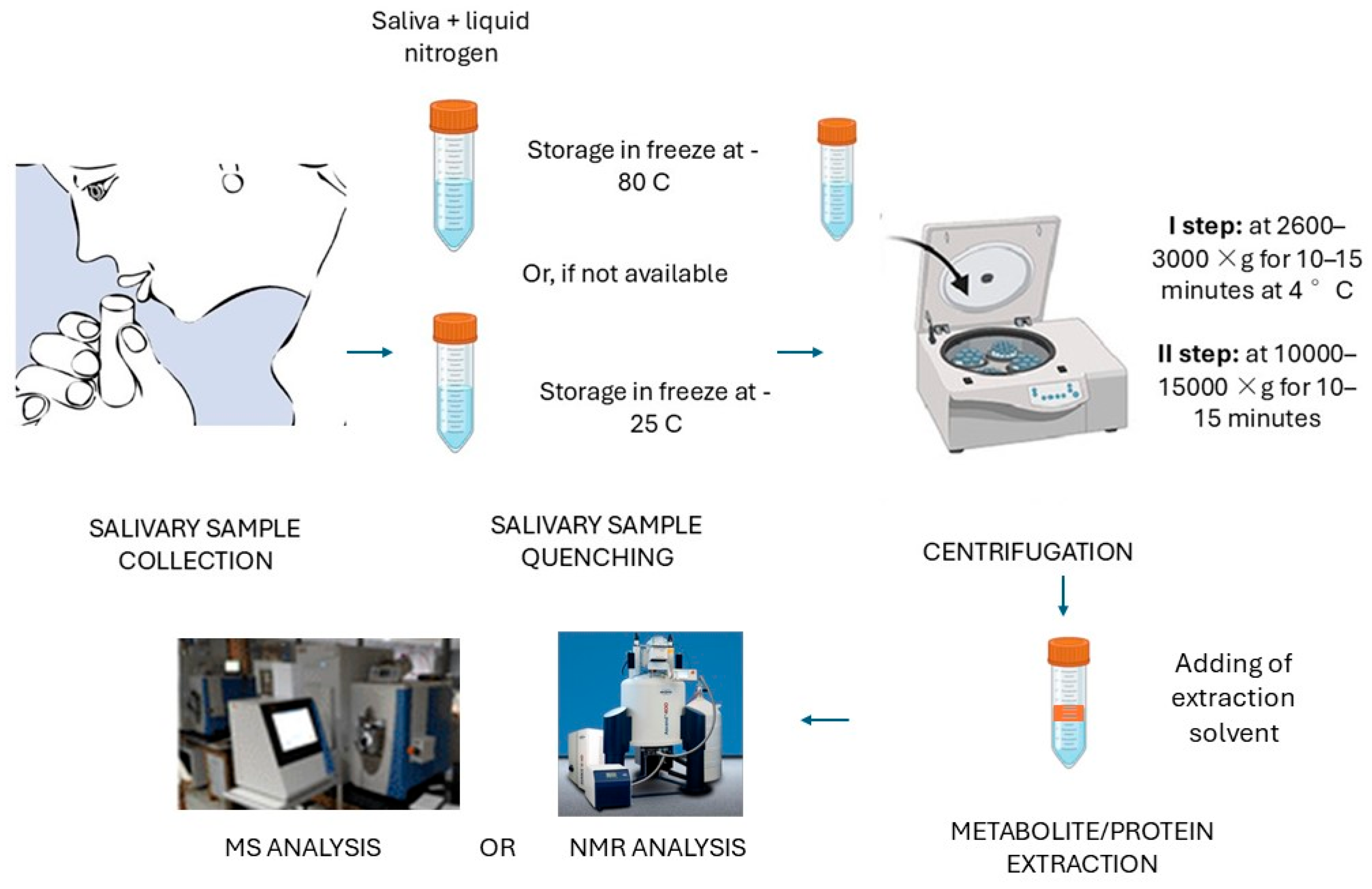
4. Salivary Samples: The Importance of the Correct Collection and Preparation
5. Salivary Metabolomics: Techniques and Applications
6. Salivary Proteomics: Techniques and Applications
7. Targeted and Untargeted Approaches in Biomarker Discovery
8. Analytical Technologies and Data Interpretation
9. Challenges and Limitations
10. Future Perspectives
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. 1), S173–S182. [Google Scholar] [CrossRef] [PubMed]
- Frencken, J.E.; Sharma, P.; Stenhouse, L.; Green, D.; Laverty, D.; Dietrich, T. Global epidemiology of dental caries and severe periodontitis—A comprehensive review. J. Clin. Periodontol. 2017, 44 (Suppl. 18), S94–S105. [Google Scholar] [CrossRef]
- Kassebaum, N.J.; Bernabé, E.; Dahiya, M.; Bhandari, B.; Murray, C.J.; Marcenes, W. Global burden of severe periodontitis in 1990–2010: A systematic review and meta-regression. J. Dent. Res. 2014, 93, 1045–1053. [Google Scholar] [CrossRef]
- Di Stefano, M.; Polizzi, A.; Santonocito, S.; Romano, A.; Lombardi, T.; Isola, G. Impact of Oral Microbiome in Periodontal Health and Periodontitis: A Critical Review on Prevention and Treatment. Int. J. Mol. Sci. 2022, 23, 5142. [Google Scholar] [CrossRef]
- Chen, Z.; Zhong, Y.; Chen, L.; Liu, W.; Lin, C.; Chen, Y.; Wang, X. HGF Aggravated Periodontitis-Associated Gut Barrier and Microbial Dysfunction: Implications for Oral–Gut Axis Regulation. Biology 2025, 14, 496. [Google Scholar] [CrossRef] [PubMed]
- Carra, M.C.; Rangé, H.; Caligiuri, G.; Bouchard, P. Periodontitis and atherosclerotic cardiovascular disease: A critical appraisal. Periodontol 2000 2023. Online ahead of print. [Google Scholar] [CrossRef]
- Miller, C.S.; Foley, J.D.; Bailey, A.L.; Campell, C.L.; Humphries, R.L.; Christodoulides, N.; Floriano, P.N.; Simmons, G.; Bhagwandin, B.; Jacobson, J.W.; et al. Current developments in salivary diagnostics. Biomark. Med. 2010, 4, 171–189. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, J.; Lin, C.C.; Abemayor, E.; Wang, M.B.; Wong, D.T. The emerging landscape of salivary diagnostics. Periodontol 2000 2016, 70, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, Z. Saliva biomarkers in oral disease. Clin. Chim. Acta 2023, 548, 117503. [Google Scholar] [CrossRef]
- Liao, C.; Chen, X.; Fu, Y. Salivary analysis: An emerging paradigm for non-invasive healthcare diagnosis and monitoring. Interdiscip. Med. 2023, 1, e20230009. [Google Scholar] [CrossRef]
- Corana, M.; Baima, G.; Iaderosa, G.; Franco, F.; Zhang, J.; Berta, G.N.; Romano, F.; Aimetti, M. Salivary Proteomics for Detecting Novel Biomarkers of Periodontitis: A Systematic Review. J. Periodontal. Res. 2024. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, E.; Savolainen, M.; Mikkonen, J.J.W.; Kullaa, A.M. Salivary Metabolomics for Diagnosis and Monitoring Diseases: Challenges and Possibilities. Metabolites 2021, 11, 587. [Google Scholar] [CrossRef] [PubMed]
- Dede, F.; Ozden, F.O.; Avcı, B. 8-hydroxy-deoxyguanosine levels in gingival crevicular fluid and saliva in patients with chronic periodontitis after initial periodontal treatment. J. Periodontol. 2013, 84, 821–828. [Google Scholar] [CrossRef]
- Kim, H.D.; Kim, S.; Jeon, S.; Kim, S.J.; Cho, H.J.; Choi, Y.N. Diagnostic and Prognostic ability of salivary MMP-9 and S100A8 for periodontitis. J. Clin. Periodontol. 2020, 47, 1191–1200. [Google Scholar] [CrossRef]
- Hu, S.; Loo, J.A.; Wong, D.T. Human saliva proteome analysis and disease biomarker discovery. Expert. Rev. Proteom. 2007, 4, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Mikkonen, J.J.; Raittila, J.; Rieppo, L.; Lappalainen, R.; Kullaa, A.M.; Myllymaa, S. Fourier Transform Infrared Spectroscopy and Photoacoustic Spectroscopy for Saliva Analysis. Appl. Spectrosc. 2016, 70, 1502–1510. [Google Scholar] [CrossRef]
- Marshall, D.D.; Powers, R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 100, 1–16. [Google Scholar] [CrossRef]
- Shin, M.S.; Kim, Y.G.; Shin, Y.J.; Ko, B.J.; Kim, S.; Kim, H.D. Deep sequencing salivary proteins for periodontitis using proteomics. Clin. Oral Investig. 2019, 23, 3571–3580. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef]
- Abdulkareem, A.A.; Al-Taweel, F.B.; Al-Sharqi, A.J.B.; Gul, S.S.; Sha, A.; Chapple, I.L.C. Current concepts in the pathogenesis of periodontitis: From symbiosis to dysbiosis. J. Oral Microbiol. 2023, 15, 2197779. [Google Scholar] [CrossRef]
- Page, R.C.; Kornman, K.S. The pathogenesis of human periodontitis: An introduction. Periodontol 2000 1997, 14, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Graves, D. Cytokines that promote periodontal tissue destruction. J. Periodontol. 2008, 79 (Suppl. 8), 1585–1591. [Google Scholar] [CrossRef] [PubMed]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef] [PubMed]
- Preshaw, P.M.; Alba, A.L.; Herrera, D.; Jepsen, S.; Konstantinidis, A.; Makrilakis, K.; Taylor, R. Periodontitis and diabetes: A two-way relationship. Diabetologia 2012, 55, 21–31. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Van Dyke, T.E. Periodontitis and atherosclerotic cardiovascular disease: Consensus report of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J. Periodontol. 2013, 84 (Suppl. 4), S24–S29. [Google Scholar] [CrossRef]
- Chapple, I.L.; Van der Weijden, F.; Doerfer, C.; Herrera, D.; Shapira, L.; Polak, D.; Madianos, P.; Louropoulou, A.; Machtei, E.; Donos, N.; et al. Primary prevention of periodontitis: Managing gingivitis. J. Clin. Periodontol. 2015, 42 (Suppl. 16), S71–S76. [Google Scholar] [CrossRef]
- Korte, D.L.; Kinney, J. Personalized medicine: An update of salivary biomarkers for periodontal diseases. Periodontology 2000 2016, 70, 26–37. [Google Scholar] [CrossRef]
- Giannobile, W.V.; Beikler, T.; Kinney, J.S.; Ramseier, C.A.; Morelli, T.; Wong, D.T. Saliva as a diagnostic tool for periodontal disease: Current state and future directions. Periodontol 2000 2009, 50, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Ramseier, C.A.; Kinney, J.S.; Herr, A.E.; Braun, T.; Sugai, J.V.; Shelburne, C.A.; Rayburn, L.A.; Tran, H.M.; Singh, A.K.; Giannobile, W.V. Identification of pathogen and host-response markers correlated with periodontal disease. J. Periodontol. 2009, 80, 436–446. [Google Scholar] [CrossRef]
- Baima, G.; Iaderosa, G.; Citterio, F.; Grossi, S.; Romano, F.; Berta, G.N.; Buduneli, N.; Aimetti, M. Salivary metabolomics for the diagnosis of periodontal diseases: A systematic review with methodological quality assessment. Metabolomics 2021, 17, 1. [Google Scholar] [CrossRef]
- Wallner-Liebmann, S.; Tenori, L.; Mazzoleni, A.; Dieber-Rotheneder, M.; Konrad, M.; Hofmann, P.; Luchinat, C.; Turano, P.; Zatloukal, K. Individual Human Metabolic Phenotype Analyzed by (1)H NMR of Saliva Samples. J. Proteome Res. 2016, 15, 1787–1793. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.J.; Preshaw, P.M. Gingival crevicular fluid and saliva. Periodontology 2000 2016, 70, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Li, F.; Xie, Y.; Liu, J.; Yu, T.; Feng, X. Microbial and metabolomic analysis of gingival crevicular fluid in general chronic periodontitis patients: Lessons for a predictive, preventive, and personalized medical approach. Epma J. 2020, 11, 197–215. [Google Scholar] [CrossRef]
- Papagerakis, P.; Zheng, L.; Kim, D.; Said, R.; Ehlert, A.A.; Chung, K.K.M.; Papagerakis, S. Saliva and Gingival Crevicular Fluid (GCF) Collection for Biomarker Screening. Methods Mol. Biol. 2019, 1922, 549–562. [Google Scholar] [CrossRef]
- Bruce, S.J.; Tavazzi, I.; Parisod, V.; Rezzi, S.; Kochhar, S.; Guy, P.A. Investigation of human blood plasma sample preparation for performing metabolomics using ultrahigh performance liquid chromatography/mass spectrometry. Anal. Chem. 2009, 81, 3285–3296. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.R.; Kim, H.-R.; Chae, H.-J. Compliance with saliva collection protocol in healthy volunteers: Strategies for managing risk and errors. Int. J. Med. Sci. 2018, 15, 823. [Google Scholar] [CrossRef]
- Tan, S.Z.; Begley, P.; Mullard, G.; Hollywood, K.A.; Bishop, P.N. Introduction to metabolomics and its applications in ophthalmology. Eye 2016, 30, 773–783. [Google Scholar] [CrossRef]
- Burgess, K.; Rankin, N.; Weidt, S. Metabolomics. In Handbook of Pharmacogenomics and Stratified Medicine; Elsevier: Amsterdam, The Netherlands, 2014; pp. 181–205. [Google Scholar]
- Sake, C.L.; Newman, D.M.; Boyle, N.R. Evaluation of quenching methods for metabolite recovery in photoautotrophic Synechococcus sp. PCC 7002. Biotechnol. Prog. 2020, 36, e3015. [Google Scholar] [CrossRef]
- García-Villaescusa, A.; Morales-Tatay, J.M.; Monleón-Salvadó, D.; González-Darder, J.M.; Bellot-Arcis, C.; Montiel-Company, J.M.; Almerich-Silla, J.M. Using NMR in saliva to identify possible biomarkers of glioblastoma and chronic periodontitis. PLoS ONE 2018, 13, e0188710. [Google Scholar] [CrossRef]
- Kumari, S.; Goyal, V.; Kumaran, S.S.; Dwivedi, S.N.; Srivastava, A.; Jagannathan, N.R. Quantitative metabolomics of saliva using proton NMR spectroscopy in patients with Parkinson’s disease and healthy controls. Neurol. Sci. 2020, 41, 1201–1210. [Google Scholar] [CrossRef]
- Rzeznik, M.; Triba, M.N.; Levy, P.; Jungo, S.; Botosoa, E.; Duchemann, B.; Le Moyec, L.; Bernaudin, J.F.; Savarin, P.; Guez, D. Identification of a discriminative metabolomic fingerprint of potential clinical relevance in saliva of patients with periodontitis using 1H nuclear magnetic resonance (NMR) spectroscopy. PLoS ONE 2017, 12, e0182767. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Córdoba, B.; Santiago-Garcia, J. Saliva: A fluid of study for OMICS. Omics J. Integr. Biol. 2014, 18, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, X. Saliva metabolomics opens door to biomarker discovery, disease diagnosis, and treatment. Appl. Biochem. Biotechnol. 2012, 168, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Romano, F.; Meoni, G.; Manavella, V.; Baima, G.; Mariani, G.M.; Cacciatore, S.; Tenori, L.; Aimetti, M. Effect of non-surgical periodontal therapy on salivary metabolic fingerprint of generalized chronic periodontitis using nuclear magnetic resonance spectroscopy. Arch. Oral Biol. 2019, 97, 208–214. [Google Scholar] [CrossRef]
- Singh, M.P.; Saxena, M.; Saimbi, C.S.; Arif, J.M.; Roy, R. Metabolic profiling by 1 H NMR spectroscopy of saliva shows clear distinction between control and diseased case of periodontitis. Metabolomics 2017, 13, 137. [Google Scholar] [CrossRef]
- Mathon, C.; Bovard, D.; Dutertre, Q.; Sendyk, S.; Bentley, M.; Hoeng, J.; Knorr, A. Impact of sample preparation upon intracellular metabolite measurements in 3D cell culture systems. Metabolomics 2019, 15, 92. [Google Scholar] [CrossRef]
- Vuckovic, D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 1523–1548. [Google Scholar] [CrossRef]
- Kageyama, G.; Saegusa, J.; Irino, Y.; Tanaka, S.; Tsuda, K.; Takahashi, S.; Sendo, S.; Morinobu, A. Metabolomics analysis of saliva from patients with primary Sjögren’s syndrome. Clin. Exp. Immunol. 2015, 182, 149–153. [Google Scholar] [CrossRef]
- Hurskainen, M.O.; Sarin, J.K.; Myllymaa, S.; González-Arriagada, W.A.; Kullaa, A.; Lappalainen, R. Feasibility of Near-Infrared Spectroscopy for Identification of L-Fucose and L-Proline—Towards Detecting Cancer Biomarkers from Saliva. Appl. Sci. 2021, 11, 9662. [Google Scholar] [CrossRef]
- Graves, D.T.; Corrêa, J.D.; Silva, T.A. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 2019, 98, 148–156. [Google Scholar] [CrossRef]
- Weber, R.J.; Lawson, T.N.; Salek, R.M.; Ebbels, T.M.; Glen, R.C.; Goodacre, R.; Griffin, J.L.; Haug, K.; Koulman, A.; Moreno, P. Computational tools and workflows in metabolomics: An international survey highlights the opportunity for harmonisation through Galaxy. Metabolomics 2017, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Metabonomics Methods Protoc. 2015, 1277, 161–193. [Google Scholar]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Koo, I.; Wei, X.; Zhang, X. Analysis of metabolomic profiling data acquired on GC–MS. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 543, pp. 315–324. [Google Scholar]
- Horning, E.; Horning, M. Human metabolic profiles obtained by GC and GC/MS. J. Chromatogr. Sci. 1971, 9, 129–140. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ju, W.; Yao, W.; Shi, C.; Yang, X.; Qian, W. The salivary metabolomics analyses reveal the variable metabolites in distinct staging of periodontitis. BMC Oral Health 2025, 25, 480. [Google Scholar] [CrossRef]
- Gika, H.G.; Wilson, I.D.; Theodoridis, G.A. LC–MS-based holistic metabolic profiling. Problems, limitations, advantages, and future perspectives. J. Chromatogr. B 2014, 966, 1–6. [Google Scholar] [CrossRef]
- Kosho, M.X.; Ciurli, A.; Giera, M.; Neefjes, J.; Loos, B.G. Metabolomic Profiles of Oral Rinse Samples to Distinguish Severe Periodontitis Patients From Non-Periodontitis Controls. J. Periodontal Res. 2025. Online ahead of print. [Google Scholar] [CrossRef]
- Fonseca, T.A.; Von Rekowski, C.P.; Araújo, R.; Oliveira, M.C.; Justino, G.C.; Bento, L.; Calado, C.R. The impact of the serum extraction protocol on metabolomic profiling using UPLC-MS/MS and FTIR Spectroscopy. ACS Omega 2023, 8, 20755–20766. [Google Scholar] [CrossRef] [PubMed]
- Albahri, J.; Allison, H.; Whitehead, K.A.; Muhamadali, H. The role of salivary metabolomics in chronic periodontitis: Bridging oral and systemic diseases. Metabolomics 2025, 21, 24. [Google Scholar] [CrossRef]
- Rodrigues, J.; Amin, A.; Chandra, S.; Mulla, N.J.; Nayak, G.S.; Rai, S.; Ray, S.; Mahato, K.K. Machine learning enabled photoacoustic spectroscopy for noninvasive assessment of breast tumor progression in vivo: A preclinical study. ACS Sens. 2024, 9, 589–601. [Google Scholar] [CrossRef]
- Mikkonen, J. Infrared and Nuclear Magnetic Resonance Spectroscopic Methods for Salivary Analysis; Itä-Suomen yliopisto: Joensuu, Finland, 2019. [Google Scholar]
- Dame, Z.T.; Aziat, F.; Mandal, R.; Krishnamurthy, R.; Bouatra, S.; Borzouie, S.; Guo, A.C.; Sajed, T.; Deng, L.; Lin, H.; et al. The human saliva metabolome. Metabolomics 2015, 11, 1864–1883. [Google Scholar] [CrossRef]
- Takeda, I.; Stretch, C.; Barnaby, P.; Bhatnager, K.; Rankin, K.; Fu, H.; Weljie, A.; Jha, N.; Slupsky, C. Understanding the human salivary metabolome. NMR Biomed. 2009, 22, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Kesavalu, L.; Vasudevan, B.; Raghu, B.; Browning, E.; Dawson, D.; Novak, J.M.; Correll, M.C.; Steffen, M.J.; Bhattacharya, A.; Fernandes, G.; et al. Omega-3 Fatty Acid Effect on Alveolar Bone Loss in Rats. J. Dent. Res. 2006, 85, 648–652. [Google Scholar] [CrossRef]
- Sorkin, B.C.; Niederman, R. Short chain carboxylic acids decrease human gingival keratinocyte proliferation and increase apoptosis and necrosis. J. Clin. Periodontol. 1998, 25, 311–315. [Google Scholar] [CrossRef]
- Courtois, P.; Labbé, M.; Pourtois, M.; Mandelbaum, I.M. Anaerobes and short-chain fatty acids in crevicular fluid from adults with chronic periodontitis. Bull. Group. Int. Rech. Sci. Stomatol. Odontol. 1989, 32, 19–22. [Google Scholar]
- Hatanaka, K.; Shirahase, Y.; Yoshida, T.; Kono, M.; Toya, N.; Sakasegawa, S.I.; Konishi, K.; Yamamoto, T.; Ochiai, K.; Takashiba, S. Enzymatic measurement of short-chain fatty acids and application in periodontal disease diagnosis. PLoS ONE 2022, 17, e0268671. [Google Scholar] [CrossRef]
- Takahashi, N. Oral Microbiome Metabolism: From “Who Are They?” to “What Are They Doing?”. J. Dent. Res. 2015, 94, 1628–1637. [Google Scholar] [CrossRef]
- Na, H.S.; Kim, S.Y.; Han, H.; Kim, H.-J.; Lee, J.-Y.; Lee, J.-H.; Chung, J. Identification of potential oral microbial biomarkers for the diagnosis of periodontitis. J. Clin. Med. 2020, 9, 1549. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N. Microbial ecosystem in the oral cavity: Metabolic diversity in an ecological niche and its relationship with oral diseases. In International Congress Series; Elsevier: Amsterdam, The Netherlands, 2005; pp. 103–112. [Google Scholar]
- Citterio, F.; Romano, F.; Meoni, G.; Iaderosa, G.; Grossi, S.; Sobrero, A.; Dego, F.; Corana, M.; Berta, G.N.; Tenori, L.; et al. Changes in the Salivary Metabolic Profile of Generalized Periodontitis Patients after Non-surgical Periodontal Therapy: A Metabolomic Analysis Using Nuclear Magnetic Resonance Spectroscopy. J. Clin. Med. 2020, 9, 3977. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.J.; Song, Y.; Lee, H.A.; Kim, S.; Chung, J. Metabolic phenotyping of saliva to identify possible biomarkers of periodontitis using proton nuclear magnetic resonance. J. Clin. Periodontol. 2021, 48, 1240–1249. [Google Scholar] [CrossRef]
- Kim, C.; Cha, Y.N. Taurine chloramine produced from taurine under inflammation provides anti-inflammatory and cytoprotective effects. Amino Acids 2014, 46, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Marcinkiewicz, J.; Kontny, E. Taurine and inflammatory diseases. Amino Acids 2014, 46, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Shirasugi, M.; Nakagawa, M.; Nishioka, K.; Yamamoto, T.; Nakaya, T.; Kanamura, N. Relationship between periodontal disease and butyric acid produced by periodontopathic bacteria. Inflamm. Regen. 2018, 38, 23. [Google Scholar] [CrossRef]
- Holme, P.; Huss, M.; Jeong, H. Subnetwork hierarchies of biochemical pathways. Bioinformatics 2003, 19, 532–538. [Google Scholar] [CrossRef]
- Aimetti, M.; Cacciatore, S.; Graziano, A.; Tenori, L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics 2012, 8, 465–474. [Google Scholar] [CrossRef]
- Romano, F.; Meoni, G.; Manavella, V.; Baima, G.; Tenori, L.; Cacciatore, S.; Aimetti, M. Analysis of salivary phenotypes of generalized aggressive and chronic periodontitis through nuclear magnetic resonance-based metabolomics. J. Periodontol. 2018, 89, 1452–1460. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Meng, H.; Yu, J.; Lu, H.; Li, W.; Lu, R.; Zhao, Y.; Li, Q.; Su, L. Butyrate rather than LPS subverts gingival epithelial homeostasis by downregulation of intercellular junctions and triggering pyroptosis. J. Clin. Periodontol. 2019, 46, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Gawron, K.; Wojtowicz, W.; Łazarz-Bartyzel, K.; Łamasz, A.; Qasem, B.; Mydel, P.; Chomyszyn-Gajewska, M.; Potempa, J.; Mlynarz, P. Metabolomic Status of The Oral Cavity in Chronic Periodontitis. In Vivo 2019, 33, 1165–1174. [Google Scholar] [CrossRef]
- Zhang, S.; Zeng, X.; Ren, M.; Mao, X.; Qiao, S. Novel metabolic and physiological functions of branched chain amino acids: A review. J. Anim. Sci. Biotechnol. 2017, 8, 10. [Google Scholar] [CrossRef]
- Barnes, V.M.; Teles, R.; Trivedi, H.M.; Devizio, W.; Xu, T.; Mitchell, M.W.; Milburn, M.V.; Guo, L. Acceleration of purine degradation by periodontal diseases. J. Dent. Res. 2009, 88, 851–855. [Google Scholar] [CrossRef]
- Barnes, V.M.; Teles, R.; Trivedi, H.M.; Devizio, W.; Xu, T.; Lee, D.P.; Mitchell, M.W.; Wulff, J.E.; Milburn, M.V.; Guo, L. Assessment of the effects of dentifrice on periodontal disease biomarkers in gingival crevicular fluid. J. Periodontol. 2010, 81, 1273–1279. [Google Scholar] [CrossRef] [PubMed]
- Alamri, M.M.; Williams, B.; Le Guennec, A.; Mainas, G.; Santamaria, P.; Moyes, D.L.; Nibali, L. Metabolomics analysis in saliva from periodontally healthy, gingivitis and periodontitis patients. J. Periodontal Res. 2023, 58, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Belstrøm, D.; Jersie-Christensen, R.R.; Lyon, D.; Damgaard, C.; Jensen, L.J.; Holmstrup, P.; Olsen, J.V. Metaproteomics of saliva identifies human protein markers specific for individuals with periodontitis and dental caries compared to orally healthy controls. PeerJ 2016, 4, e2433. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Aebersold, R. Interpretation of shotgun proteomic data: The protein inference problem. Mol. Cell Proteom. 2005, 4, 1419–1440. [Google Scholar] [CrossRef]
- Proctor, G.B. The physiology of salivary secretion. Periodontology 2000 2016, 70, 11–25. [Google Scholar] [CrossRef]
- Pedersen, A.; Sørensen, C.; Proctor, G.; Carpenter, G.; Ekström, J. Salivary secretion in health and disease. J. Oral Rehabil. 2018, 45, 730–746. [Google Scholar] [CrossRef]
- Siqueira, W.L.; Dawes, C. The salivary proteome: Challenges and perspectives. Proteom.-Clin. Appl. 2011, 5, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Maddu, N. Functions of saliva. In Saliva and Salivary Diagnostics; IntechOpen: London, UK, 2019. [Google Scholar]
- Gorr, S.-U.; Venkatesh, S.; Darling, D. Parotid secretory granules: Crossroads of secretory pathways and protein storage. J. Dent. Res. 2005, 84, 500–509. [Google Scholar] [CrossRef]
- Kinney, J. Saliva/pathogen biomarker signatures and periodontal disease progression. J. Dent. Res. 2011, 90, 752–758. [Google Scholar] [CrossRef]
- Salazar, M.G.; Jehmlich, N.; Murr, A.; Dhople, V.M.; Holtfreter, B.; Hammer, E.; Völker, U.; Kocher, T. Identification of periodontitis associated changes in the proteome of whole human saliva by mass spectrometric analysis. J. Clin. Periodontol. 2013, 40, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends Immunol. 2014, 35, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.J.; Yoon, M.J.; Kim, D.H.; Kim, T.U.; Kang, Y.J. Profilin-1; a novel regulator of DNA damage response and repair machinery in keratinocytes. Mol. Biol. Rep. 2021, 48, 1439–1452. [Google Scholar] [CrossRef]
- Witke, W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004, 14, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Pintos, T.; Regueira-Iglesias, A.; Relvas, M.; Alonso-Sampedro, M.; Bravo, S.B.; Balsa-Castro, C.; Tomás, I. Diagnostic Accuracy of Novel Protein Biomarkers in Saliva to Detect Periodontitis Using Untargeted ‘SWATH’ Mass Spectrometry. J. Clin. Periodontol. 2025, 52, 199–214. [Google Scholar] [CrossRef]
- Ryckman, C.; Vandal, K.; Rouleau, P.; Talbot, M.; Tessier, P.A. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 2003, 170, 3233–3242. [Google Scholar] [CrossRef]
- Passey, R.J.; Xu, K.; Hume, D.A.; Geczy, C.L. S100A8: Emerging functions and regulation. J. Leukoc. Biol. 1999, 66, 549–556. [Google Scholar] [CrossRef]
- Grant, M.M.; Taylor, J.J.; Jaedicke, K.; Creese, A.; Gowland, C.; Burke, B.; Doudin, K.; Patel, U.; Weston, P.; Milward, M.; et al. Discovery, validation, and diagnostic ability of multiple protein-based biomarkers in saliva and gingival crevicular fluid to distinguish between health and periodontal diseases. J. Clin. Periodontol. 2022, 49, 622–632. [Google Scholar] [CrossRef]
- Loos, B.G. Systemic markers of inflammation in periodontitis. J. Periodontol. 2005, 76 (Suppl. 11), 2106–2115. [Google Scholar] [CrossRef]
- Weisel, J.W. Fibrinogen and fibrin. Adv. Protein Chem. 2005, 70, 247–299. [Google Scholar] [CrossRef]
- Romano, F.; Franco, F.; Corana, M.; Abbadessa, G.; Di Scipio, F.; Pergolizzi, B.; Castrignano, C.; Aimetti, M.; Berta, G.N. Cystatin SN (CST1) as a Novel Salivary Biomarker of Periodontitis. Int. J. Mol. Sci. 2023, 24, 13834. [Google Scholar] [CrossRef] [PubMed]
- Bostanci, N.; Selevsek, N.; Wolski, W.; Grossmann, J.; Bao, K.; Wahlander, A.; Trachsel, C.; Schlapbach, R.; Öztürk, V.; Afacan, B.; et al. Targeted Proteomics Guided by Label-free Quantitative Proteome Analysis in Saliva Reveal Transition Signatures from Health to Periodontal Disease. Mol. Cell Proteom. 2018, 17, 1392–1409. [Google Scholar] [CrossRef] [PubMed]
- Ochieng, J.; Chaudhuri, G. Cystatin superfamily. J. Health Care Poor Underserved. 2010, 21 (Suppl. 1), 51–70. [Google Scholar] [CrossRef]
- Mertens, B.; Orti, V.; Vialaret, J.; Gibert, P.; Relaño-Ginés, A.; Lehmann, S.; Deville de Périère, D.; Hirtz, C. Assessing a multiplex-targeted proteomics approach for the clinical diagnosis of periodontitis using saliva samples. Bioanalysis 2018, 10, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Yuan, C.; Ma, Z.; Zhu, C.; Tong, P.; Gallagher, J.E.; Sun, X.; Zheng, S. The potentiality of salivary peptide biomarkers for screening patients with periodontal diseases by mass spectrometry. Clin. Chim. Acta 2019, 495, 278–286. [Google Scholar] [CrossRef]
- Antezack, A.; Chaudet, H.; Tissot-Dupont, H.; Brouqui, P.; Monnet-Corti, V. Rapid diagnosis of periodontitis, a feasibility study using MALDI-TOF mass spectrometry. PLoS ONE 2020, 15, e0230334. [Google Scholar] [CrossRef]
- Hartenbach, F.; Velasquez, É.; Nogueira, F.C.S.; Domont, G.B.; Ferreira, E.; Colombo, A.P.V. Proteomic analysis of whole saliva in chronic periodontitis. J. Proteom. 2020, 213, 103602. [Google Scholar] [CrossRef]
- Casarin, R.C.V.; Salmon, C.R.; Stolf, C.S.; Paz, H.E.S.; Rangel, T.P.; Domingues, R.R.; Pauletti, B.A.; Paes-Leme, A.F.; Araújo, C.; Santamaria, M.P.; et al. Salivary annexin A1: A candidate biomarker for periodontitis. J. Clin. Periodontol. 2023, 50, 942–951. [Google Scholar] [CrossRef]
- Gonçalves Lda, R.; Soares, M.R.; Nogueira, F.C.; Garcia, C.; Camisasca, D.R.; Domont, G.; Feitosa, A.C.; Pereira Dde, A.; Zingali, R.B.; Alves, G. Comparative proteomic analysis of whole saliva from chronic periodontitis patients. J. Proteom. 2010, 73, 1334–1341. [Google Scholar] [CrossRef]
- Chaiyarit, P.; Taweechaisupapong, S.; Jaresitthikunchai, J.; Phaonakrop, N.; Roytrakul, S. Comparative evaluation of 5-15-kDa salivary proteins from patients with different oral diseases by MALDI-TOF/TOF mass spectrometry. Clin. Oral Investig. 2015, 19, 729–737. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Mas, S.; Akesson, M.; Smedsgaard, J.; Nielsen, J. Mass spectrometry in metabolome analysis. Mass. Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef] [PubMed]
- Bingol, K. Recent Advances in Targeted and Untargeted Metabolomics by NMR and MS/NMR Methods. High. Throughput 2018, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Gil de la Fuente, A.; Grace Armitage, E.; Otero, A.; Barbas, C.; Godzien, J. Differentiating signals to make biological sense—A guide through databases for MS-based non-targeted metabolomics. Electrophoresis 2017, 38, 2242–2256. [Google Scholar] [CrossRef]
- Zhan, X. Metabolomics: Methodology and Applications in Medical Sciences and Life Sciences; BoD–Books on Demand: Norderstedt, Germany, 2021. [Google Scholar]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef]
- Pavlou, M.P.; Diamandis, E.P. The cancer cell secretome: A good source for discovering biomarkers? J. Proteom. 2010, 73, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30.2.1–30.2.24. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Letertre, M.P.; Dervilly, G.; Giraudeau, P. Combined nuclear magnetic resonance spectroscopy and mass spectrometry approaches for metabolomics. Anal. Chem. 2020, 93, 500–518. [Google Scholar] [CrossRef]
- Castagnola, M.; Cabras, T.; Iavarone, F.; Fanali, C.; Nemolato, S.; Peluso, G.; Laura Bosello, S.; Faa, G.; Ferraccioli, G.; Messana, I. The human salivary proteome: A critical overview of the results obtained by different proteomic platforms. Expert. Rev. Proteom. 2012, 9, 33–46. [Google Scholar] [CrossRef]
- Mitulović, G.; Vukajlović, J.M. High-Throughput Chromatography for Clinical Proteomics Applications. In Relevant Applications of High-Performance Liquid Chromatography in Food, Environmental, Clinical and Biological Fields; IntechOpen: London, UK, 2024. [Google Scholar]
- Holzgrabe, U.; Deubner, R.; Schollmayer, C.; Waibel, B. Quantitative NMR spectroscopy—Applications in drug analysis. J. Pharm. Biomed. Anal. 2005, 38, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.L.; Baker, J.M.; Miller, S.J.; Deborde, C.; Maucourt, M.; Biais, B.; Rolin, D.; Moing, A.; Moco, S.; Vervoort, J. An inter-laboratory comparison demonstrates that [1 H]-NMR metabolite fingerprinting is a robust technique for collaborative plant metabolomic data collection. Metabolomics 2010, 6, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Van, Q.N.; Issaq, H.J.; Jiang, Q.; Li, Q.; Muschik, G.M.; Waybright, T.J.; Lou, H.; Dean, M.; Uitto, J.; Veenstra, T.D. Comparison of 1D and 2D NMR spectroscopy for metabolic profiling. J. Proteome Res. 2008, 7, 630–639. [Google Scholar] [CrossRef]
- Robinette, S.L.; Ajredini, R.; Rasheed, H.; Zeinomar, A.; Schroeder, F.C.; Dossey, A.T.; Edison, A.S. Hierarchical alignment and full resolution pattern recognition of 2D NMR spectra: Application to nematode chemical ecology. Anal. Chem. 2011, 83, 1649–1657. [Google Scholar] [CrossRef]
- Orellana, G.; Vanden Bussche, J.; Van Meulebroek, L.; Vandegehuchte, M.; Janssen, C.; Vanhaecke, L. Validation of a confirmatory method for lipophilic marine toxins in shellfish using UHPLC-HR-Orbitrap MS. Anal. Bioanal. Chem. 2014, 406, 5303–5312. [Google Scholar] [CrossRef] [PubMed]
- Nye, L.C.; Williams, J.P.; Munjoma, N.C.; Letertre, M.P.; Coen, M.; Bouwmeester, R.; Martens, L.; Swann, J.R.; Nicholson, J.K.; Plumb, R.S. A comparison of collision cross section values obtained via travelling wave ion mobility-mass spectrometry and ultra high performance liquid chromatography-ion mobility-mass spectrometry: Application to the characterisation of metabolites in rat urine. J. Chromatogr. A 2019, 1602, 386–396. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Xu, F.; Laguna, L.; Sarkar, A. Aging-related changes in quantity and quality of saliva: Where do we stand in our understanding? J. Texture Stud. 2019, 50, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Tzimas, K.; Pappa, E. Saliva Metabolomic Profile in Dental Medicine Research: A Narrative Review. Metabolites 2023, 13, 379. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Escalante-Herrera, A.; Marin, L.M.; Siqueira, W.L. Progression from healthy periodontium to gingivitis and periodontitis: Insights from bioinformatics-driven proteomics–A systematic review with meta-analysis. J. Periodontal. Res. 2025, 60, 8–29. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Sedghi, L.; Ganther, S.; Malone, E.; Kamarajan, P.; Kapila, Y.L. Host-microbe interactions: Profiles in the transcriptome, the proteome, and the metabolome. Periodontology 2000 2020, 82, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Haga, S.B.; Beskow, L.M. Ethical, legal, and social implications of biobanks for genetics research. Adv. Genet. 2008, 60, 505–544. [Google Scholar] [CrossRef]
- Mikkelsen, R.B.; Gjerris, M.; Waldemar, G.; Sandøe, P. Broad consent for biobanks is best—Provided it is also deep. BMC Med. Ethics 2019, 20, 71. [Google Scholar] [CrossRef]
- Arnerić, S.P.; Batrla-Utermann, R.; Beckett, L.; Bittner, T.; Blennow, K.; Carter, L.; Dean, R.; Engelborghs, S.; Genius, J.; Gordon, M.F.; et al. Cerebrospinal Fluid Biomarkers for Alzheimer’s Disease: A View of the Regulatory Science Qualification Landscape from the Coalition Against Major Diseases CSF Biomarker Team. J. Alzheimers Dis. 2017, 55, 19–35. [Google Scholar] [CrossRef]
- Ng, T.K.S.; Udeh-Momoh, C.; Lim, M.A.; Gleerup, H.S.; Leifert, W.; Ajalo, C.; Ashton, N.; Zetterberg, H.; Rissman, R.A.; Winston, C.N.; et al. Guidelines for the standardization of pre-analytical variables for salivary biomarker studies in Alzheimer’s disease research: An updated review and consensus of the Salivary Biomarkers for Dementia Research Working Group. Alzheimers Dement. 2025, 21, e14420. [Google Scholar] [CrossRef]
- Rathnayake, N.; Akerman, S.; Klinge, B.; Lundegren, N.; Jansson, H.; Tryselius, Y.; Sorsa, T.; Gustafsson, A. Salivary biomarkers for detection of systemic diseases. PLoS ONE 2013, 8, e61356. [Google Scholar] [CrossRef]
- Kumari, S.; Samara, M.; Ampadi Ramachandran, R.; Gosh, S.; George, H.; Wang, R.; Pesavento, R.P.; Mathew, M.T. A Review on Saliva-Based Health Diagnostics: Biomarker Selection and Future Directions. Biomed. Mater. Devices 2023, 2, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Ornelas-González, A.; Ortiz-Martínez, M.; González-González, M.; Rito-Palomares, M. Enzymatic Methods for Salivary Biomarkers Detection: Overview and Current Challenges. Molecules 2021, 26, 7026. [Google Scholar] [CrossRef]
- Nonaka, T.; Wong, D.T.W. Saliva diagnostics: Salivaomics, saliva exosomics, and saliva liquid biopsy. J. Am. Dent. Assoc. 2023, 154, 696–704. [Google Scholar] [CrossRef]
- Kayid, A. The Role of Artificial Intelligence in Future Technology; Department of Computer Science, The German University in Cairo: Cairo, Egypt, 2020. [Google Scholar]
- Ko, J.; Baldassano, S.N.; Loh, P.L.; Kording, K.; Litt, B.; Issadore, D. Machine learning to detect signatures of disease in liquid biopsies—A user’s guide. Lab. Chip 2018, 18, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, X.; Petinrin, O.O.; Zhang, W.; Rahaman, S.; Tang, Z.R.; Wong, K.C. Machine Learning Protocols in Early Cancer Detection Based on Liquid Biopsy: A Survey. Life 2021, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, T.; Cui, J.; Borole, P.; Benjamin, A.; Kording, K.; Issadore, D. A web-based automated machine learning platform to analyze liquid biopsy data. Lab. Chip 2020, 20, 2166–2174. [Google Scholar] [CrossRef]
- Amelio, I.; Bertolo, R.; Bove, P.; Buonomo, O.C.; Candi, E.; Chiocchi, M.; Cipriani, C.; Di Daniele, N.; Ganini, C.; Juhl, H.; et al. Liquid biopsies and cancer omics. Cell Death Discov. 2020, 6, 131. [Google Scholar] [CrossRef]
- Biswas, N.; Chakrabarti, S. Artificial Intelligence (AI)-Based Systems Biology Approaches in Multi-Omics Data Analysis of Cancer. Front. Oncol. 2020, 10, 588221. [Google Scholar] [CrossRef]
- Hunter, B.; Hindocha, S.; Lee, R.W. The Role of Artificial Intelligence in Early Cancer Diagnosis. Cancers 2022, 14, 1524. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Arora, A.; Kaur, D.; Patiyal, S.; Kaur, D.; Tomer, R.; Raghava, G.P.S. SalivaDB-a comprehensive database for salivary biomarkers in humans. Database 2023, 2023, baad002. [Google Scholar] [CrossRef]
- Ai, J.Y.; Smith, B.; Wong, D.T. Bioinformatics advances in saliva diagnostics. Int. J. Oral Sci. 2012, 4, 85–87. [Google Scholar] [CrossRef][Green Version]
- Leonardi, R.; Vaiid, N. Artificial Intelligence in Orthodontics: Concerns, Conjectures, and Ethical Dilemmas. Int. Dent. J. 2025, 75, 20–22. [Google Scholar] [CrossRef]
- Mohammed, H.A.; Abdulkareem, A.A.; Zardawi, F.M.; Gul, S.S. Determination of the Accuracy of Salivary Biomarkers for Periodontal Diagnosis. Diagnostics 2022, 12, 2485. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Bai, H.; Zhang, P.; Zhou, X.; Ying, B. Promising applications of human-derived saliva biomarker testing in clinical diagnostics. Int. J. Oral Sci. 2023, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Garcia, P.N.; de Souza, M.M.; Izidoro, M.A.; Juliano, L.; Lourenço, S.V.; Camillo, C.M.C. Saliva metabolomics: Concepts and applications in oral disorders. Clin. Oral Investig. 2024, 28, 579. [Google Scholar] [CrossRef]
- Divaris, K.; Moss, K.; Beck, J.D. Biologically informed stratification of periodontal disease holds the key to achieving precision oral health. J. Periodontol. 2020, 91, S50–S55. [Google Scholar] [CrossRef]
- Lyman, G.H.; Moses, H.L. Biomarker tests for molecularly targeted therapies—The key to unlocking precision medicine. N. Engl. J. Med. 2016, 375, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Kuboniwa, M.; Sakanaka, A.; Hashino, E.; Bamba, T.; Fukusaki, E.; Amano, A. Prediction of Periodontal Inflammation via Metabolic Profiling of Saliva. J. Dent. Res. 2016, 95, 1381–1386. [Google Scholar] [CrossRef]
- Haririan, H.; Andrukhov, O.; Laky, M.; Rausch-Fan, X. Saliva as a Source of Biomarkers for Periodontitis and Periimplantitis. Front. Dent. Med. 2021, 2, 687638. [Google Scholar] [CrossRef]

| Metabolomics Sequencing Techniques | Methodologies | Strengths | Limitations | Potential Role in Early Diagnosis of Periodontitis | References |
|---|---|---|---|---|---|
| NMR Spectroscopy | Detects nuclear magnetic resonance signals (commonly 1H, 13C); non-destructive and quantitative. | - High reproducibility - Non-destructive - Absolute quantification | - Low sensitivity - Signal overlap - Costly instruments | Reliable for saliva metabolite fingerprinting and longitudinal studies. | [42,46,53,54] |
| GC-MS | Separates volatile/derivatized compounds; detects by MS. | - High sensitivity - Good for small molecules - Established protocol | - Requires derivatization - Limited to volatile analytes | Useful for SCFAs and volatile microbial metabolites. | [55,56,57] |
| LC-MS | Separates polar/non-polar metabolites in liquid phase; detects ions by MS. | - Broad coverage - High sensitivity - Versatile | - Ion suppression - Requires complex QC | Gold standard for untargeted saliva metabolomics. | [58,59] |
| FTIR Spectroscopy | Measures infrared absorption of molecular bonds; generates spectral fingerprints. | - Rapid, label-free - Minimal preparation | - Lower specificity - Mostly qualitative - Sensitive to water | Distinguishes healthy from diseased saliva via biochemical fingerprinting. | [16,60,61] |
| PAS (Photoacoustic Spectroscopy) | Measures acoustic waves generated after absorption of modulated light; used to assess biochemical changes. | - Non-invasive - High sensitivity to molecular vibrations - Can be miniaturized | - Lower metabolite specificity - Limited commercial systems - Sensitive to water | Emerging tool for real-time screening of periodontal changes in saliva. | [12,62,63] |
| Biomarkers | Molecular Categories | Biological Pathways | Functions and Significance | References |
|---|---|---|---|---|
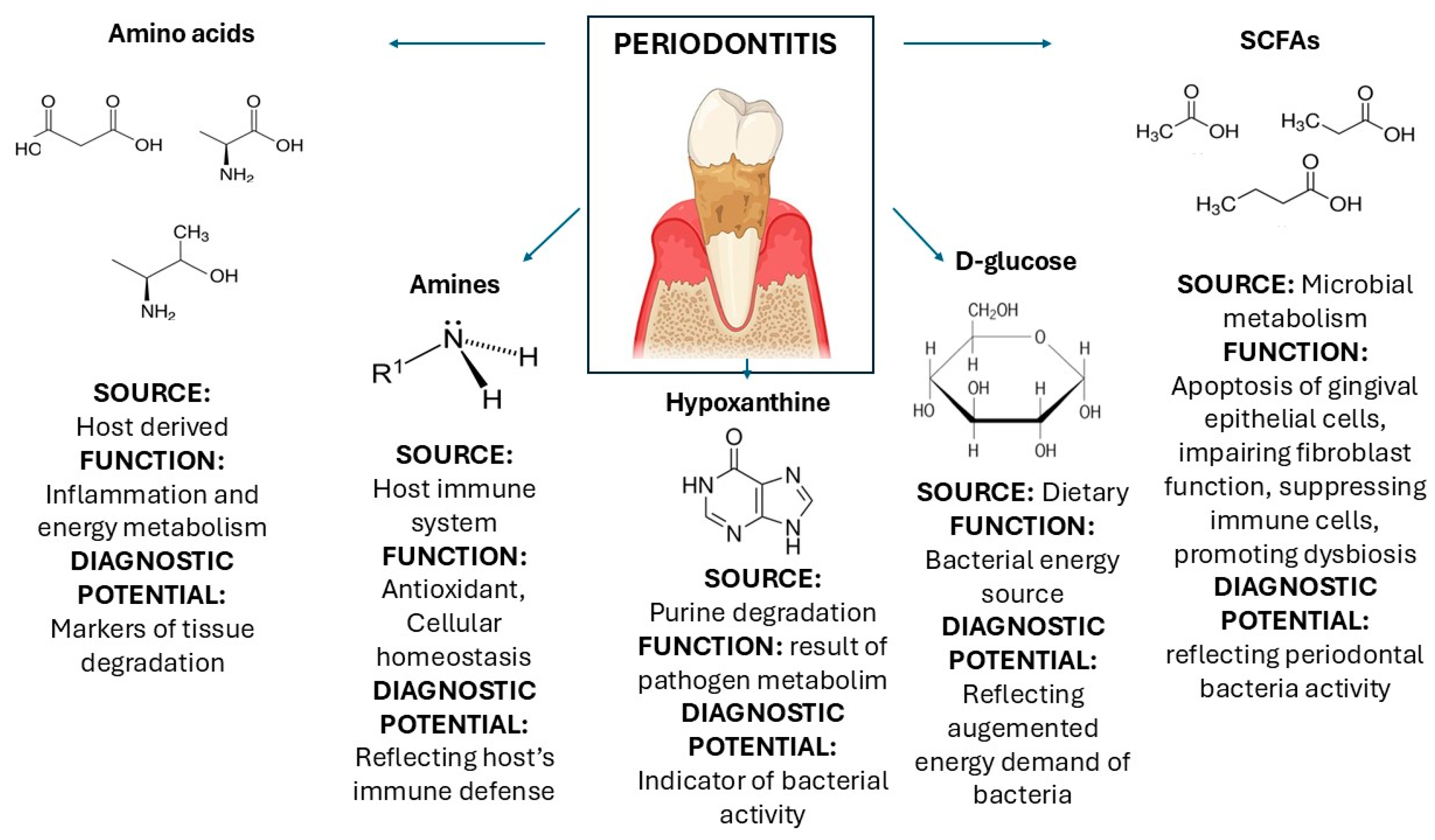
| Acetic, acetoacetic, butyric, citric, formic, isovalerate, formate, glycolic, lactic, propionic, and pyruvic acids. | SCFAs | They result from the fermentation of sugars and proteins from anaerobic bacteria (Porphyromonas gingivalis, Fusobacterium nucleatum, and Prevotella intermedia), and some of them, like formate, may result from purine breakdown under oxidative stress. | Inducing apoptosis of gingival epithelial cells, impairing fibroblast function, suppressing immune cells, promoting dysbiosis and neutrophil recruitment, and decreasing pH. | [64,66,67,68,69,70,71] |
| L-alanine, d-glutamic acid, l-valine, l-methionine, l-threonine, l-leucine, isoleucine, l-tyrosine, and l-phenylalanine. | Amino acids | They are energy sources for anaerobic bacteria (Porphyromonas gingivalis, Fusobacterium nucleatum, and Prevotella intermedia, and they result from the inflammation and tissue damage process. | Markers of tissue degradation, elevated in inflammation, reflecting the host’s immune response involved in energy metabolism, and bacteria. | [64,72,73] |
| Taurine, ethanolamine, dimethylamine, and methylamine. | Amines | They are released by the host immune system. | Antioxidant effect, in particular, Taurine. Maintaining cellular homeostasis under inflammatory conditions. | [64,74,75,76] |
| D-glucose | Sugar | They are an energetic source for periodontal bacteria. | Augmented energy demand of bacteria. | [64,77] |
| Hypoxanthine | Purine nucleobase | It is a byproduct of purine degradation mediated by bacteria. | Its presence indicates periodontal pathogens’ activity. | [78] |
| References | Study Design | Sample Size | Method of Analysis | Variation in Metabolomic Profile |
|---|---|---|---|---|
| Aimetti et al. [79] | Cross-sectional study | 32 cases, 22 controls | Nuclear Magnetic Resonance | Higher concentrations of acetate, γ-aminobutyrate, n-butyrate, succinate, trimethylamine, propionate, phenylalanine, and valine, and decreased concentrations of pyruvate and N-acetyl in GCP patients compared with controls. |
| Rzeznic et al. [42] | Cross-sectional study | 26 cases, 25 controls | Nuclear Magnetic Resonance | Higher concentrations of short-chain fatty acids and lower concentrations of lactate, γ-amino-butyrate, methanol, and threonine in periodontitis. |
| Romano et al. [80] | Cross-sectional study | 33 cases with GCP, 28 cases with GAgP; 39 controls. | Nuclear Magnetic Resonance | Higher concentrations of pyruvate, N-acetyl groups, and lactate, and higher levels of proline, phenylalanine, and tyrosine in GCP and GAgP patients compared with controls. |
| Kim et al. [74] | Cross-sectional study | 129 cases, 92 controls. | Nuclear Magnetic Resonance | Higher concentrations of taurine, isovalerate, butyrate, and glucose in periodontitis. |
| García-Villaescusa et al. [39] | Case-control study | 91 cases, 39 controls. | Nuclear Magnetic Resonance | Higher concentrations of caproate, isocaproate, butyrate, isovalerate, isopropanol, methanol, 4-aminobutyrate, choline, sucrose, sucrose-glucose-lysine, lactate-proline, lactate, and proline in periodontitis. |
| Biomarker | Molecular Category | Biological Pathway | Function and Significance | Reference |
|---|---|---|---|---|
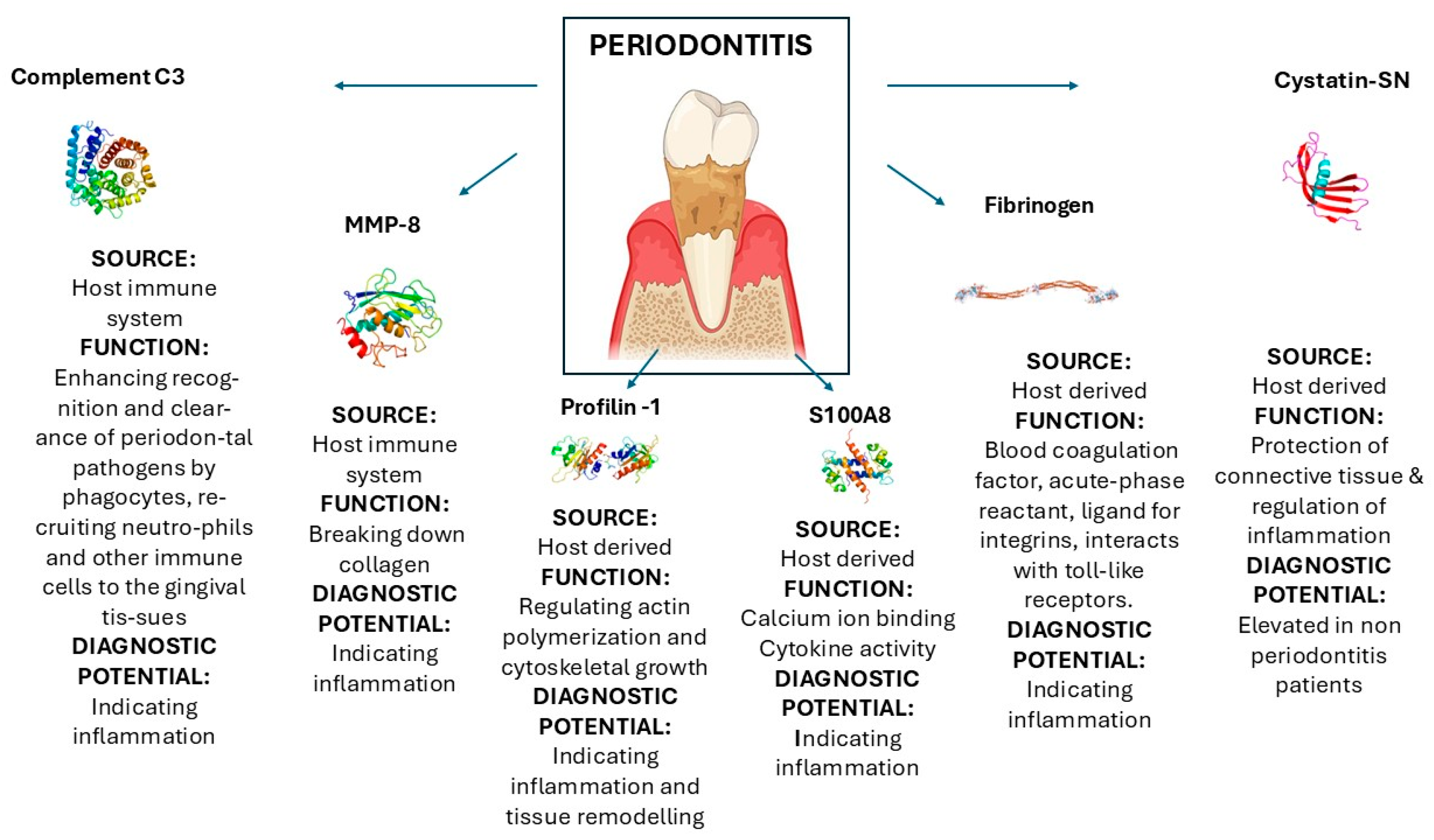
| Matrix metalloproteinase-8 | Enzyme | It is released by neutrophils and fibroblasts | Breaking down collagen | [95] |
| Complement C3 | Protein part of the complement system | It is triggered by immune complexes | Enhancing recognition and clearance of periodontal pathogens by phagocytes, recruiting neutrophils and other immune cells to the gingival tissues, leading to chronic inflammation | [96,97] |
| Profilin-1 | Actin-binding protein | Rho GTPase signaling → Profilin-1 → Actin polymerization → Cell movement and adhesion. | Regulating actin polymerization and cytoskeletal growth | [98,99,100] |
| S100A8 | Pro-inflammatory mediator | S100A8/A9 → TLR4/RAGE → NF-κB → Inflammation | Calcium ion binding Cytokine activity (in inflammatory states) | [18,101,102,103] |
| Fibrinogen | Glycoprotein | Fibrinogen → Thrombin cleavage → Fibrin → Clot formation & tissue scaffolding | Blood coagulation factor, acute-phase reactant, ligand for integrins (e.g., Mac-1, αIIbβ3), interacts with toll-like receptors (e.g., TLR4). | [96,104,105] |
| Cystatin-SN | Cysteine protease inhibitor | Cystatin-SN → Inhibition of cathepsins/proteases → Protection of connective tissue & regulation of inflammation | Cysteine-type endopeptidase inhibitor activity, protease binding | [106,107,108] |
| References | Study Design | Sample Size | Method of Analysis | Variation in the Proteomic Profile |
|---|---|---|---|---|
| Shin et al. [18] | Cross-sectional study | 36 cases, 36 controls | Mass spectrometry-based untargeted proteomics | S100A8 and S100A9 higher in periodontitis patients. |
| Belstrøm et al. [87] | Cross-sectional study | 10 cases, 10 controls | Mass spectrometry-based untargeted proteomics | Proteins associated with innate immune response were higher in Periodontitis patients |
| Bostanci et al. [107] | Cross-sectional study | 17 CP, 17 AgP; 16 controls | Mass spectrometry-based untargeted proteomics | Lactoferrin, lacritin, sCD14, Mucin 5B, and Mucin 7 down-regulated in AP and SLC4A1 was upregulated. Cystatin SN higher in controls |
| Mertens et al. [109] | Cross-sectional study | 10 CP, 11 AgP; 12 controls. | Mass spectrometry-based untargeted proteomics. | Hemopexin, plasminogen, and α-fibrinogen were higher in periodontitis patients. |
| Tang et al. [110] | Cross-sectional study | 16 cases, 17 controls. | Mass spectrometry-based untargeted proteomics. | Two peptide peaks had a lower level of intensity in the CP group, while the rest of the differentially expressed peptides had a higher level of intensity. |
| Antezack et al. [111] | Cross-sectional study | 67 cases, 74 controls | Mass spectrometry-based untargeted proteomics | No identification of specific proteins |
| Hartenbach et al. [112] | Cross-sectional study | 30 cases, 10 controls. | Mass spectrometry-based untargeted proteomics. | Salivary acidic proline-rich phosphoprotein, submaxillary gland androgen-regulated protein, histatin-1, fatty acid binding protein, thioredoxin, and cystatin-SA higher in periodontitis patients. |
| Grant et al. [103] | Cross-sectional study | 10 MMP, 10 AP; 10 controls. | Mass spectrometry-based untargeted proteomics. | MMP9, S100A8, A1AGP, and pyruvate kinase higher in periodontitis patients. |
| Casarin et al. [113] | Cross-sectional study | 12 cases, 13 controls | Mass spectrometry-based untargeted proteomics | GHG1, CSTB, KRT9, SMR3B, IGHG4, and SERPINA1 were higher in periodontitis patients. |
| Romano et al. [106] | Cross-sectional study | 15 UP, 15 TP; 15 controls. | Mass spectrometry-based untargeted proteomics. | Cystatin SN higher in healthy patients and in patients under periodontal active treatment. |
| Gonçalves et al. [114] | Cross-sectional study | 10 cases, 10 controls | Mass spectrometry-based untargeted proteomics | Periodontitis patients had higher levels of albumin, hemoglobin, and immunoglobulin, and they had a lower abundance of cystatin compared to the control group. |
| Salazar et al. [96] | Cross-sectional study | 20 cases, 20 controls. | Mass spectrometry-based untargeted proteomics | alpha-2-macroglobulin, ceruloplasmin, complement C3, alpha-2-HS-glycoprotein, fibrinogen alpha chain higher in periodontitis patients- |
| Chaiyarit et al. [115] | Cross-sectional study | 30 cases, 30 controls. | Mass spectrometry-based untargeted proteomics | No identification of specific proteins |
| Targeted Analysis | Untargeted Analysis |
|---|---|
| Hypothesis-driven | Hypothesis-generating |
| Subset analysis | Global/Comprehensive analysis |
| Correlated to reference standards | Correlated to the database/libraries |
| Identification already know | Qualitative identification |
| Absolute quantification | Relative quantification |
| Feature | NMR Spectroscopy | Mass Spectrometry (MS) | References |
|---|---|---|---|
| Sensitivity | Low (µM range), although improving with hyperpolarization techniques. | Very high (nM–pM range), especially with HR-MS. | [125,128,129,130,131] |
| Quantification | Highly accurate, can quantify multiple analytes. | Quantification is relative unless internal standards are used | [125,128,129,130,131] |
| Reproducibility | High, robust across time and between labs. | Moderate; targeted methods are reproducible, and untargeted methods need strict quality control. | [125,132,133] |
| Data Complexity | Moderate; relatively easier to interpret. | High; requires extensive data processing and normalization. | [125] |
| Throughput | Moderate (10–20 samples/day); limited by long acquisition times in 2D. | High (50–200+ samples/day), depending on the platform. | [125] |
| Method Development | Stable protocols; recent improvements in 2D NMR and hyperpolarization. | Rapidly evolving, flexible with multiple ionization and chromatographic modes. | [125] |
| Limitations | Low sensitivity–Long acquisition times for complex spectra. | - Ion suppression - Requires strict QC - Less robust than NMR | [125,128,129,130,131] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amato, M.; Polizzi, A.; Blasi, A.; Grippaudo, C.; Isola, G. Untargeted Salivary Metabolomics and Proteomics: Paving the Way for Early Detection of Periodontitis. Appl. Sci. 2025, 15, 6642. https://doi.org/10.3390/app15126642
Amato M, Polizzi A, Blasi A, Grippaudo C, Isola G. Untargeted Salivary Metabolomics and Proteomics: Paving the Way for Early Detection of Periodontitis. Applied Sciences. 2025; 15(12):6642. https://doi.org/10.3390/app15126642
Chicago/Turabian StyleAmato, Mariacristina, Alessandro Polizzi, Andrea Blasi, Cristina Grippaudo, and Gaetano Isola. 2025. "Untargeted Salivary Metabolomics and Proteomics: Paving the Way for Early Detection of Periodontitis" Applied Sciences 15, no. 12: 6642. https://doi.org/10.3390/app15126642
APA StyleAmato, M., Polizzi, A., Blasi, A., Grippaudo, C., & Isola, G. (2025). Untargeted Salivary Metabolomics and Proteomics: Paving the Way for Early Detection of Periodontitis. Applied Sciences, 15(12), 6642. https://doi.org/10.3390/app15126642








