A Novel Tool for Biodiversity Studies: Earthworm Classification via NGS and Neural Networks
Abstract
1. Introduction
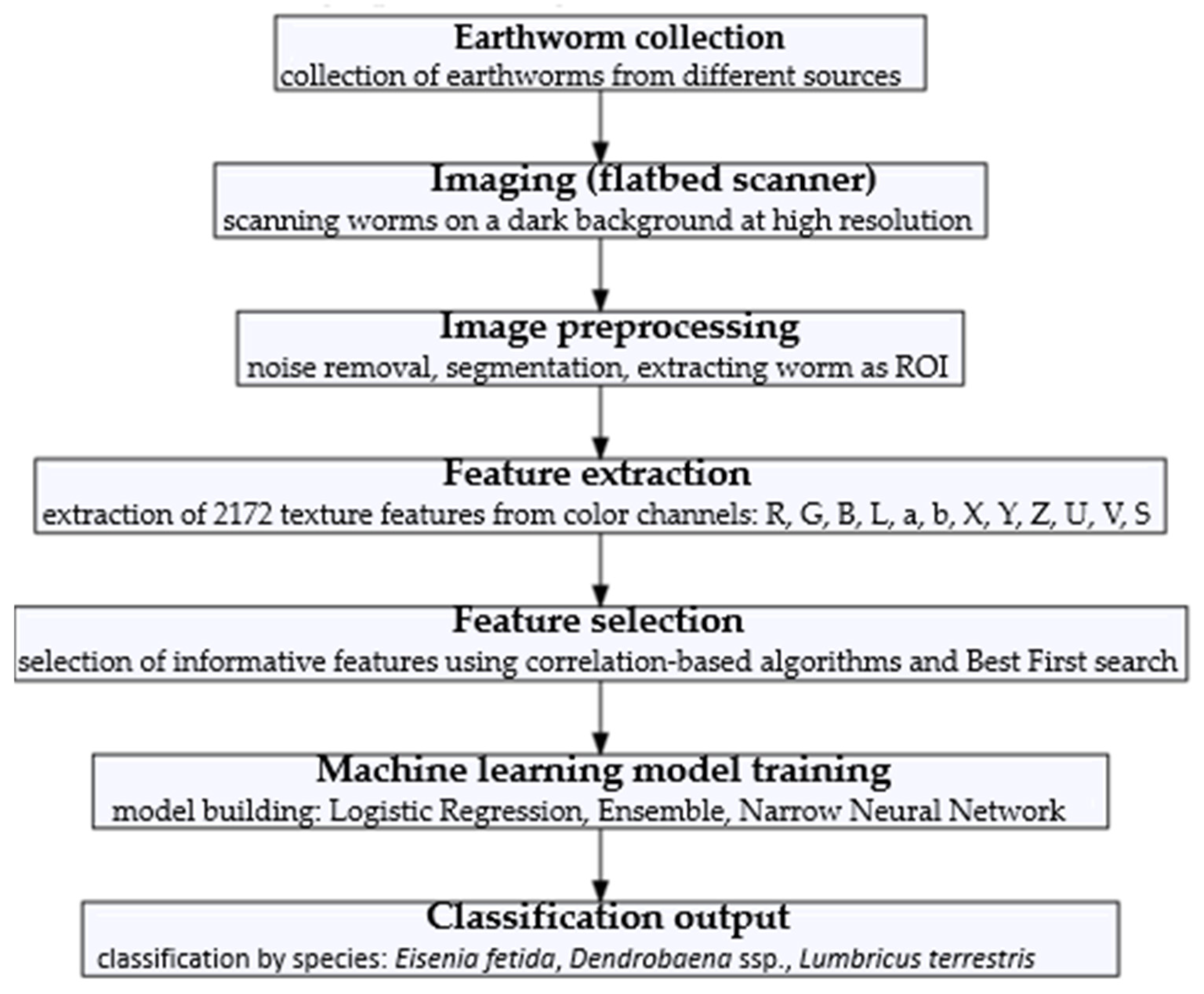
2. Materials and Methods
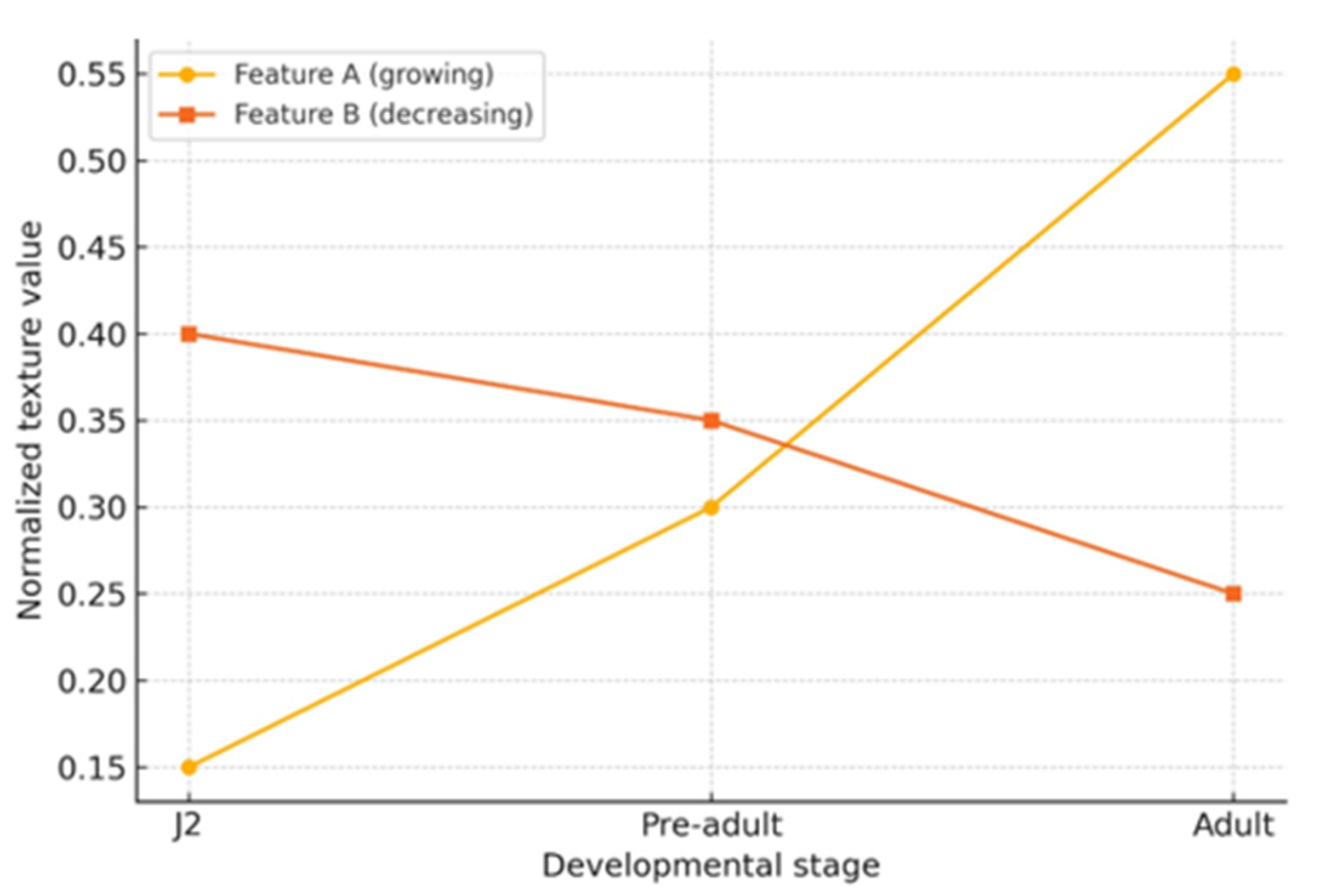
2.1. Image Analysis
2.2. Earthworm Classification
- -
- Naive Bayes—debug: False; batchSize: 100; doNotCheckCapabilities: False; useKernelEstimator: False; useSupervisedDiscretization: False;
- -
- Logistic—debug: False; batchSize: 100; doNotCheckCapabilities: False; maxIts: −1; ridge: 1.0 × 10−8; useConjugateGradientDescent: False.
- -
- Simple Logistic—debug: False; batchSize: 100; doNotCheckCapabilities: False; heuristicStop: 50; maxBoostingIterations: 500; useCrossValidation: True;
- -
- KStar—debug: False; batchSize: 100; doNotCheckCapabilities: False; entropicAutoBlend: False; globalBlend: 20; missingMode: Average column entropy curves;
- -
- Random Forest—bagSizePercent: 100; batchSize: 100; debug: False; doNotCheckCapabilities: False; calcOutOfBag: False; breakTiesRandomly: False; numExecutionSlots: 1; numIterations: 100; seeds: 1; storeOutOfBagPredictions: False.
- -
- SVM—preset: linear SVM; box constraint level: 1; kernel scale: automatic; kernel function: linear; standardize data: yes; multiclass method: one-vs-one;
- -
- KNN—preset: fine; distance metric: Euclidean; number of neighbors: 1; standardize data: yes; distance weight: equal;
- -
- Ensemble—preset: Subspace Discriminant; Ensemble method: subspace; learner type: discriminant;
- -
- Neural Network—preset: Narrow Neural Network; number of fully connected layers: 1; first layer size: 10; iteration limit: 1000; activation: ReLU; standardize data: yes; regularization strength (Lambda): 0.
2.3. RNA Extraction
2.4. Direct RNA Libraries Preparation and Sequencing
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Solgi, A.; Najafi, A.; Page-Dumroese, D.S.; Zenner, E.K. Assessment of topsoil disturbance caused by different skidding machine types beyond the margins of the machine operating trail. Geoderma 2020, 367, 114238. [Google Scholar] [CrossRef]
- Drewry, J.J. Natural recovery of soil physical properties from treading damage of pastoral soils in New Zealand and Australia: A review. Agric. Ecosyst. Environ. 2006, 114, 159–169. [Google Scholar] [CrossRef]
- Kooch, Y.; Jalilvand, H. Earthworms as Ecosystem Engineers and the Most Important Detritivors in Forest Soils. Pak. J. Biol. Sci. 2008, 11, 819–825. [Google Scholar] [CrossRef]
- Jouquet, P.; Blanchart, E.; Capowiez, Y. Utilization of earthworms and termites for the restoration of ecosystem functioning. Appl. Soil Ecol. 2014, 73, 34–40. [Google Scholar] [CrossRef]
- Feledyn-Szewczyk, B.; Kleofas Berbeć, A.; Radzikowski, P. Rola dżdżownic w kształtowaniu jakości gleb oraz wpływ różnych zabiegów agrotechnicznych na ich występowanie. Stud. I Rap. IUNG-PIB 2017, 54, 57–71. [Google Scholar]
- Ducasse, V.; Darboux, F.; Auclerc, A.; Legout, A.; Ranger, J.; Capowiez, Y. Can Lumbricus terrestris be released in forest soils degraded by compaction? Preliminary results from laboratory and field experiments. Appl. Soil Ecol. 2021, 168, 104131. [Google Scholar] [CrossRef]
- Sinha, R.K.; Valani, D.; Chauhan, K.; Agarwal, S. Embarking on a second green revolution for sustainable agriculture by vermiculture biotechnology using earthworms: Reviving the dreams of Sir Charles Darwin. J. Agric. Biotechnol. Sustain. Dev. 2010, 2, 113–128. [Google Scholar]
- Pfiffner, L. Earthworms- architects of fertile soils. In Their Significance and Recommendations for Their Promotion in Agriculture. Technical Guide on Earthworms; Order no 1629, international edition; Research Institute of Organic Agriculture FIBL and TILMAN-ORG project Consortium: Frick, Switzerland, 2014; Available online: https://organic-farmknowledge.org/tool/30567 (accessed on 1 August 2024).
- Edwards, C.A. Earthworm Ecology, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar] [CrossRef]
- Przemieniecki, S.W.; Zapałowska, A.; Skwiercz, A.; Damszel, M.; Telesiński, A.; Sierota, Z.; Gorczyca, A. An evaluation of selected chemical, biochemical, and biological parameters of soil enriched with vermicompost. Environ. Sci. Pollut. Res. 2021, 28, 8117–8127. [Google Scholar] [CrossRef]
- Zapałowska, A.; Skwiercz, A.; Puchalski, C.; Malewski, T. Influence of Eisenia fetida on the Nematode Populations during Vermicomposting Process. Appl. Sci. 2024, 14, 1576. [Google Scholar] [CrossRef]
- Paoletti, M.G. The role of earthworms for assessment of sustainability and as bioindicators. Agric. Ecosyst. Environ. 1999, 74, 137–155. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, T.; Tian, G.; Zhang, L.; Bian, B. Pilot-scale vermicomposting of sewage sludge mixed with mature vermicompost using earthworm reactor of frame composite structure. Sci. Total Environ. 2021, 767, 144217. [Google Scholar] [CrossRef] [PubMed]
- Devi, C.; Khwairakpam, M. Bioconversion of Lantana camara by vermicomposting with two different earthworm species in monoculture. Bioresour. Technol. 2020, 296, 122308. [Google Scholar] [CrossRef]
- Domnguez, J.; Gmez-Brand, M. Vermicomposting Composting with earthworms to recycle organic wastes. In Management of Organic Waste; Intech Open: London, UK, 2012. [Google Scholar] [CrossRef]
- Mane, V.B.; Kanase, S.S.; Sawale, N.S.; Bandsode, A.K.; Suryawanshi, M.A. Application of earthworm Eisenia fetida in waste management in municipal solid waste. Environ. Dev. Sustain. 2024, 1–19. [Google Scholar] [CrossRef]
- Keniya, B.; Patel, H.; Patel, K.; Bhatt, S.; Patel, T. Vermistabilization of mango tree pruning waste with five earthworm species: A biochemical and heavy metal assessment. Heliyon 2023, 9, e19908. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khalid, H.; Ikhlaq, A.; Pervaiz, U.; Wie, Y.-M.; Lee, E.-J.; Lee, K.-H. Municipal Waste Degradation by Vermicomposting Using a Combination of Eisenia fetida and Lumbricus rubellus Species. Agronomy 2023, 13, 1370. [Google Scholar] [CrossRef]
- Sinha, R.K.; Bharambe, G.; Chaudhari, U. Sewage treatment by vermifiltration with synchronous treatment of sludge by earthworms: A low cost sustainable technology over conventional systems with potential for decentralization. Environmentalist 2008, 28, 409–420. [Google Scholar] [CrossRef]
- Gupta, R.; Garg, V.K. Stabilization of primary sewage sludge during vermicomposting. J. Hazard. Mater 2008, 162, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Khwairakpam, M.; Bhargava, R. Vermitechnology for sewage sludge recycling. J. Hazard. Mater 2009, 161, 948–954. [Google Scholar] [CrossRef]
- Loh, T.C.; Lee, Y.C.; Liang, J.B.; Tan, D. Vermicomposting of cattle and goat manures by Eisenia Foetida and their growth and reproduction preference. Bioresour. Technol. 2005, 96, 111–114. [Google Scholar] [CrossRef]
- Plaza, C.; Nogales, R.; Senesi, N.; Benitez, E.; Polo, A. Organic matter humification by vermicomposting of cattle manure alone and mixed with two-phase olive pomace. Bioresour. Technol. 2007, 9, 5085–5089. [Google Scholar]
- Ghosh, M.; Chattopadhyay, G.N.; Baral, K. Transformation of phosphorus during vermicomposting. Bioresour. Technol. 1999, 69, 149–154. [Google Scholar] [CrossRef]
- Garg, V.K.; Kaushik, P. Vermistabilization of textile mill sludge spiked with poultry droppings by an epigeic earthworm Eisenia foetida. Bioresour. Technol. 2005, 96, 1063–1071. [Google Scholar] [CrossRef]
- Suthar, S. Bioremediation of agriculture wastes through vermicomposting. Bioremed. J. 2009, 13, 1–8. [Google Scholar] [CrossRef]
- Pramanik, P. Changes in microbial properties and nutrient dynamics in bagasse and coir during vermicomposting: Quantification of fungal biomass through ergosterol estimation in vermicompost. Waste Manag. 2010, 30, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Verma, D.; Singh, B.L.; Kumar, U.; Shweta. Composting of sugarcane waste by-products through treatment with microorganisms and subsequent vermicomposting. Bioresour. Technol. 2010, 101, 6707–6711. [Google Scholar] [CrossRef] [PubMed]
- Sen, B.; Chandra, T.S. Chemolytic and solid-state spectroscopic evaluation of organic matter transformation during vermicomposting of sugar industry waste. Bioresour. Technol. 2006, 98, 1680–1683. [Google Scholar] [CrossRef]
- Sangwan, P.; Kaushik, C.P.; Garg, V.K. Vermiconversion of industrial sludge for recycling the nutrients. Bioresour. Technol. 2008, 99, 8699–8704. [Google Scholar] [CrossRef]
- Yadav, A.; Garg, V.K. Feasibility of nutrient recovery from industrial sludge by vermicomposting technology. J. Hazard. Mater. 2009, 168, 262–268. [Google Scholar] [CrossRef]
- Subramanian, S.; Sivarajan, M.; Saravanapriya, S. Chemical changes during vermicomposting of sago industry solid wastes. J. Hazard. Mater. 2010, 179, 318–322. [Google Scholar] [CrossRef]
- Yadav, K.D.; Tare, V.; Ahammed, M.M. Vermicomposting of source-separated human faeces for nutrient recycling. Waste Manag. 2010, 30, 50–56. [Google Scholar] [CrossRef]
- Jaenike, J. Eisenia foetida is two biological species. Megadrilogica 1982, 4, 6–7. [Google Scholar]
- Sherlock, E. Key to the Earthworms of the UK and Ireland, 2nd ed.; Field Studies Council: Amersham, UK, 2018; p. 96. ISBN 9781908819406. [Google Scholar]
- Plisko, J.D. Lumbricidae. Dżdżownice (Annelida: Oligochaeta). In Fauna Polski; PAN, Instytut Zoologiczny: Warsaw, Poland, 1973; Volume 1, p. 156. [Google Scholar]
- Poier, K.R.; Richter, J. Spatial distribution of earthworms and soil properties in an arable loess soil. Soil Biol. Biochem. 1992, 24, 1601–1608. [Google Scholar] [CrossRef]
- Albani, J.R.; Demuynck, S.; Grumiaux, F.; Lepretre, A. Fluorescence fingerprints of Eisenia fetida and Eisenia andrei. Photochem. Photobiol. 2003, 78, 599–602. [Google Scholar] [CrossRef]
- Römbke, J.; Aira, M.; Backeljau, T.; Breugelmans, K.; Domínguez, J.; Funke, E.; Graf, N.; Hajibabaei, M.; Pérez-Losada, M.; Porto, P.G.; et al. DNA barcoding of earthworms (Eisenia fetida/andrei complex) from 28 ecotoxicological test laboratories. Appl. Soil Ecol. 2016, 104, 3–11. [Google Scholar] [CrossRef]
- Katsiamides, A.; Stürzenbaum, S.R. Cryptic speciation and blurred species boundaries of the earthworm: A challenge for soil-based toxicological risk assessments. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 2021, 239, 108880. [Google Scholar] [CrossRef]
- Szczypiński, P.M.; Strzelecki, M.; Materka, A. Mazda-a software for texture analysis. In Proceedings of the International Symposium on Information Technology Convergence (ISITC 2007), Jeonju, Korea, 23–24 November 2007; pp. 245–249. [Google Scholar]
- Szczypiński, P.M.; Strzelecki, M.; Materka, A.; Klepaczko, A. MaZda—A software package for image texture analysis. Comput. Methods Programs Biomed. 2009, 94, 66–76. [Google Scholar] [CrossRef]
- Strzelecki, M.; Szczypiński, P.; Materka, A.; Klepaczko, A. A software tool for automatic classification and segmentation of 2D/3D medical images. Nucl. Instrum. Methods Phys. Res. Sect. A: Accel. Spectrometers Detect. Assoc. Equip. 2013, 702, 137–140. [Google Scholar] [CrossRef]
- Frank, E.; Hall, M.A.; Witten, I.H. The WEKA Workbench. Online Appendix for Data Mining: Practical Machine Learning Tools and Techniques, 4th ed.; The University of Waikato: Hamilton, New Zealand, 2016. [Google Scholar]
- Bouckaert, R.R.; Frank, E.; Hall, M.; Kirkby, R.; Reutemann, P.; Seewald, A.; Scuse, D. WEKA Manual for Version 3-9-1; University of Waikato: Hamilton, New Zealand, 2016. [Google Scholar]
- Witten, I.H.; Frank, E. Data Mining: Practical Machine Learning Tools and Techniques, 2nd ed.; Elsevier: San Francisco, CA, USA, 2005; p. 525. [Google Scholar]
- Ropelewska, E.; Szwejda-Grzybowska, J. A comparative analysis of the discrimination of pepper (Capsicum annuum L.) based on the cross-section and seed textures determined using image processing. J. Food Process Eng. 2021, 44, e13694. [Google Scholar] [CrossRef]
- Ropelewska, E. Distinguishing lacto-fermented and fresh carrot slice images using the Multilayer Perceptron neural network and other machine learning algorithms from the groups of Functions, Meta, Trees, Lazy, Bayes and Rules. Eur. Food Res. Technol. 2022, 248, 2421–2429. [Google Scholar] [CrossRef]
- Poursalavati, A.; Larafa, A.; Fall, M.L. dsRNA-based viromics: A novel tool unveiled hidden soil viral diversity and richness. bioRxiv 2023. [Google Scholar] [CrossRef]
- Lee, S.; Nguyen, L.T.; Hayes, B.J.; Ross, E.M. Prowler: A novel trimming algorithm for Oxford Nanopore sequence data. Bioinformatics 2021, 37, 3936–3937. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Clausen, P.T.L.C.; Buchmann, J.P.; Wille, M.; Iredell, J.R.; Meyer, W.; Lund, O.; Sorrell, T.C.; Holmes, E.C. CCMetagen: Comprehensive and accurate identification of eukaryotes and prokaryotes in metagenomic data. Genome Biol. 2021, 21, 1–15. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 103. [Google Scholar] [CrossRef]
- Vaupel, A.; Hommel, B.; Beule, L. High-resolution melting (HRM) curve analysis as a potential tool for the identification of earthworm species and haplotypes. PeerJ 2022, 10, e13661. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Y.; Xu, W.; Fang, Y.; Ruan, H. Characterization of Five New Earthworm Mitogenomes (Oligochaeta: Lumbricidae): Mitochondrial Phylogeny of Lumbricidae. Diversity 2021, 13, 580. [Google Scholar] [CrossRef]
- Kumar, P.; Raghupathi, M.; Bolan, N.S.; Miklavcic, S. Phenotyping Earthworm by Image Analysis. In Proceedings of the 13th International Conference on Control, Automation, Robotics & Vision (ICARCV 2014), Marina Bay Sands, Singapore, 10–12 December 2014; pp. 205–210. [Google Scholar]
- Andleeb, S.; Abbasi, W.A.; Mustafa, R.G.; Islam, G.; Naseer, A.; Shafique, I.; Parween, A.; Shaheen, B.; Shafiq, M.; Altaf, M.; et al. ESIDE: A computationally intelligent method to identify earthworm species (E. fetida) from digital images: Application in taxonomy. PLoS ONE 2021, 16, e0255674. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Kvist, S.; Lenihan, J.; Giribet, G.; Ziegler, A. Sine systemate chaos? A versatile tool for earthworm taxonomy: Non-destructive imaging of freshly fixed and museum specimens using micro-computed tomography. PLoS ONE 2014, 9, e96617. [Google Scholar] [CrossRef] [PubMed]
- Çelik, A.; Uğuz, S. A deep learning based system for real-time detection and sorting of earthworm cocoons. Turk. J. Electr. Eng. Comput. Sci. 2022, 30, 20. [Google Scholar] [CrossRef]
- Djerdj, T.; Hackenberger, D.K.; Hackenberger, D.K.; Hackenberger, B.K. Observing earthworm behavior using deep learning. Geoderma 2020, 358, 113977. [Google Scholar] [CrossRef]
- Fernández, L.; Castillero, C.; Aguilera, J.M. An application of image analysis to dehydration of apple discs. J. Food Eng. 2005, 67, 185–193. [Google Scholar] [CrossRef]
- Armi, L.; Fekri-Ershad, S. Texture image analysis and texture classification methods—A review. Int. Online J. Image Process. Pattern Recogn. 2019, 2, 1–29. [Google Scholar]

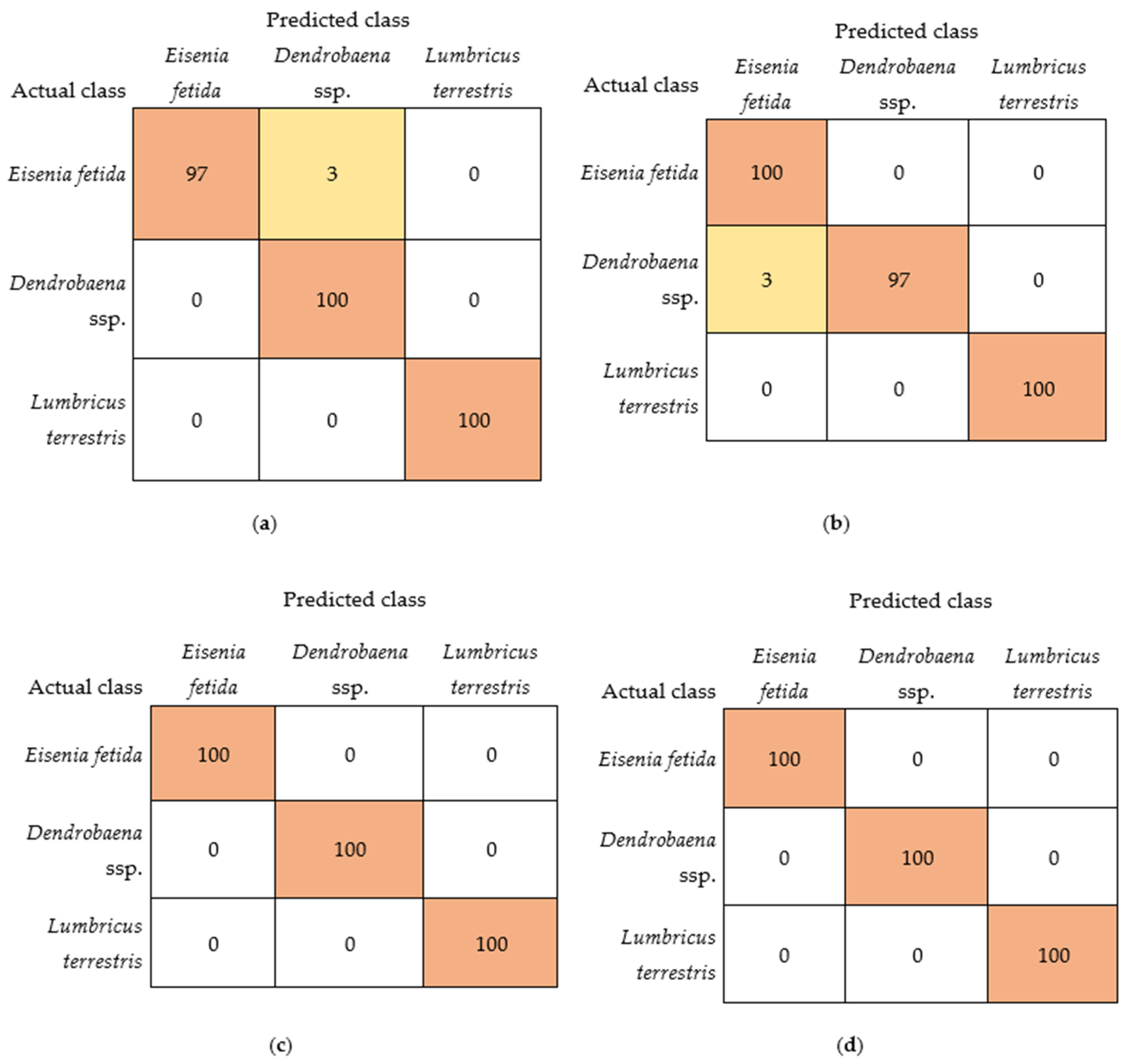
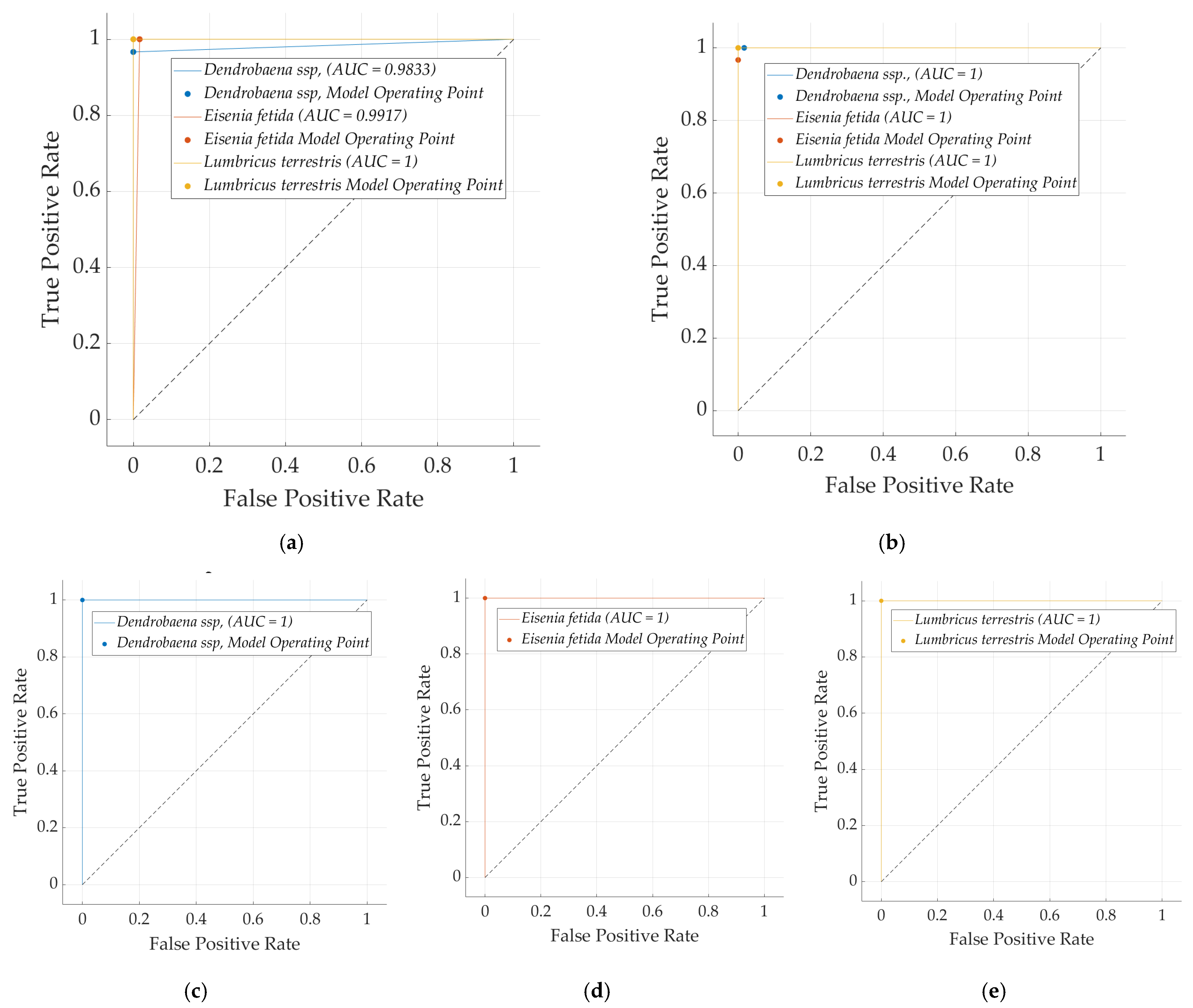
| Algorithm | Predicted Class (%) | Actual Class | Average Accuracy (%) | Kappa Statistic | ||
|---|---|---|---|---|---|---|
| Eisenia fetida | Dendrobaena ssp. | Lumbricus terrestris | ||||
| Naive Bayes (Bayes) | 97 | 3 | 0 | Eisenia fetida | 99 | 0.9833 |
| 0 | 100 | 0 | Dendrobaena ssp. | |||
| 0 | 0 | 100 | Lumbricus terrestris | |||
| Logistic (Functions) | 100 | 0 | 0 | Eisenia fetida | 100 | 1.0000 |
| 0 | 100 | 0 | Dendrobaena ssp. | |||
| 0 | 0 | 100 | Lumbricus terrestris | |||
| Simple Logistic (Functions) | 93 | 7 | 0 | Eisenia fetida | 97 | 0.9500 |
| 3 | 97 | 0 | Dendrobaena ssp. | |||
| 0 | 0 | 100 | Lumbricus terrestris | |||
| KStar (Lazy) | 97 | 3 | 0 | Eisenia fetida | 94 | 0.9167 |
| 7 | 93 | 0 | Dendrobaena ssp. | |||
| 0 | 7 | 93 | Lumbricus terrestris | |||
| Random Forest (Trees) | 100 | 0 | 0 | Eisenia fetida | 99 | 0.9833 |
| 3 | 97 | 0 | Dendrobaena ssp. | |||
| 0 | 0 | 100 | Lumbricus terrestris | |||
| Algorithm | Class | TP Rate | FP Rate | Precision | Recall | F-Measure | MCC | ROC Area | PRC Area |
|---|---|---|---|---|---|---|---|---|---|
| Naive Bayes (Bayes) | Eisenia fetida | 0.967 | 0.000 | 1.000 | 0.967 | 0.983 | 0.975 | 0.999 | 0.999 |
| Dendrobaena ssp. | 1.000 | 0.017 | 0.968 | 1.000 | 0.984 | 0.976 | 0.999 | 0.999 | |
| Lumbricus terrestris | 1.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Logistic (Functions) | Eisenia fetida | 1.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Dendrobaena ssp. | 1.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Lumbricus terrestris | 1.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Simple Logistic (Functions) | Eisenia fetida | 0.933 | 0.017 | 0.966 | 0.933 | 0.949 | 0.925 | 0.997 | 0.993 |
| Dendrobaena ssp. | 0.967 | 0.033 | 0.935 | 0.967 | 0.951 | 0.926 | 0.995 | 0.991 | |
| Lumbricus terrestris | 1.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| KStar (Lazy) | Eisenia fetida | 0.967 | 0.033 | 0.935 | 0.967 | 0.951 | 0.926 | 0.999 | 0.998 |
| Dendrobaena ssp. | 0.933 | 0.050 | 0.903 | 0.933 | 0.918 | 0.876 | 0.988 | 0.960 | |
| Lumbricus terrestris | 0.933 | 0.000 | 1.000 | 0.933 | 0.966 | 0.950 | 0.999 | 0.998 | |
| Random Forest (Trees) | Eisenia fetida | 1.000 | 0.017 | 0.968 | 1.000 | 0.984 | 0.976 | 1.000 | 1.000 |
| Dendrobaena ssp. | 0.967 | 0.000 | 1.000 | 0.967 | 0.983 | 0.975 | 0.999 | 0.999 | |
| Lumbricus terrestris | 1.000 | 0.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Template (Species; GenBank Accession Number; Gene) | Template Length | Number of Sequence Reads | Query Identity | p Value |
|---|---|---|---|---|
| Eisenia fetida; AY874481.1; 28S rDNA | 2031 | 110 | 94.23 | 1.0 × 10−26 |
| Eisenia fetida; AF212166.1; 28S rDNA | 3633 | 68 | 94.59 | 1.0 × 10−26 |
| Eisenia fetida; EF534709.1; 18S rDNA partial seq., ITS1, 5.8S, ITS2, and 28S partial seq. | 2972 | 88 | 95.09 | 1.0 × 10−26 |
| Hexaplex trunculus; AM920318; 5S rRNA and intergenic sequence | 258 | 7 | 89.29 | 1.0× 10−26 |
| Urechis unicinctus; X00998.1; 5S rRNA | 120 | 6 | 94.68 | 1.0 × 10−14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malewski, T.; Ropelewska, E.; Skwiercz, A.; Lutsiuk, A.; Zapałowska, A. A Novel Tool for Biodiversity Studies: Earthworm Classification via NGS and Neural Networks. Appl. Sci. 2025, 15, 6597. https://doi.org/10.3390/app15126597
Malewski T, Ropelewska E, Skwiercz A, Lutsiuk A, Zapałowska A. A Novel Tool for Biodiversity Studies: Earthworm Classification via NGS and Neural Networks. Applied Sciences. 2025; 15(12):6597. https://doi.org/10.3390/app15126597
Chicago/Turabian StyleMalewski, Tadeusz, Ewa Ropelewska, Andrzej Skwiercz, Anastasiia Lutsiuk, and Anita Zapałowska. 2025. "A Novel Tool for Biodiversity Studies: Earthworm Classification via NGS and Neural Networks" Applied Sciences 15, no. 12: 6597. https://doi.org/10.3390/app15126597
APA StyleMalewski, T., Ropelewska, E., Skwiercz, A., Lutsiuk, A., & Zapałowska, A. (2025). A Novel Tool for Biodiversity Studies: Earthworm Classification via NGS and Neural Networks. Applied Sciences, 15(12), 6597. https://doi.org/10.3390/app15126597










