1. Introduction
The medaka fish [
1] (
Oryzias latipes) is a well-established model organism in biomedical research, particularly valued for its substantial genetic homology with mammals. Its popularity stems from several advantageous traits, including low maintenance cost, a short developmental cycle, and transparent embryos, which facilitate in vivo experimental observations. These features have consolidated its role in diverse research domains, including genetics, toxicology, and disease modeling. In cardiovascular studies, medaka is especially useful due to its heart structure, which shares highly conserved anatomical features with the human heart, such as atrial and ventricular partitioning and valvular systems [
2].
Accurate segmentation of the medaka ventricle is essential for quantifying cardiac mechanical parameters such as heart rate and ejection fraction (EF). These indicators are widely used for evaluating cardiac function, diagnosing heart failure, and assessing treatment responses. EF, in particular, is a well-established predictor of mortality and major adverse cardiac events. In preclinical models such as medaka and zebrafish, EF serves as a critical quantitative measure in both developmental biology and pharmacological screening. Therefore, high-throughput and reproducible segmentation of the ventricle is vital for supporting large-scale analysis and drug discovery.
To address this need, Naderi et al. [
3] explored traditional image processing techniques such as edge detection, background subtraction, color filtering, and histogram-based segmentation to identify ventricular contours in zebrafish videos. However, these approaches lack robustness when applied to medaka data, especially under conditions of poor image quality or indistinct ventricular boundaries.
In recent years, deep learning methods have shown tremendous promise in biomedical image segmentation. For instance, Akerberg et al. [
4] developed the CFIN framework based on SegNet, which automatically segments cardiac regions and estimates functional parameters such as ventricular volume using high-resolution fluorescent microscopy. The U-Net [
5] architecture has also demonstrated excellent segmentation performance. Schutera et al. [
6] employed U-Net in the HeartSeg framework for ventricular segmentation and size quantification in medaka. Similarly, Naderi et al. [
3] introduced the ZACAF (Zebrafish Automatic Cardiovascular Assessment Framework) framework, which applies U-Net to grayscale videos acquired through standard microscopy for automatic segmentation and evaluation of EF and fractional shortening (FS) in zebrafish embryos. Nevertheless, these models do not explicitly incorporate temporal dependencies between frames, resulting in segmentation inconsistencies during dynamic cardiac cycles. Moreover, they typically rely on high-quality data captured under controlled laboratory conditions, limiting their generalizability to diverse imaging environments.
Zeng et al. [
7] were the first to apply video segmentation techniques [
8] to medaka ventricular analysis. Their STAVOS (Spatiotemporal Attention-based Ventricular Object Segmentation) model introduced a frame-matching mechanism to establish temporal associations across frames, significantly improving segmentation stability during phases of intense ventricular deformation. They also released the first publicly available pixel-level annotated video dataset of medaka embryonic hearts, providing a valuable benchmark for subsequent research.
However, the dataset was recorded using the same imaging device under identical conditions, resulting in high visual similarity across videos. This led to overfitting during training and limited generalization to videos with varying illumination, equipment, or ventricular scale. Furthermore, as a matching-based approach, STAVOS involves a complex network with a large parameter set, resulting in slow inference speed, which restricts its applicability in real-time scenarios.
To overcome these limitations, we propose LAAVOS (Local Attention Asymptotic Video Object Segmentation), a video segmentation model designed to enhance accuracy, robustness, and efficiency across diverse imaging settings. First, we constructed a more complex medaka ventricular video dataset that includes 19 sequences recorded under varied conditions of color, brightness, and equipment, totaling over 4200 pixel-level annotated frames. Second, we introduce a DeAOT-based architecture that features two key innovations: (1) an attention-enhanced encoder, LA-ResNet, which incorporates local attention into ResNet bottlenecks to emphasize ventricular features and suppress artifacts such as blur and occlusion; and (2) a progressive multi-scale decoder, AFPViS, which constructs hierarchical feature fusion paths and a gradual decoding mechanism to resolve misalignments in semantic detail across spatial scales. Benefiting from the memory-efficient propagation design of DeAOT, LAAVOS not only achieves improved segmentation accuracy but also significantly enhances inference speed compared to matching-based models like STAVOS, making it more suitable for real-time and large-scale screening applications.
The main contributions of this paper are as follows: (1) We created a diverse, pixel-level video segmentation dataset for medaka ventricular segmentation. It comprises 19 videos captured under various imaging environments, containing over 4200 precisely annotated frames. (2) We proposed LA-ResNet, a novel attention-enhanced encoder that integrates local attention into the ResNet bottleneck structure, enabling the model to focus more effectively on ventricular features while resisting image degradation and organ occlusion. (3) We designed AFPViS, a new progressive multi-scale decoder. By introducing hierarchical multi-scale feature fusion and constructing cross-layer feature interaction pathways, this decoder addresses the limitations of conventional FPN architectures in aligning semantic detail, thereby enhancing segmentation performance across ventricular scales.
2. The LAAVOS Model
We name the proposed semi-automatic video segmentation model LAAVOS, which comprises three major components: an LA-ResNet encoder, a feature propagation module incorporating Transformer [
9], and an APFViS decoder.
As illustrated in
Figure 1, specifically, the current frame (frame t), the preceding frame (frame t − 1), and a reference frame (frame 1) are sequentially fed into the LA-ResNet encoder to extract multi-scale visual features enhanced by local attention mechanisms.
These attention-refined features are then passed into the intermediate feature propagation module, which consists of two branches: a visual branch and an ID branch. The visual branch is responsible for computing cross-frame attention maps and temporally propagating visual features across frames. In contrast, the ID branch reuses the attention maps generated by the visual branch to propagate object identity embeddings, enabling consistent identity tracking throughout the sequence.
Finally, the enhanced features from both branches are fused and fed into the APFViS decoder to produce fine-grained segmentation masks for each frame.
This architectural design effectively enhances segmentation robustness and enables accurate extraction of ventricular regions in medaka larval videos, maintaining stable performance even under adverse conditions such as motion blur, occlusion, and color distortion.
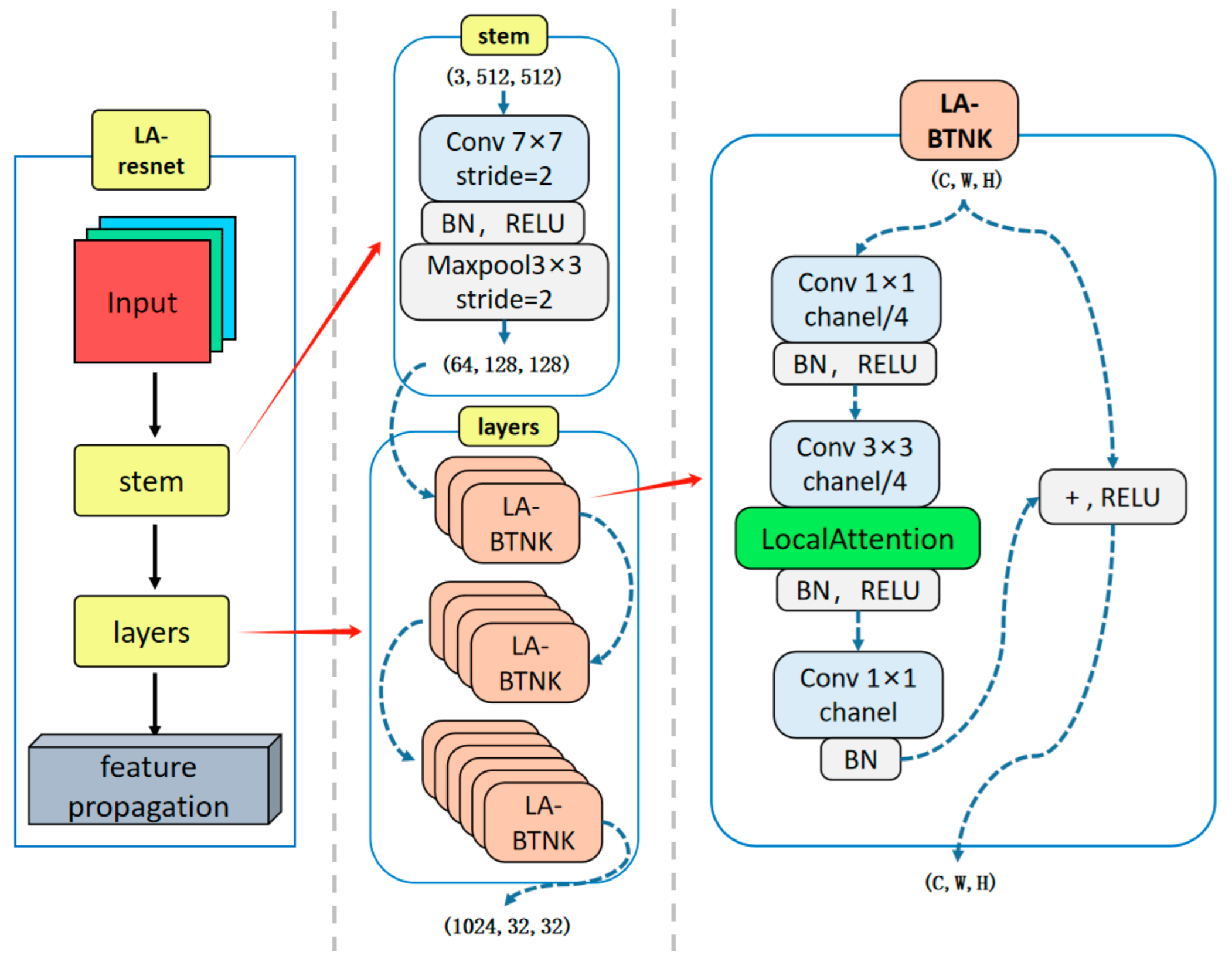
2.1. The LA-ResNet Encoder
In the task of medaka heart video segmentation, segmentation models are prone to being influenced by various factors. For instance, the lighting and background vary across different videos. Additionally, the color changes as blood flows through blood vessels, atria, and ventricles. Moreover, organs and tissues similar to the ventricle, as well as overlapping organs in some video frames, further complicate the task. The close proximity of the ventricle and atrium in medaka leads to unclear boundaries. Additionally, some video frames suffer from blurring due to imaging issues or the fish’s movement. These factors significantly interfere with the model’s feature extraction and interpretation.
To address these challenges, the model requires stronger feature extraction capabilities. Therefore, we embedded an attention-enhanced encoder into the model’s architecture. This enables the network to prioritize and concentrate on key target regions during feature extraction while reducing the impact of various types of noise.
This attention-enhanced encoder incorporates the local importance attention mechanism into the bottleneck module of ResNet-50 [
10]. We refer to the bottleneck module embedded with the LA attention mechanism [
11] as LA-BTNK and the improved encoder as LA-ResNet.
As illustrated in
Figure 2, the encoding process begins with a stem module consisting of a 7 × 7 convolution with stride 2, followed by batch normalization, ReLU activation, and a 3 × 3 max pooling layer, effectively reducing the spatial resolution while retaining essential low-level features. Following this, the layers module comprises a sequence of Local Attention Bottleneck Blocks (LA-BTNKs), each of which contains a dimensionality-reducing 1 × 1 convolution, a 3 × 3 convolution applied to the reduced channels, and a final 1 × 1 convolution to restore the original channel dimensions. A Local Attention module is embedded within each LA-BTNK to generate spatial importance maps by emphasizing task-relevant regions and suppressing noise and background clutter. The attention-enhanced features are then reintegrated into the residual path via a skip connection and non-linear activation. Through this hierarchical structure, the LA-ResNet encoder effectively captures semantic representations, which serve as robust feature maps for subsequent feature propagation and decoding stages.
The Local Importance Attention (LA) Module
The core of LA attention mechanisms is to adaptively enhance useful information while suppressing irrelevant information based on the importance of input correlations. In existing research, importance maps are calculated using subnetworks or matrix multiplications. Examples include ESA (first order: element-wise multiplication) [
12] and self-attention (second order: two matrix multiplications) [
13]. While ESA suffers from weak performance, self-attention has high spatial complexity.
Other approaches, such as the LA-layer [
14], adopt deformable local attention with learnable offsets and Q–K–V-based aggregation, aiming to enhance flexibility and receptive field adaptation. However, this design introduces additional computational and memory overheads, along with parameter tuning, which still impacts real-time performance.
In contrast, our LA attention mechanism adopts a simplified second-order spatial attention strategy that achieves a better balance between efficiency and segmentation accuracy.
The Local Attention module adopts a three-branch architecture that integrates the main feature path, the attention generation path, and the guided gating path. This design enhances the model’s ability to focus on informative regions while suppressing irrelevant background responses. As illustrated in
Figure 3, the left part of the diagram presents the overall structure composed of three key computational branches, while the right part highlights the SoftPool operation used in the attention generation path.
Specifically, the main feature path directly forwards the original input feature map without any transformation, aiming to preserve the complete semantic and contextual information as the foundation for subsequent attention modulation.
The attention generation path obtains local importance through regional softmax. It calculates the importance value of the pixel
within its surrounding region
, as shown below:
where
represents the local importance at coordinates
.
denotes the neighborhood centered at
, and
represents learnable weights that can be used to refine the computed importance values.
We instantiate the formula by stacking SoftPool [
15] and 3 × 3 convolutions to improve efficiency and applicability. Similar to first-order attention mechanisms like ESA, we utilize stride and squeeze convolutions to reduce computation and expand the receptive field. Additionally, we employ sigmoid and bilinear interpolation as activation and resolution recovery functions to maintain feature map consistency.
To address the distortion of local importance caused by down-sampling and to avoid artifacts introduced by strided convolutions and bilinear interpolation, we employ a gating mechanism [
16,
17] to optimize the local importance
. Unlike traditional gating mechanisms that use additional networks, we select the first channel
of the input map as the gate to avoid introducing extra parameters.
Finally, the outputs of the three paths are integrated through element-wise multiplication. We define the complete local information enhancement process as follows:
where
represents the sigmoid activation function, while
signifies the resolution recovery function implemented through bilinear interpolation.
2.2. The AFPViS Decoder
2.2.1. Progressive Multi-Scale Feature Fusion
In medaka videos, the size of the ventricular target exhibits significant dynamic variation. Specifically, differences in the shooting environments of different videos lead to notable diversity in the scale of the ventricular target. Within the same video, the size of the ventricular target changes regularly due to the periodic beating of the medaka heart. This multi-scale dynamic characteristic introduces additional challenges for the video segmentation task, demanding that the model possess stronger scale adaptability and temporal modeling capabilities.
In applications such as object localization, semantic segmentation, and video object segmentation, feature pyramid structures are commonly employed to address scale variations. Among these, FPN [
18] is the most widely used feature pyramid structure. FPN employs a top-down pathway to propagate semantic-rich features from higher levels to lower levels, facilitating multi-level feature integration. However, this process often fails to achieve true fusion between high-level and low-level features, leading to the degradation and loss of semantic information. This ultimately results in blurred edges in the segmentation of the medaka ventricle, sometimes causing the model to fail to distinguish between the ventricle and adjacent tissues.
To resolve this limitation, the proposed approach introduces a progressive feature pyramid network (AFPViS) decoder in the decoder part of the LAAVOS network. Inspired by AFPN [
19], the AFPViS structure introduces a denser feature fusion mechanism. As illustrated in
Figure 4, the feature propagation layer of LAAVOS generates a collection of multi-scale features, encompassing low-level, mid-level, and high-level representations, denoted as {F1, F2, F3}. These features are then progressively integrated by AFPViS, which first fuses the low-level and mid-level features before incorporating the high-level features. Through this fusion mechanism, the semantic disparity across multi-level features is progressively minimized, preventing the degradation and loss of semantic information.
After the feature fusion step, the enhanced multi-scale features {P3, P4, P5} are progressively integrated through a three-level feature refinement process. First, the high-level feature P5 is compressed in channels via a 1 × 1 convolution and added to the global semantic feature. The semantic information is then refined through a 3 × 3 convolution. Next, bilinear interpolation is used to up-sample to the resolution of P4, aligning channels and performing spatial-weighted fusion with the mid-level features from the adapter to restore detailed features. Finally, another up-sampling to the resolution of P4 is performed, and the result is added to the low-level high-resolution features for further refinement. A 3 × 3 convolution improves the local detail representation, and a 1 × 1 convolution outputs the pixel-wise classification prediction map.
2.2.2. The Adaptive Spatial Feature Fusion (ASFF) Module
Compared to other feature pyramid network encoders, AFPViS requires more feature fusion, highlighting the need to prioritize key levels and mitigate inconsistencies caused by multi-level features. We introduced the Adaptive Spatial Feature Fusion (ASFF) module [
20], which dynamically allocates spatial weights based on the hierarchical level of features.
Figure 5 shows the process of hierarchical feature fusion by AFPViS through the ASFF module.
Specifically, for the three-level fusion process, let
denote the feature vector at position
from level
to level
. The result obtained through adaptive spatial feature fusion is represented by
, which is a linear combination of
,
, and
, as shown in the formula below:
where
,
and
denote the spatial weights assigned to the three-level features within a specific level
, constrained by
+
+
= 1. Due to variations in the quantity of fused features across each stage of AFPViS, the ASFF module adaptively operates based on the number of input feature vectors in a specific stage.
3. Experimental Data and Processing
This section outlines the experimental design and evaluation framework adopted in this study. First, the medaka heart video dataset, utilized for both training and testing, is described. It covers its sources, preprocessing techniques, and data augmentation strategies. Next, the experimental framework and optimization algorithms are detailed. The hardware and software configurations utilized in the experiments are then introduced.
Finally, the performance metrics, including mIoU, Jaccard index, and F-measure, are described to enable a thorough evaluation of the model’s segmentation outcomes.
3.1. Experimental Dataset
Currently, the field of Medaka research is confronted with a paucity of datasets for ventricular segmentation. At present, the field of Medaka research lacks datasets that can be adapted to practical research (existing datasets are overly simplistic). To address this challenge, we have undertaken the creation of a novel dataset tailored for the task of video object segmentation of the Medaka ventricle. To enhance the robustness of the Medaka ventricular segmentation model to adapt to videos captured in different environments in practical scenarios, we have collected video data from several different environments from two distinct sources.
One dataset originates from the Biological Research Center at Heidelberg University in Germany. The video data is freely accessible and can be obtained from the Open Science Framework (OSF), a platform that underpins scientific research across disciplines such as social sciences, medicine, and engineering. The dataset is available for download at
https://osf.io/6svkf (accessed on 20 March 2025). Each video spans approximately 11 to 13 s, recorded at a frame rate of 15 frames per second (fps) with a resolution of 640 × 480. Some videos exhibit suboptimal quality, with issues such as blurring or shadow occlusion in certain frames. Consequently, we have selected nine high-quality videos from them.
The other dataset is sourced from the College of Fisheries at Shanghai Ocean University. Each video in this collection lasts between 8 to 10 s, recorded at a frame rate of 30 fps and a resolution of 1216 × 1028.
All video frames are captured from the ventral perspective of the Medaka, and
Figure 6 shows several representative video frames from different environments.
The process of creating our dataset is as follows: First, we extract frames from the medaka heartbeat videos, with each video containing a varying number of frames, ranging approximately from 165 to 280 frames. Next, we use LabelMe (version 5.5.0), a pixel-level image annotation tool developed by MIT, to annotate the medaka frames frame by frame, generating JSON annotation files that contain the coordinates of the medaka ventricular edges and corresponding PNG images of the ventricular masks. Finally, we organize the dataset according to the folder structure requirements of the DAVIS dataset. The training set includes 13 videos, comprising 2746 video frames; the validation set includes 3 videos, with 716 video frames; and the test set includes 3 videos, containing 763 video frames.
3.2. Experimental Environment
This study implemented the DeAOT model based on the PyTorch deep learning framework, an open-source video object segmentation framework. The experimental environment was configured as follows: PyTorch version 1.8.1 and Python version 3.8. The hardware setup included a 14-core Intel(R) Xeon(R) Platinum 8362 CPU (Intel, Santa Clara, CA, USA) (base frequency 2.80 GHz) and one NVIDIA RTX 3090 GPU (24 GB VRAM) (NVIDIA, Santa Clara, CA, USA), supported by 45 GB of system memory. To facilitate GPU-accelerated deep learning tasks, CUDA 11.0 and the cuDNN library were installed to optimize parallel computing performance.
3.3. Parameter Settings
During the model training process, the AdamW optimizer was employed for parameter optimization. The initial learning rate was set to 2 × 10−4, with a weight decay coefficient of 0.07 to regularize parameter magnitudes and mitigate overfitting. The momentum factor was fixed at 0.9 to expedite gradient descent convergence and reduce oscillations during parameter updates. The total number of training epochs was set to 20,000, with a learning rate decay exponent of p = 0.9 to control the rate of learning rate reduction. To ensure training stability, the minimum learning rate was set to 2 × 10−5, and a linear warm-up strategy was applied for the first 5000 epochs, gradually increasing the learning rate from the minimum value to the initial value. The warm-up phase accounted for 5% of the total training epochs. The batch size was set to 8, and the gradient clipping threshold was set to 5.0 to prevent gradient explosion issues. Additionally, an exponential moving average (EMA) model smoothing strategy was employed with a decay rate of 0.1 to improve the model’s generalization performance.
3.4. Data Augmentation and Preprocessing
The input data underwent multi-scale dynamic augmentation to enhance the model’s robustness. First, the short side of the image was fixed to 480 pixels, followed by random cropping to 465 × 465 pixels to align with the model’s receptive field. Data augmentation strategies included horizontal flipping (probability 0.5) and temporal sampling (interval of 12 frames from the DAVIS dataset), with a dynamic feature fusion probability of 0.3 to improve the model’s ability to model spatiotemporal features. The backbone network utilized ResNet-50 weights pre-trained on the ImageNet dataset, and the parameters of the first two layers of the encoder were frozen to accelerate model convergence.
3.5. Evaluation Metrics
In previous studies on medaka ventricular segmentation, image segmentation methods were commonly used, and thus the Intersection over Union (IoU) and mean Intersection over Union (mIoU) were widely adopted as the primary evaluation metrics. Among these, mIoU calculates the average IoU of all predicted regions and their corresponding ground truth annotations, and its formula can be expressed as follows:
where
represents the total number of categories, and
and
denote the number of true positives, false positives, and false negatives for the
c-th category, respectively.
However, to comprehensively evaluate the performance of our proposed video segmentation model, this study selected two metrics specified in the DAVIS dataset challenge during model training: “Jaccard (J) per sequence” and “Boundary (F) per sequence.” These metrics are variants of the IoU (Jaccard index) and F-Measure, focusing on the performance of each individual video sequence rather than the overall performance of the entire dataset.
The formula for calculating Jaccard (J) per sequence is as follows:
where
represents the predicted segmentation region for the
t-th frame,
is the corresponding ground truth annotation, and
is the total number of frames in the video.
This metric quantifies the segmentation consistency of the model across the temporal dimension by calculating the average IoU between predicted and ground truth regions frame by frame. Its value range is [0, 1], and a value approaching 1 indicates that the model has excellent cross-frame stable segmentation capabilities.
The formula for calculating Boundary (F) per sequence is as follows:
This metric focuses on the topological fidelity of segmentation edges by calculating the harmonic mean of precision and recall. Its computation involves constructing an ε-neighborhood (typically ε = 3 pixels) for both the predicted contours and the ground truth annotations and then measuring boundary alignment accuracy through a bidirectional contour matching algorithm. This approach is highly sensitive to dynamic interference factors such as motion blur and deformation.
This dual-metric evaluation system constructs an evaluation space from two orthogonal dimensions: region coverage (J) and edge alignment (F). The J metric reflects the global accuracy of the segmentation results, while the F metric characterizes the fine-grained details of local edges.
4. Experimental Results and Analysis
This section evaluates the performance of multiple traditional fish heart image segmentation approaches and advanced semi-supervised video segmentation techniques on the medaka heart dataset, including Unet, TBD, DeAOT, and the proposed LAAVOS.
4.1. Training Result
In this experiment, to comprehensively evaluate the performance of LAAVOS during the training process, we systematically recorded its Intersection over Union (IoU) metrics on the validation set and conducted a comparative analysis with the DeAOT model. Specifically, after every 20 training iterations, both LAAVOS and DeAOT were tested on the validation set.
Figure 7 illustrates the variation of the IoU metrics with respect to the number of iterations.
The experimental results demonstrate that LAAVOS exhibits rapid convergence capability in the early stages of training, with its IoU values increasing significantly within fewer iterations and stabilizing after the 5000th iteration. By the end of the training process, LAAVOS achieved an IoU value of 99.6% for Medaka ventricle segmentation on the validation set. In contrast, while the IoU values of the DeAOT model also showed an upward trend, its final stabilized value was slightly lower than that of LAAVOS. These findings indicate that LAAVOS not only surpasses DeAOT in training efficiency but also demonstrates greater robustness in high-precision segmentation tasks.
4.2. Comparative Experiments
4.2.1. Quantitative Results
This experiment evaluates the segmentation performance of the proposed LAAVOS model against several widely used semi-automatic video object segmentation methods, including U-Net, TBD [
21], STAVOS, and DeAOT.
Table 1 presents a comprehensive comparison in terms of segmentation accuracy (mIoU, J-Mean, F-Mean, J&F-Mean) and inference speed (FPS).
In our experimental setup, all evaluations were conducted on the self-constructed medaka ventricle video dataset at 480p resolution, with individual video frame sizes matched to those in the DAVIS2017 validation set. It is important to note that J-Mean is equivalent to mIoU in our binary-class segmentation task and can be used interchangeably for performance comparison.
The LAAVOS model achieves a J-Mean of 92.89%, significantly surpassing U-Net’s mIoU of 79.41%, highlighting its capacity for fine-grained segmentation in challenging video conditions. Compared to other state-of-the-art matching-based models, LAAVOS demonstrates consistently superior accuracy. Specifically, it improves upon TBD by 13.01%, 8.72%, and 10.86% in J-Mean, F-Mean, and J&F-Mean, respectively. When compared with STAVOS, which integrates spatiotemporal attention but suffers from higher latency, LAAVOS still achieves notable gains of 10.54%, 8.19%, and 9.36% across the same metrics. Furthermore, relative to the more lightweight and efficient DeAOT model, LAAVOS yields improvements of 4.83%, 3.71%, and 4.26%, while maintaining competitive inference speed (25 FPS vs. 27 FPS).
These results confirm that LAAVOS achieves a strong trade-off between segmentation accuracy and computational efficiency, owing to the integration of its attention-enhanced encoder (LA-ResNet) and progressive decoding architecture (AFPViS). Its high accuracy and real-time speed make it well suited for medaka cardiac analysis scenarios requiring both precision and throughput.
4.2.2. Qualitative Results
To provide a more intuitive comparison of the segmentation performance across different models, we selected one systolic (ES) frame and one diastolic (ED) frame from test videos captured under two distinct imaging environments. As illustrated in
Figure 8, these representative samples display the original images, ground truth masks, and the segmentation outputs produced by the models listed in the previous table. Among them, LAAVOS consistently delivers more accurate and spatially coherent segmentation results across all cardiac phases, demonstrating strong robustness under varying imaging conditions.
4.3. Ablation Experiments
To validate the efficacy of the proposed LAAVOS method for the medaka heart segmentation task, we conducted ablation experiments to evaluate the contributions of its various enhancements. These experiments were designed to comprehensively assess the impact of each enhancement on overall performance and investigate their practical implications across diverse tasks. To ensure fairness and consistency, all comparative experiments were performed under identical experimental conditions and parameter settings.
First, we evaluated the performance of the original DeAOT model without AFPViS, using different encoders to investigate the impact of LA-ResNet on segmentation accuracy and model complexity. We conducted a comparative analysis across several mainstream backbone architectures, including MobileNetV2, MobileNetV3, ResNet-50/102/152, and the SwinB Transformer [
22,
23]. As summarized in
Table 2, LA-ResNet achieved a Jaccard index of 90.25% and an F-score of 94.31%, resulting in a combined J&F metric of 92.28%, outperforming the baseline ResNet-50 by 2.27 percentage points. Despite its significant performance gain, LA-ResNet maintained a lightweight parameter count of 26.3 M, representing only 29.9% of the SwinB encoder’s 88 M parameters, while delivering comparable accuracy (a J&F difference of merely 0.16%). Compared to deeper ResNet variants such as ResNet-102 (44.5 M) and ResNet-152 (60.2 M), LA-ResNet offered better or equivalent segmentation performance with nearly half the model size. This demonstrates LA-ResNet’s superior ability to achieve an optimal trade-off between segmentation precision and computational efficiency.
These results validate that the integration of local attention mechanisms in LA-ResNet substantially enhances the model’s ability to extract fine-grained features of medaka ventricles, while improving robustness to image degradation factors such as blur, occlusion, and color variance. The detailed performance comparison is presented in
Table 2.
Secondly, we designed four experiments to evaluate the impact of the AFPViS decoder on the performance of the LAAVOS model, as well as the interaction between AFPViS and LA-ResNet, while keeping all other parameters consistent. The experimental results, presented in
Table 3, clearly demonstrate the contribution of each improvement to the model’s performance.
In Experiment 2, the introduction of the AFPViS decoder alone resulted in an increase of 1.84% in the Jaccard coefficient and 1.79% in the F-score compared to the original model. This demonstrates that the AFPViS decoder effectively enhances the fusion capability of multi-scale features and underscores its importance in improving segmentation performance.
In Experiment 4, when AFPViS and LA-ResNet were combined, the model’s performance improved significantly: the Jaccard coefficient increased by 4.83% compared to Experiment 1, and the F-score reached 95.66%, surpassing the best performance achieved by either module alone. These results indicate that the combination of the two modules significantly enhances the model’s segmentation performance, effectively addressing the challenges of the complex medaka ventricular segmentation problem.
5. Conclusions
Currently, there is a lack of reliable datasets for medaka embryonic ventricular segmentation tasks. Additionally, the semantic segmentation methods commonly used in this field demonstrate insufficient accuracy in ventricular localization and boundary segmentation when processing medaka heart videos due to the complexity of the task. To address these issues, this study first constructed a medaka heart video dataset containing 4200 frames with pixel-level annotations, which includes videos of medaka ventral views captured under several different environmental conditions. Furthermore, this paper proposes a semi-supervised learning-based video segmentation model, LAAVOS, which significantly improves the accuracy and robustness of medaka ventricular segmentation by introducing a locally attention-enhanced LA-ResNet encoder and a progressive multi-scale feature fusion decoder, AFPViS.
The experimental results indicate that the proposed LAAVOS model significantly outperforms the conventional U-Net-based image segmentation approach, achieving a notable improvement of 13.48% in the mean Intersection over Union (mIoU) metric. Moreover, in comparison with DeAOT—a state-of-the-art semi-supervised video segmentation method—LAAVOS demonstrates a competitive advantage, attaining a 4.83% enhancement in the Jaccard and F-measure (J&F) metric. Specifically, the LA-ResNet encoder, by integrating the ResNet-50 backbone with a local attention mechanism, effectively enhances the model’s ability to extract ventricular features and significantly suppresses noise interference such as blurred images and organ occlusion. The AFPViS decoder, through a progressive multi-scale feature fusion mechanism, reduces semantic information degradation and loss during feature fusion in traditional Feature Pyramid Networks (FPNs), further improving the precision of segmentation.
In summary, this study provides a more accurate and environmentally adaptive segmentation method for medaka heart development research, establishing a technical foundation for studying the pathological mechanisms of heart diseases and developing treatment strategies.
However, this study still has the following limitations: the segmentation accuracy may decline due to blurred cardiac boundary images when blood pumping from the atria to the ventricles causes dynamic tissue deformation; and the model exhibits insufficient generalization capability for pathological features when dealing with embryonic specimens with ventricular malformations or severe lesions.
Future research directions include expanding the dataset further and investigating more powerful deep learning models to enhance domain generalization and adaptation capabilities, thereby achieving performance that can accurately segment more complex cases such as malformed embryos. Meanwhile, developing more efficient and flexible tools based on deep learning models for heart development research can improve biologists’ research efficiency.
To promote collaboration and advance research in this field, we have made all the materials utilized in this study publicly available. This includes the dataset and model implementation. These resources can be accessed at raokai645761/LAAVOS (
https://github.com/raokai645761/LAAVOS, accessed on 3 June 2025), enabling researchers to replicate and build upon our work. We call upon more biologists to share relevant imaging data to facilitate further development of medaka computer vision research.