1. Introduction
The expansion of urban areas and the resulting decline in natural spaces has led to new challenges, including ecosystem degradation, species loss, and increased vulnerability to climate hazards, among others. Urban biodiversity is essential for sustainable cities, as it helps address challenges that negatively affect human health and well-being. ECOLOPES (ECOlogical building enveloPES), an H2020 FET Project, aims to develop a design approach for multi-species as stakeholders to achieve regenerative urban ecosystems [
1]. For multi-species, we mean four different types of inhabitants that can co-occupy the envelope: plants, animals, microbiota, and humans.
Designing with biodiversity in mind involves addressing the challenge of accommodating the diverse requirements and constraints of multiple species while also considering architectural factors. These requirements and constraints must be clearly defined, formalised, and placed in the appropriate context to ensure effective integration within urban and architectural planning. Ontologies [
2] provide a structured approach to representing knowledge by defining concepts and relationships within and across various domains. Additionally, they enable axiomatic reasoning, allowing implicit facts to be inferred from explicitly stated information within a knowledge base [
3]. Implementing a knowledge base using knowledge graphs [
4] offers greater flexibility in data integration, as they are built upon well-established Semantic Web and Linked Data principles—such as IRIs for uniquely identifying entities and RDF for representing facts in the form of triples, ensuring interoperability and semantic richness in data integration processes.
Integrating the diverse data required for stakeholders and beyond—spanning life sciences, geography, and architecture—and utilising it for design presents a significant challenge. This paper introduces an ontology-driven approach that utilises ontology-based data management (OBDM) [
5] as a framework that integrates diverse data sources, enabling ecologists and architects to design sites and buildings that foster urban biodiversity. The proposed ontology-aided design methodology builds upon the concept of performance-oriented design [
6,
7] and the conceptual framework focused on data-driven approaches to understanding and designing environments [
8].
Data sources range from publicly available datasets, which can be easily accessed and retrieved via web APIs—such as GLoBI (Global Biotic Interactions for species) [
9]—to private datasets generated within ECOLOPES, including Plant Functional Groups (PFGs) [
10], Animal Functional Groups (AFGs), and voxel data [
11] that capture site-specific variables like coordinates, aspect, slope, or solar radiation. Plant and animal functional groups, defined by shared traits or ecological roles rather than taxonomic classification, play a crucial role in ecosystem processes, resilience, and biodiversity conservation. A voxel model contains useful aggregated information applicable to various use cases, such as computing the
Habitat Suitability Index (HSI), a model which evaluates how well a habitat suits a given species. Therefore, depending on the
weighting scheme employed by the respective specific publication or study on the reported species (e.g., rabbit, fox), one can calculate and infer for each point in a CAD environment: aspect, slope, elevation, solar radiation, and so forth, how suitable it is for the designated species with respect to environmental conditions in a voxel model. Another dimension of data emerges from the designer’s interaction with CAD and Grasshopper environments. In this environment, based on the project brief requirements, the designer creates a
network configuration by using predefined ‘circle shapes’ of
EcoNodes (denoting species) and
ArchiNodes (denoting buildings or other architectural objects).
The resulting disparate and heterogeneous data requires integration, alignment, and consolidation into a unified RDF-based graph data model, with ontological terms used to define the mappings. The result is a knowledge graph [
4,
12] that contains curated and contextualised knowledge, enabling holistic querying and answer retrieval through the use of domain ontologies. In order to implement it, we have employed OBDM—an extension of ontology-based data access (OBDA) [
13] with support for updates [
14] in addition to queries–as a data integration framework. This approach led to some data being virtualised (e.g., voxel model), with the remaining data materialised, i.e., stored and consequently indexed by the triple store. This approach is pragmatic, as the data are not moved, and no data duplication is created, albeit with the general downside of reduced performance in query evaluation. Nonetheless, subject to performance indicators or providing a seamless workflow for the designer, a subset of voxel data can be materialised in order to meet the required interaction criteria [
15]. For instance, in our case, the
Euclidian distance between two nodes is a candidate for pre-materialisation to increase the performance of the query, typically viable in cases where changes to the position of nodes do not occur frequently.
The scope of the knowledge graph is often determined by the set of competency questions (CQs) [
2] it is designed to answer, such as
“Provide me with the local species that are known to have the protection status threatened”. To achieve this, new data must be ingested, such as public data on local species provided by municipalities, citizen science (e.g., GBIF (
https://www.gbif.org, accessed on 17 March 2025)), and threatened species data from the International Union for Conservation of Nature (IUCN) (
https://iucn.org, accessed on 17 March 2025). Additionally, this data often requires alignment across datasets—for instance, by linking data referring to the same entity using
owl:sameAs assertions or custom relationships defined within the ontology. In this case, the species’ Latin name alongside the species’ taxonomic rank (if applicable) can be leveraged in order to perform the disambiguation process and, therefore, aid in the alignment process between entities.
Ontologies, particularly those built using OWL (
https://www.w3.org/TR/owl2-overview/, accessed on 17 March 2025) (Web Ontology Language), not only provide a comprehensive description of a domain but also enable the inference of new implicit facts (i.e., RDF triples) based on the explicit facts asserted in the knowledge graph. Domain experts and ontologists often aim to reuse concepts from existing ontologies or establish alignments to create more expressive and granular structures, which can be leveraged on demand or to support reasoning capabilities using Open World Assumption (OWA). In our case, we have modelled ECOLOPES ontology using a top-down conceptualisation, a bottom-up approach informed by the data, and by reusing structures from well-established ontologies, namely OBO Relations [
16] or Darwin Core
https://dwc.tdwg.org (accessed on 17 March 2025) following ontology construction best practices.
The proposed ontology, developed in collaboration with domain experts and adhering to Semantic Web and Linked Data best practices, serves as a mediator between life sciences data (e.g., species distribution and habitats) and geometric information (e.g., maps, voxel models of building structures). The ontology is an interface with CAD and aids the design of ecological building envelopes addressing the critical “data-to-design gap” [
17,
18] in computational architecture and multi-species architectural design.
Constraints, unlike ontologies–which are designed for logical, deductive reasoning–adhere to the Closed World Assumption (CWA), which precisely defines how data should be structured by imposing specific restrictions on the data model. In our case, constraints are represented as explicit facts in the knowledge graph, such as the solar requirements of plants or the minimum and maximum proximity distances between species. By using standards like SHACL
https://www.w3.org/TR/shacl/ (accessed on 17 March 2025), one can define and validate the expected “shapes” and structure of the data, ensuring data quality and completeness while also enabling the enforcement of consistency across the dataset. In our case, validation is currently performed using a set of SPARQL queries. The constraints in our problem setting can be translated to SPARQL queries, e.g.,
“Provide me the locations in site where I can place the plant Abies alba (EcoNode) such that its solar requirements are met?”. Given the spatial nature of this query, typically, one would employ a GeoSPARQL query to answer it; nonetheless, in our case, the voxel model containing x, y, and z coordinates boils down to
equality checking with respect to CAD coordinates.
The computational model that is used to generate design outcomes based on the designer’s input is rule-based. This approach computes the design outcomes based on the selected rules that can be run in a designated order. Our current implementation mimics rule-based reasoning by employing a set of SPARQL update queries [
19] in the context of OBDM that are run in a sequence that is orchestrated by the Grasshopper environment.
This integration enables the adaptation of sites, buildings, and geometries, respectively, to create habitats that attract and support urban wildlife, contributing to ecological sustainability. The paper illustrates the practical utility of the ontology through a (case study) demonstrator, highlighting its role in guiding building designs that promote species attractiveness and urban biodiversity.
Our contributions are the following:
A new EIM (ECOLOPES Information Model) ontology and knowledge graph in the domains of architecture and ecology.
A method that utilises OBDM to map and integrate different data sources—stemming from different environments from the respective domains—through an ontology, effectively creating the knowledge graph. OBDM enables query and update operations to be performed directly on the integrated data.
A designer workflow, in which the designer obtains feedback (e.g., the fulfilment of solar radiation or proximity constraints) or asks CQs against the knowledge graph, enabled by OBDM.
The rest of the paper is structured as follows. In
Section 2, we provide the necessary background, motivation, and problem statement. In
Section 3, we discuss related work with respect to using ontologies in design and ecology. In
Section 4, we explain the ontology design, reuse, and alignment. In
Section 5, we discuss the data integration framework used for integrating heterogeneous data sources. In
Section 6, we describe the demonstrator, and we provide the results achieved in that context. Finally, in
Section 7, we conclude the paper with conclusions and future work.
2. Motivation and Problem Statement
The
ontology-aided generative computational design process facilitates design generation, namely for a search space populated with alternative design solutions that can be analysed and evaluated. The overall process of design generation entails synthesising heterogeneous ecological, environmental, and architectural data, voxel-based data structuring, context information at regional/urban and local/architectural scales, and modelling information according to decision needs. A detailed description of the conceptual framework that informed the development of the research presented in this paper is published in a separate study [
20].
The role of ontologies is to model information regarding the entities and relationships that need to be represented and to aid the design of ecological building envelopes. We distinguish between ontologies using schema for TBox and instance data for ABox. More formally, for a given data source and its variables
x, we create ABox by applying a set of mappings:
, where
E is a TBox class or property, and
is a set of IRI templates, with each applied to the variable
x in order to create a literal or IRI. A knowledge graph KG is a union of ABox and TBox (herein, we also distinguish taxonomies or controlled vocabularies as a part of KG, sometimes referred to as CBox. They are essential to capture hierarchical representations, e.g., species taxonomic rank) represented in RDF and typically stored (materialised) in a triple store. To avoid the complexity of reification statements in RDF, we chose to model statements using RDF-star for convenience, namely for expressing proximity constraints. A part of ABox can be virtualised instead of being materialised, depending on the use case requirements. In this particular case, the mappings ⇝ that are represented in a specific mapping language are used to translate the queries on the fly back to the original data sources. TBox axioms are used to infer new implicit triples in ABox based on asserted (ABox) triples, which are either stored in the triple store or, alternatively, a query is rewritten during the query time with respect to TBox axioms to account for the same (query) answers. This proposition does not hold for expressive OWL ontology languages but rather for (minimal) RDFS, consisting of
rdfs:domain/
rdfs:range,
rdfs:subClassOf, and
rdfs:subPropertyOf axioms [
14,
21]. For such pragmatic reasons, we use (minimal) RDFS in conjunction with a few OWL axioms (e.g., for aligning properties via
owl:equivalentProperty) for the task of ontology modelling. In the case of OBDA and mappings, aside from query rewriting, an additional step of
query unfolding is executed, in which SPARQL queries are translated on the fly to SQL queries in order to obtain the so-called
certain query answers [
22]. OBDM [
5] extends OBDA with support for update queries.
The role of the voxel model within the
ontology-aided design process is to describe the real-world location in which the design process is taking place by structuring and spatialising datasets. An early definition of voxel models as “knowledge representation schemata” [
23] was identified in the course of an extensive literature review of voxel models’ role in the engineering fields [
11], and this was re-established in the context of computational design. The voxel model implementation developed for this study extends the Composite Voxel Model methodology [
24] by incorporating the explicit link with knowledge representation through the ontology-aided design process. The implemented ECOLOPES Voxel Modell, described in detail in [
25], integrates geometric data describing the real-world site’s physical geometry and simulated environmental performance. Currently, voxel-based design approaches addressing anthropogenic landscape adaptations can be found [
26], and studies aiming to integrate voxel and ontological modelling are ongoing [
27]. A unique property of the presented ECOLOPES Voxel Model is the implementation within the relational database environment (PostgreSQL), which enables direct access to voxel data through the native “OBDA” mappings. As indicated in
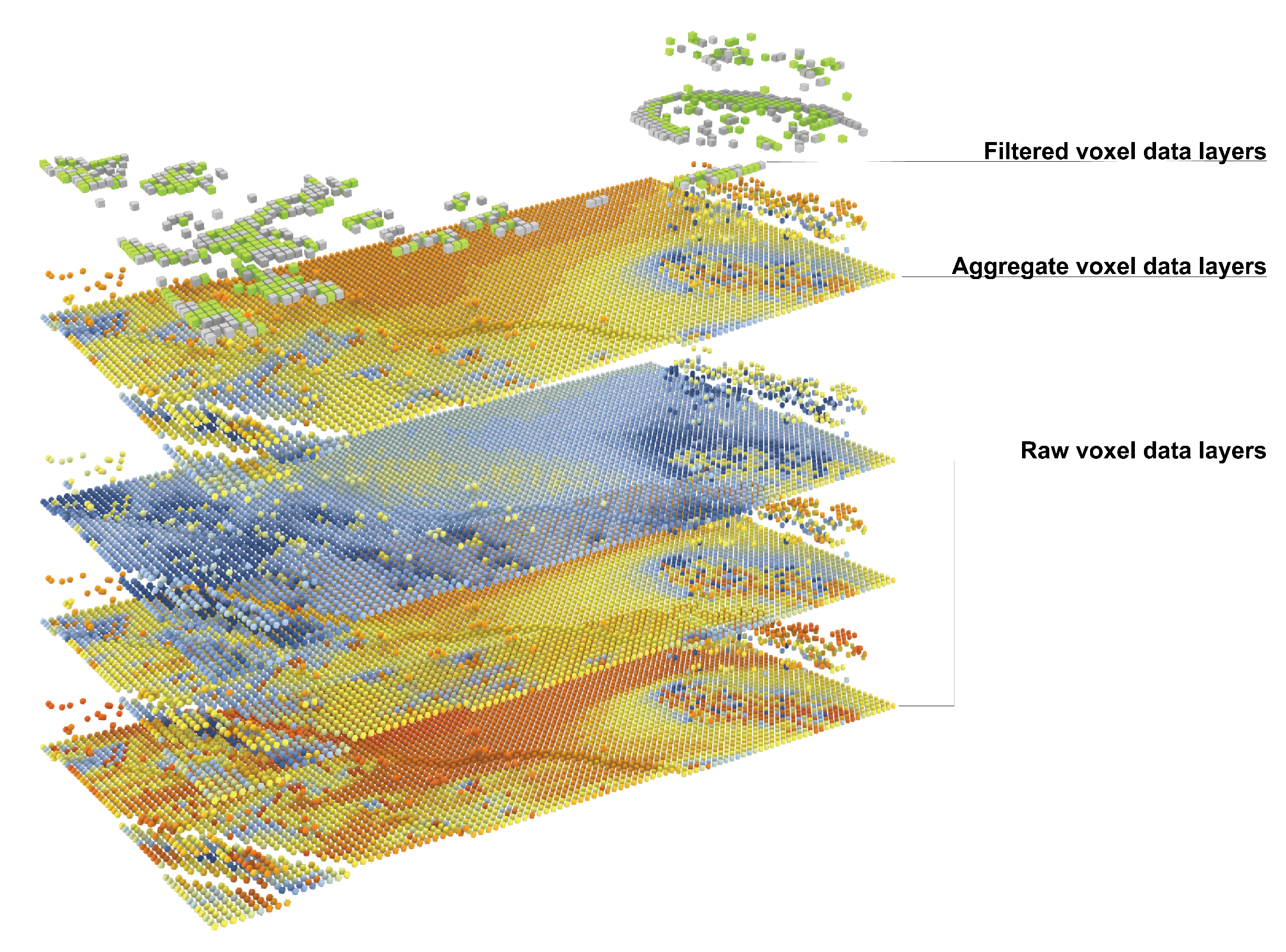
Figure 1, the concept of voxel data layers is used to structure datasets provided by the ECOLOPES Voxel Model. By utilising the OBDA virtualisation technique, the EIM Ontologies can currently access the raw, aggregate, and filtered data layer, enabling the interactive temporal resolution change of the environmental simulation data contained in the ECOLOPES Voxel Model (cf.
Figure 2). For example, average sunlight exposure (sunlight hours) in summer, autumn, winter, spring, and the growing season can be aggregated from the raw voxel data representing monthly average sunlight exposure. Multi-scalar voxel data representing geometry and all available parameters stored in different voxel levels can be retrieved by designers and accessed through the SPARQL endpoint (see
Section 5).
Two processes are combined in the ontology-aided generative computational design process: (i) the translational and (ii) the generative processes.
In the
translational process, the designer analyses, correlates, and locates spatially architectural and ecological requirements contained in the design brief and design-specific determinations while taking into consideration relevant constraints (i.e., as given by planning regulations). In order to do so, the designer uses a set of predefined nodes and places them in the CAD environment in order to satisfy ecological and architectural requirements. Herein, the designer can query the knowledge graph in order to obtain feedback regarding the solution space for permissible design outputs in terms of satisfying the proximity distances between nodes (e.g., species) or solar radiation requirements. Nodes that fulfil the constraints are depicted as green circles, whereas those that do not are depicted as red crosses. The Grasshopper environment is used as a component for doing the orchestration process of the validation. A knowledge graph also acts as a mediator with the voxel model (cf.
Figure 1) that contains prepared datasets regarding geometric and environmental variables. In this paper, we focus only on the translation process. The intended goal of the translational process is shown in
Figure 3.
The generative process consists of two distinct stages. In the first stage, the variations of spatial organisation are generated and evaluated. In the second one, specific surface geometries are generated for selected outputs of the previous stage and are evaluated according to defined criteria, i.e., key performance indicators. The generative process falls outside the scope of this work and is planned for future exploration.
3. Related Work
The use of Semantic Web technologies and OWL to represent ecological network specifications is explored in [
30]. Ecological networks capture the structure of existing ecosystems and provide a basis for planning their expansion, conservation, and enhancement. An OWL ontology is employed to model these networks, enabling reasoning about potential improvements and identifying violations in proposed network expansions. The approach incorporates a language called GeCoLan to define ecological network specifications, which are subsequently translated into GeoSPARQL queries. This methodology enhances the planning and management of ecological networks by supporting automated reasoning and integrating diverse data sources effectively.
Global Biotic Interactions (GLoBI) [
9] is an open-access platform designed for researchers, aggregating diverse datasets to provide comprehensive information on species interactions, such as predator–prey, pollinator–plant, and parasite–host relationships, among others. The platform serves as a valuable resource for understanding ecological networks, predicting the effects of environmental changes, and guiding conservation strategies. It consolidates and harmonises data on species and their occurrences and interactions from a variety of publications and observational databases. GLoBI offers both an API and a SPARQL endpoint (with partial data coverage) for accessing species interaction data in CSV or RDF formats. Species data are structured using the Darwin Core vocabulary, with equivalence between species established through
owl:sameAs relations. Additionally, the platform features a web-based frontend application that supports species searches via text and/or geographical boundaries. OBO Relations (
http://obofoundry.org/ontology/ro.html, accessed on 17 March 2025) ontology can be leveraged for reasoning to infer implicit interactions based on explicitly recorded triples.
The integration of ontologies and knowledge graphs in urban ecology has gained significant attention due to their ability to model complex interactions between built environments and natural ecosystems [
18]. Existing research spans two major domains: Architecture, Engineering, and Construction (AEC(O)) and Urban Design, Planning, and Management, both of which are crucial for advancing sustainable urban development. Ontologies and knowledge graphs have been extensively used in AEC(O) to improve data interoperability, lifecycle management, and decision support systems. In Building Modelling, semantic models such as the Industry Foundation Classes (IFC) ontology enable structured data exchange across different software platforms [
31]. In Building Construction and Renovation, ontologies assist in process optimisation, cost estimation, and resource planning by integrating heterogeneous datasets [
32]. Additionally, Building Automation has benefited from ontologies like the Brick schema, which enables the semantic representation of building sensors and control systems. Ontologies also play a key role in Building Sustainability, facilitating the assessment of energy efficiency and carbon footprints [
33]. Semantic models allow for the integration of green building certification standards (e.g., LEED, BREEAM) with real-time building performance data, enhancing decision-making for sustainable design and renovation [
34]. Ontologies have been applied in City Modelling, enhancing urban digital twins with semantic interoperability [
35]. The CityGML standard, extended with ontologies, provides a structured representation of urban elements, supporting applications in Civil Construction and infrastructure management. Urban Sustainability ontologies enable the integration of environmental indicators with urban planning tools, aiding in decision-making for reducing ecological footprints [
36]. The role of Urban Green and Blue Infrastructure ontologies is particularly relevant for urban ecology. These semantic models capture the relationships between natural elements (e.g., vegetation and water bodies) and built environments, supporting biodiversity monitoring and ecosystem services assessment [
37]. While AEC(O) and urban planning ontologies provide structured models for specific domains, their integration with ecological and functional dimensions is essential for advancing sustainable urban ecosystems. Several key research areas highlight the necessity of interlinking these domains: Simulation, where ontologies enable multi-domain simulations for energy performance [
38], microclimate analysis, and species habitat suitability within urban environments [
39]; Garden Management, where knowledge graphs integrate urban vegetation data with soil conditions, climate factors, and biodiversity metrics, improving urban green space management [
40]; and Urban Key Performance Indicators (KPIs), where ontologies facilitate KPI-based urban sustainability assessments by structuring data around air quality, noise pollution, and resource efficiency [
41]. Despite advances in these areas, challenges remain in harmonising ontologies across domains, ensuring scalability, and developing reasoning mechanisms that enhance automated decision support.
Unlike the approaches reviewed in this section, the ontology we propose is designed to bridge the gap between two distinct domains, ecology and architecture, ensuring seamless integration of heterogeneous data sources. It is built on a strong foundation of Semantic Web best practices and adheres to FAIR principles, enhancing data findability, accessibility, interoperability, and reusability. Moreover, the ontology goes beyond pure modelling; it enables semantic reasoning and decision support, allowing stakeholders to derive new insights, infer implicit knowledge, and guide informed decision-making in sustainable urban planning and biodiversity-conscious architectural design.
4. Ontology Design and Development
In this section, we outline the ontology development process for EIM following W3C best practices and FAIR principles. The process begins with defining requirements through expert input and competency questions (CQs). Next, terms and axioms are created, ensuring the reuse of existing vocabularies for interoperability. The ontology is then structured and formalised, followed by validation and visualisation to enhance accessibility and usability.
4.1. Defining the Ontology Requirements
We organised a collaborative workshop with experts from the ECOLOPES consortium, comprising architects and ecologists, to define the ontology and knowledge graph requirements. Given that most participants were not ICT specialists or knowledge engineers, identifying and conceptualising ontology requirements posed a significant challenge. To facilitate this process, we structured the workshop around ontology construction best practices, ensuring that domain experts could systematically contribute their knowledge in a way that aligns with semantic modelling principles.
The participants were divided into four interdisciplinary teams, each consisting of six people, to encourage the “cross-pollination” of ideas, leveraging diverse expertise to bridge conceptual gaps. Each team was tasked with defining a comprehensive set of competency questions (CQs) spanning both domains, a fundamental step in ontology engineering that helps capture the intended scope and use cases. These CQs were written on post-it notes to foster discussion and iterative refinement, allowing experts to progressively articulate their knowledge in a structured manner.
In parallel, the participants identified key concepts and relationships relevant to answering the CQs, mapping out the foundational elements of the ontology. However, the abstraction required for formal ontological representation proved difficult for non-ontology experts. To mitigate this, they were guided to propose synonyms, group related concepts into broader categories, and outline hierarchical relationships, effectively shaping the initial class taxonomy. Recognising that defining structured properties and attributes requires a technical perspective, we encouraged participants to describe informal connections between concepts, which were later formalised by knowledge engineers. When gaps remained in structuring the ontology, participants suggested relevant datasets or external sources that could support answering the identified CQs, ensuring that the ontology remained grounded in real-world data and met FAIR principles for interoperability and reusability.
This bottom-up, expert-driven approach resulted in a draft skeleton ontology comprising initial classes, properties, and attributes.
Requirements were captured through CQs, with some listed here to name a few:
Which species do we want/do not want to attract close to our building?
Which species are in PFG herbs_3?
Which species that colonise the building are protected?
Which species are invasive in this location?
Which species can reach or live on sloped surfaces?
What is the soil depth required for PFG herbs_3?
4.2. Ontology Requirements and Scope
One of the main ontology aims was to integrate diverse data sources to support sustainable building design decisions that enhance urban biodiversity. Its scope includes ecological data (e.g., species distribution and habitat preferences), architectural data (e.g., building geometry and materials), and geographical data (e.g., environmental conditions and maps). By facilitating data interoperability and enabling reasoning capabilities, the ontology seeks to inform ecologists and architects when designing buildings that foster ecosystem development.
Once the definition phase of the CQs had been finished, the next steps were to group them based on similarity and prioritise them based on importance. After the screening and processing of the CQs have been performed, we could extract general (i.e., upper-level) requirements from those. In the scope of this paper, we consider that processing all the CQs is out of scope; hence, herein, we focus only on general constraints and requirements that pertain to the following:
The proximity between architectural objects and species (and vice versa) or, likewise, between species only.
Solar radiation (regarding species).
The prey areas in the CAD environment, which denote areas in which species are allowed to prey on one another.
4.3. Ontology Creation Process
The ontology (
https://github.com/aahmeti/Ecolopes, accessed on 17 March 2025) was developed following Semantic Web and Linked Open Data (LOD) best practices, ensuring compliance with the FAIR (Findable, Accessible, Interoperable, Reusable) principles. The methodology employed a structured approach incorporating the following:
Iterative design and refinement, integrating feedback from domain experts and ecological datasets on biodiversity.
Modular construction, enabling scalability, maintainability, and reuse across multiple domains.
Reuse of established vocabularies and ontologies, including GeoSPARQL for spatial representation and Darwin Core for biodiversity data integration.
Ontology development was conducted using Protégé for schema and taxonomy management, while GraphDB was employed for managing the knowledge graph instance data. Modularisation was applied to facilitate ontology structuring, leading to a well-defined prefix scheme that enhances accessibility and reusability. The ontology components are categorised into schemas (
networks-schema,
ecolopes-schema), taxonomies (
networks-taxo), and instance data (
ecolopes-inst,
networks-inst). This structured approach ensures a clear separation of entities originating from different environments, improving clarity and consistency, as shown in
Table 1.
Finally, the ontologies were imported into GraphDB and stored in separate named graphs. The GraphDB repository was configured using reasoning that enabled the support of
owl:sameAs at the instance level and ontology axioms for inferencing. To maintain governance and ensure robust deployment, development and production environments were distinguished as separate repositories. Additionally, GraphDB enables the querying of inferred triples using the implicit graph
FROM http://www.ontotext.com/implicit (
https://graphdb.ontotext.com/documentation/10.8/query-behavior.html#tuning-query-behavior, accessed on 17 March 2025)), allowing for advanced semantic reasoning and enriched data retrieval.
4.4. Core Concepts and Structure
The ontology’s core concepts address the integration of ecological, architectural, and geographical data. The key classes include the following:
Classes: Archi- and EchoNode, Voxel, PlaneConstraint, SolarConstraint, and more.
Relationships: nodeHasDistanceToNode, nodeHasSpecies, nodeHasArch, nodeOverlapsPlane, and more.
Attributes: x, y, z, proximityRequirementMin, proximityRequirementMax, solarRequirementMin, solarRequirementMax, and more.
4.5. Alignment with Existing Standards
To ensure full compliance with FAIR principles, the ontology was designed to be highly interoperable, reusable, and findable by being aligned with well-established standards and vocabularies. This alignment enables seamless integration with external datasets, facilitates data interoperability, and enhances the ontology’s applicability across diverse domains. Specifically, the ontology incorporates the following:
GeoSPARQL: Used to represent geographical data and enable spatial reasoning and geospatial queries across datasets.
Darwin Core: Integrated for biodiversity data representation, ensuring compatibility with life sciences repositories such as GBIF.
OBO Relations: Employed for modelling ecological relationships, such as predator–prey interactions, ensuring consistency with existing biological ontologies.
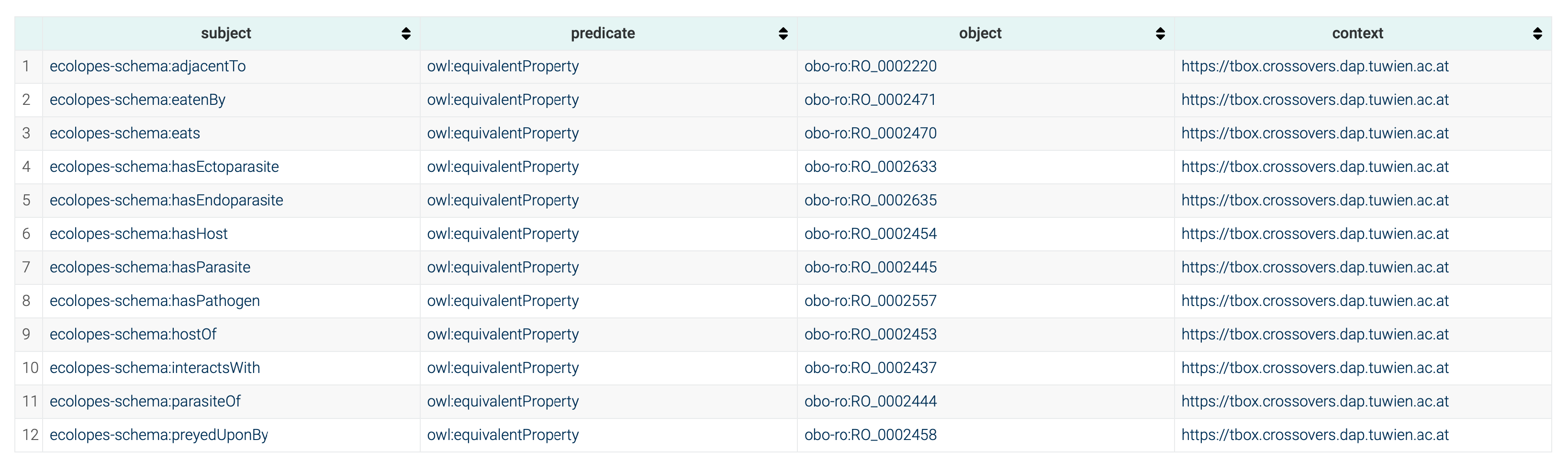
To maintain semantic consistency and interoperability, OBO Relations were aligned using
owl:equivalentProperty, allowing for seamless integration with external data sources like GLoBI while preserving existing structures without additional data transformation (cf.
Figure 4). This approach follows Linked Open Data (LOD) best practices, ensuring that the ontology extends and interacts meaningfully with established ontologies. On the other hand, the mapping of instance GLoBI data was carried out using Ontotext Refine mappings. In conjunction with the
owl:equivalentProperty alignments presented above, the reasoning would infer triples with the OBO Relations predicates, hence enabling interoperability.
Regarding geospatial data, the ontology aligns with GeoSPARQL ontology to ensure full compatibility with spatial datasets (e.g., GBIF or shapefiles, cf.
Table 2, Dataset No. 2 and 3). Shapefiles were translated to RDF, complying with GeoSPARQL vocabulary by using RML mappings generated by GeoTriples (ver. 1.1.6) [
42]. Likewise, latitude and longitude coordinates stemming from GBIF in RDF format were transformed into GeoSPARQL representations via a SPARQL update query, and the result is demonstrated in the following example.
Example 1. This example in RDF Turtle syntax represents a point close to the Nordbahnhof area using GeoSPARQL vocabulary:
ecolopes-inst:Location_48.230_16.390 a sf:Point ;
...
ecolopes-schema:hasExactGeometry [ a geo:Geometry ;
geo:asWKT "POINT(48.23012995681571 16.39000953797779)’’
]
The ontology also integrates Darwin Core for structuring species taxonomic classifications. This allows for querying across multiple biological repositories using standardised taxonomic ranks, such as
dwc:genus,
dwc:order, and
dwc:family. By adhering to Darwin Core, the ontology supports the automated classification and retrieval of species information, ensuring interoperability with existing biodiversity knowledge graphs (e.g., query federation of GBIF backbone species). Whenever possible, the species have been modelled using the Darwin Core vocabulary, i.e., the use of
dwc:scientificName and
dwc:vernacularName to represent the scientific name and common name, respectively (cf.
Section 6).
Through these measures, the ontology ensures compliance with FAIR principles, promoting interoperability, structured data reuse, and seamless integration with the broader Linked Data ecosystem.
5. Ontology-Driven Data Integration Through OBDM
We used a triple store as an implementation for OBDM, specifically GraphDB, which supports modes of virtualisation via Ontop integration [
43] and materialisation (natively). This approach is versatile, as the virtualised data can also be materialised to enhance query performance. OBDM [
5] was chosen for its ability to provide semantic interoperability, reasoning, and data virtualisation, ensuring a flexible and scalable approach to integrating heterogeneous data sources. Unlike traditional ETL-based data integration, which requires extensive preprocessing and rigid schema mappings, OBDM allows for query federation, enabling real-time access to distributed data without duplication. For these reasons, we chose to virtualise the voxel model. By aligning with FAIR principles, OBDM enhances data findability, accessibility, interoperability, and reusability, making it particularly suited to complex domains like urban ecology and sustainable architecture. Additionally, GraphDB’s mature implementation of OBDM supports efficient query execution and reasoning, further strengthening its suitability over conventional integration techniques. In GraphDB, one can configure a repository (equivalent to a database) by choosing one of the modes. Then, one can combine the results of the two modes by using query federation in SPARQL via the
SERVICE keyword. As we have explained previously (cf.
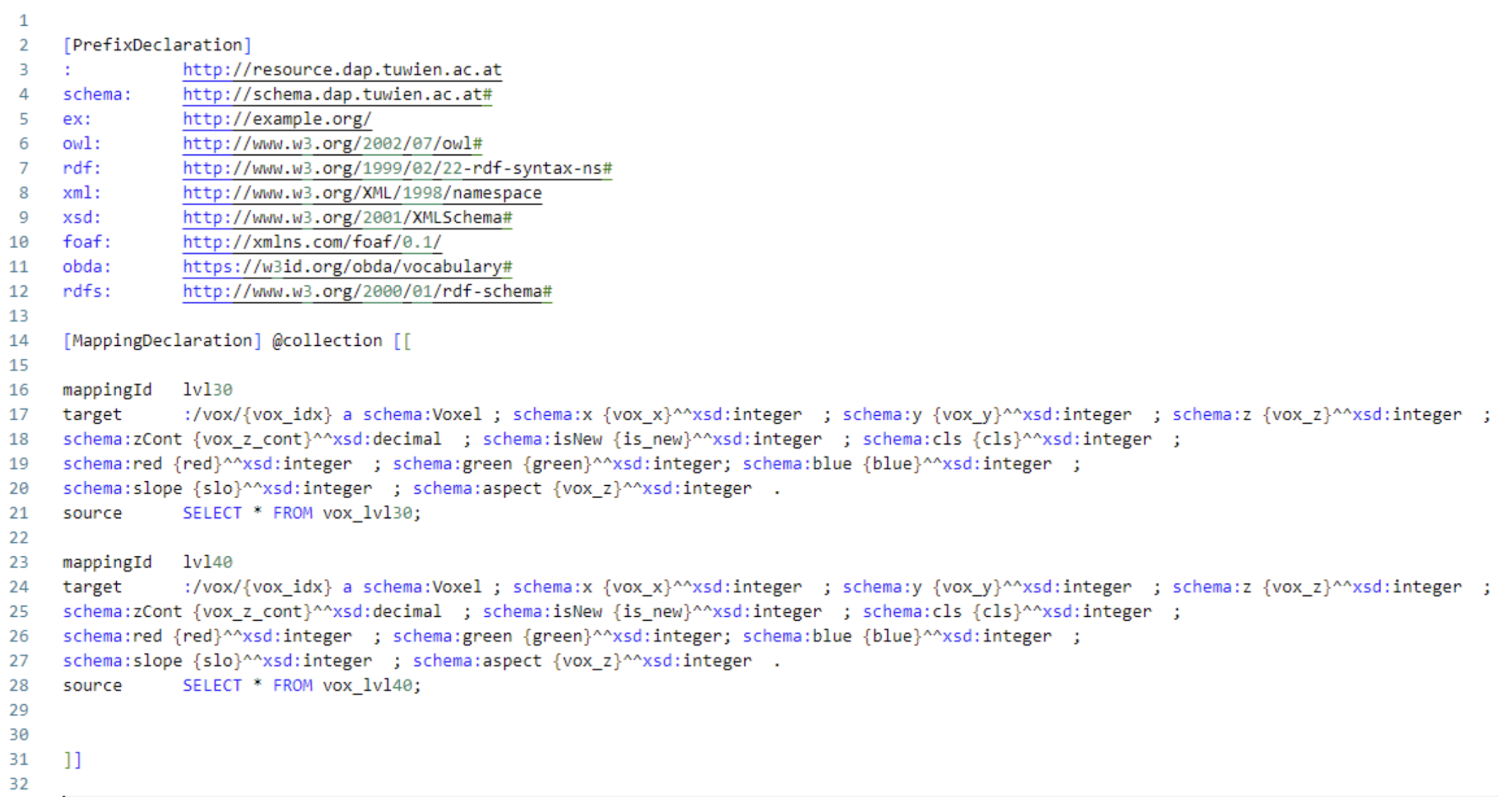
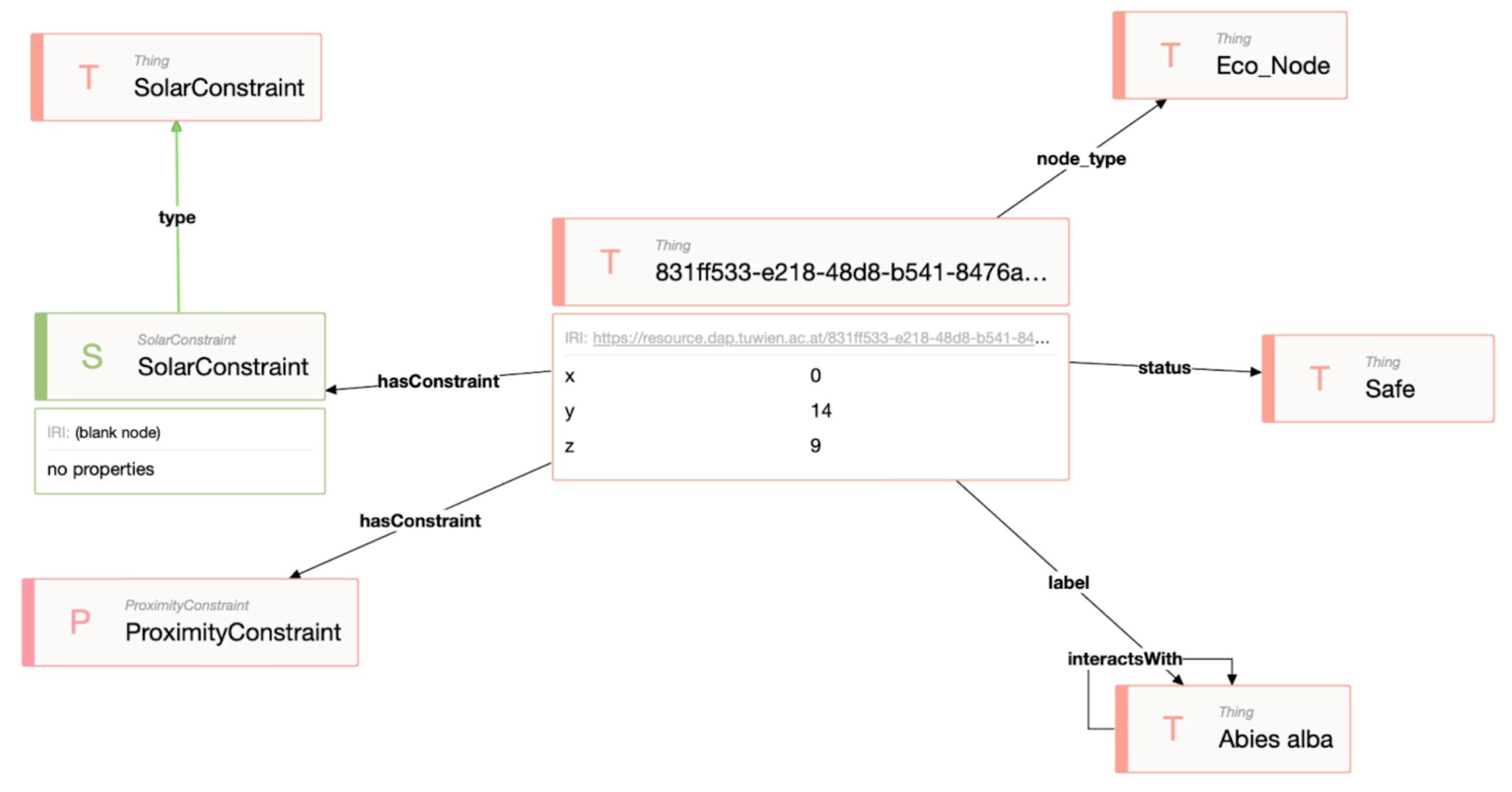
Figure 2), the voxel model is virtualised using OBDA mappings. The implementation of OBDA mappings in RDF is provided in
Figure 5. The set mappings for voxel data are provided 1-1 (one-to-one), meaning that each column from the voxel data is mapped to a unique property in RDF, i.e.,
, and so on. Keeping the mapping expressivity lean makes it future-proof for future use cases, namely in cases where updating voxel data is needed from the ontology via SPARQL update queries without the need to resolve ambiguous mappings and the side-effect of running into the
view update problem, which is known in database theory [
14].
The system allows for defining the sources (the voxel model stored and managed in the relational database) and mapping definitions (OBDA or R2RML (
https://www.w3.org/TR/r2rml/, accessed on 17 March 2025)), thereby exposing it as a separate repository in which SPARQL queries (via
SERVICE keyword) are translated into SQL queries, respectively, on the fly via query unfolding (cf.
Figure 6). For the time being, we have used the native OBDA mappings; in the future, we plan to replace this with the more future-proof R2RML mapping language.
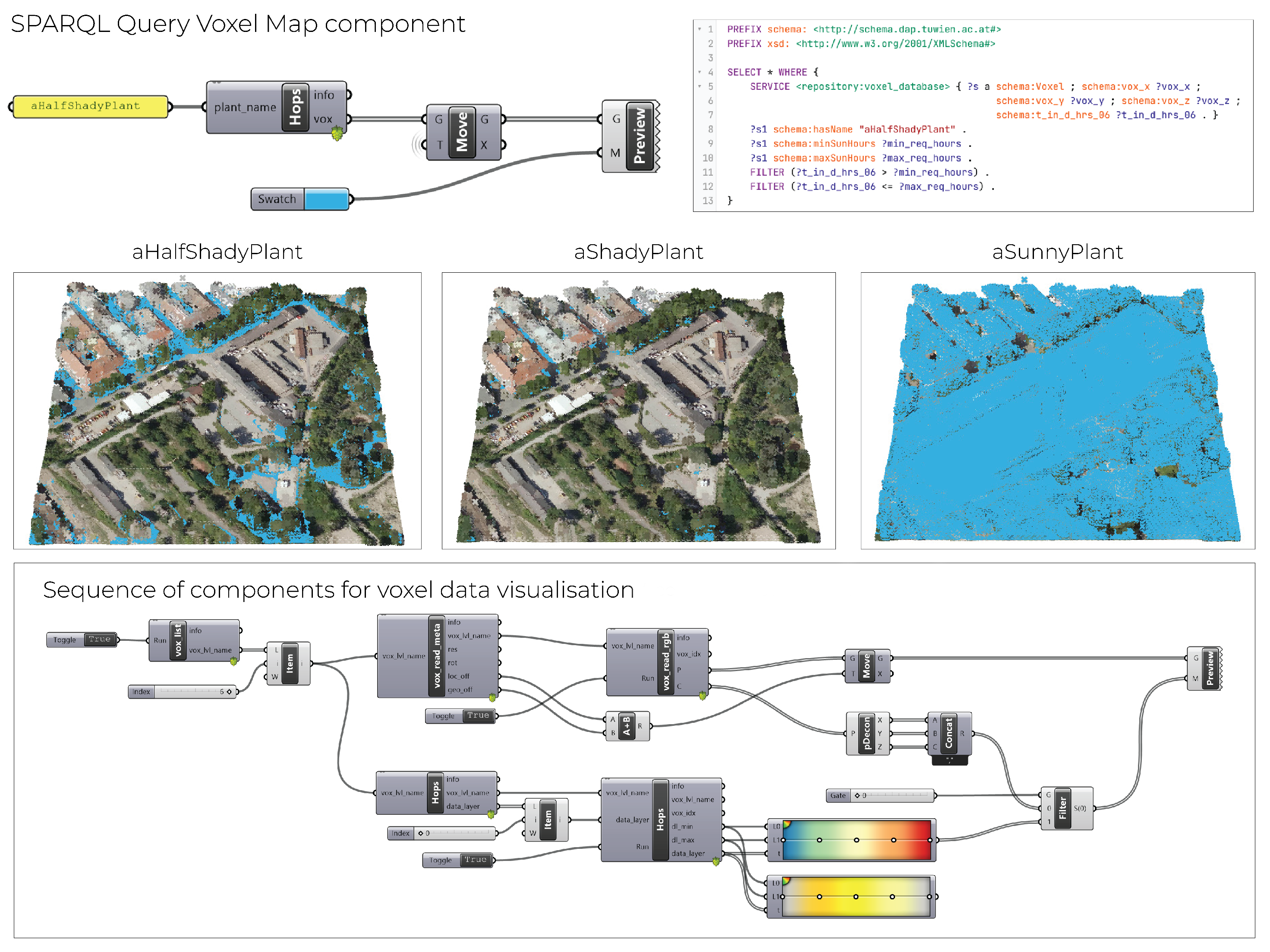
The integration scenario in
Figure 7 showcases the OBDM, namely the combination of virtualisation and materialisation in the context of the CAD and Grasshopper environments. The federated query using the
SERVICE keyword issues the query to the repository where the mappings and virtualisation are applied. The bindings are joined with the variables in the current repository that contain knowledge of species’ solar requirements. The process is computed via the Grasshopper pipeline, and the visualisation is shown in the CAD environment. The permissible places for half-shady, shady, and sunny plant species, each with their respective requirements, are shown spatially using the blue colour. OBDM, in this case, is a decision support system for the designer to make informed (spatial) decisions that are orchestrated and issued by the Grasshopper workflow. This demo showcases the integration between two datasets, namely the voxel model and plants’ solar requirements. We can iteratively increase the number of datasets that are ingested, harmonised, and linked, and, therefore, iteratively increase the number of CQs we can ask the KG or use it for more complex decision support in the design process.
Currently, a number of datasets have been identified and integrated into the KG using the defined ontological classes, properties, or attributes; see
Table 2. The triple count depicts the size and complexity of the data that can be handled from the data management perspective. Datasets No. 1 and No. 2 are local datasets which can be replaced by other (local) datasets in the case of utilising and replicating the approach for a different city.
Vienna local plants (CSV): Plants, metadata, and solar requirements.
GBIF citizen science (API): Local species filtered based on (proximity to) geographical coordinates.
Plant Functional Groups (CSV): Internal dataset generated within ECOLOPES via the
Ecological Model [
10].
Animal Functional Groups (JSON): Internal dataset generated within ECOLOPES via the Ecological Model.
Animal-aided design (PDF): Specifying the constraints between species and various architectural objects, used as a placeholder.
GLoBI (API): Global biotic interactions between species described in OBO Relations.
GBIF backbone species (RDF): Query the federation of species, including metadata details such as taxonomic rank described in Darwin Core and GBIF backbone taxonomy (
http://graph.openbiodiv.net (accessed on 17 March 2025) [
44]).
Wikidata (RDF): Query the federation of species, including metadata details, synonyms, common names, crossovers (identifiers) to many datasets (DBpedia, Catalogue of Life, and so forth) (
https://query.wikidata.org (accessed on 17 March 2025)).
Nature FIRST (RDF) [45]: Query the federation of species and habitats, including IUCN protection status.
CAD environment: Rhino CAD and Grasshopper environments.
Voxel model (RDB-RDF): Query the federation of the voxel model, storing site-specific environment variables in RDB.
Heterogeneous data are mapped and converted into a graph representation in the KG (ABox) by using different mapping languages, such as RML [
29], Ontotext Refine, and OBDA (
https://graphdb.ontotext.com/documentation/10.8/virtualization.html (accessed on 17 March 2025)). Shapefiles are converted into RDF using RML mappings that are generated by GeoTriples (ver. 1.1.6) [
42]. For this dataset (including GBIF, which can be transformed in this representation; see Example 1), the triples are indexed by the triple store and can be queried and reasoned using GeoSPARQL. The user can query the KG using (Geo)SPARQL as a unified interface that encompasses and integrates different datasets combining the results, and, if required, the user can use reasoning on top using TBox statements. Therefore, the KG integrates heterogeneous data from different sources, providing unified access to the data by using the SPARQL (or SPARQL*) query language.
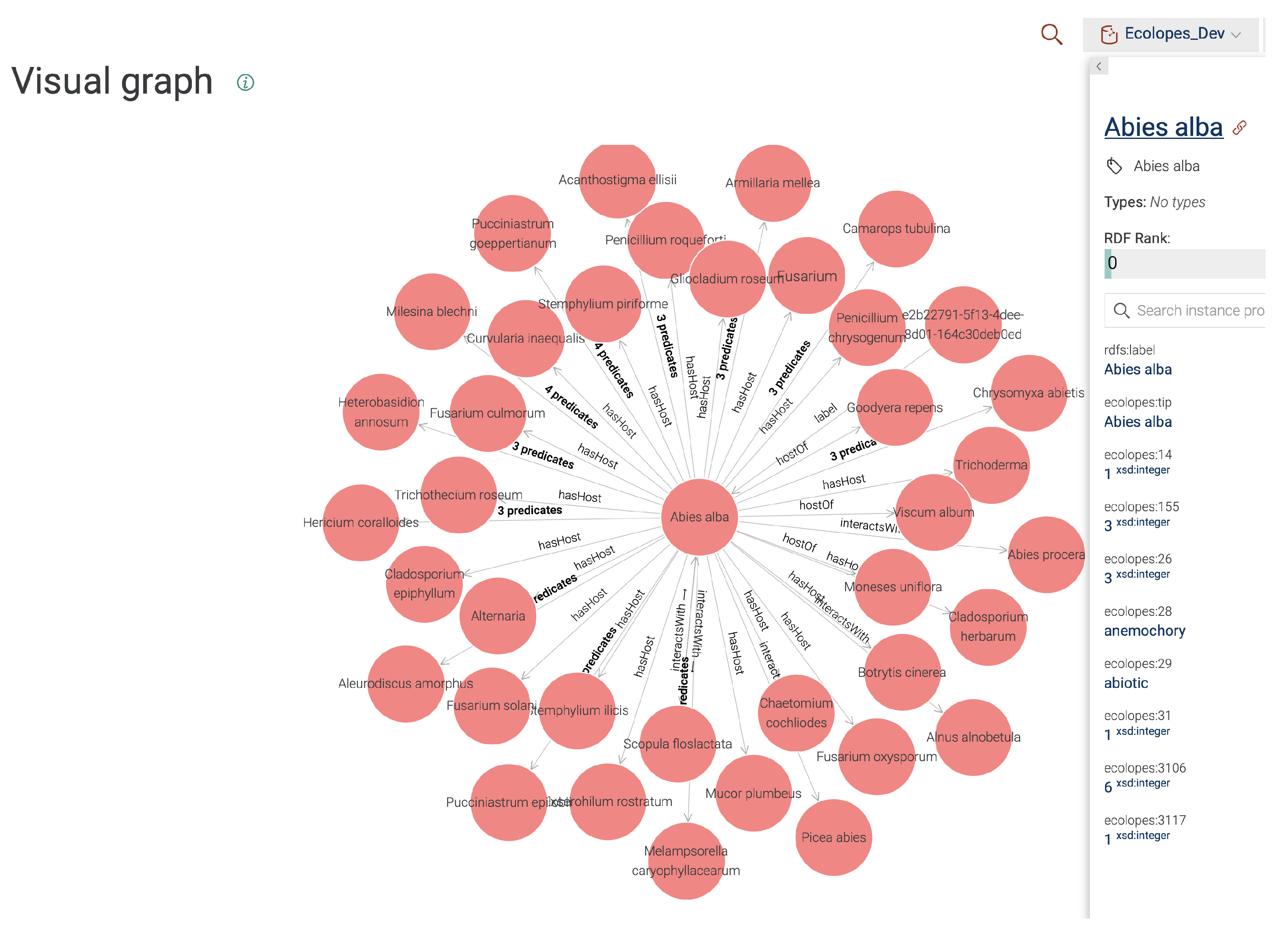
Figure 8 shows a snapshot of the core parts of the KG, which links knowledge from datasets originated in Grasshopper (left), global biotic interactions and proximity constraints (centre), and the voxel model (top) and PFGs (right). One can traverse from one context to another, given that the relations between datasets are explicitly encoded (e.g.,
owl:sameAs), or they can be inferred by SPARQL queries (e.g., if x,y,z coordinates in between datasets coincide). Whenever possible for the IRIs of species, we treat Wikidata identifiers as canonical IRIs (we mention [
46] as an approach that tackles canonical IRIs in the setting of virtual mappings and OBDM; in our case, this part of the data is materialised) and provide crossovers via
owl:sameAs to other linked datasets. This pragmatic approach allows us to start from a central entity directed towards other entities when browsing or querying species data, as well as for convenience reasons, i.e., when using reasoning support via
owl:sameAs to infer all the statements for the linked species during repository loading time (alternatively, the latter can be computed via query rewriting during query time by using path queries in SPARQL
owl:sameAs*).
Example 2. In the following, we see an example of how the requirements for the proximity between the species Abies alba and Scopula floslactata have been modelled in RDF* to represent n-ary relations in a succinct way, which are stored in GraphDB. The predicates proximityRequirementMin and proximityRequirementMax have been used to set the minimum and maximum allowed value between Abies alba and Scopula floslactata. The visualisation of RDF* triples is depicted in Figure 8 (dashed line within the centre environment/container). <<ecolopes-inst:Abies_alba ecolopes-schema:proximityDistance
ecolopes-inst:Scopula_floslactata>>
ecolopes-schema:proximityRequirementMin "20"^^xsd:integer;
ecolopes-schema:proximityRequirementMax "100"^^xsd:integer .
In the following, we describe the process used to harvest data from GLoBI to obtain all (biotic) interactions between designated species. The workflow begins by specifying a list of plant or animal species as a seed. This initial list can either be derived from geographic coordinates (e.g., by retrieving local species using the GBIF API) or manually selected based on species relevant to our context in Vienna. Once the species list is established, we utilise the GLoBI API to retrieve interaction data in the form of CSV files, with one file generated per species.
The next step involves transforming the CSV data into the RDF format. To achieve this, we use Ontotext Refine to create the necessary mappings via a graphical user interface (GUI). These mappings are subsequently exported as JSON files (cf.
Figure 9). Using the mapping file as input, along with the corresponding species CSV files, we invoke the Ontotext Refine API to generate RDF data in batch mode for each species. Given that the CSV schema returned by the GLoBI API remains consistent, we can reliably apply the same set of mappings to produce RDF representations of species interactions.
To enhance interoperability and enable reasoning over implicit triples, we aligned the interaction properties with the OBO Relations ontology (cf.
Figure 4). This alignment ensures that the RDF output adheres to established semantic standards, facilitating integration with other biodiversity and ecological datasets.
In
Figure 10, we see the visualisation of the node
Abies alba containing integrated data, along with its metadata panel on the right side, and the biotic interactions (
:hasHost,
:interactsWith) with respect to other species.
6. Demonstrator
In the following section, we demonstrate the application and utility of the ontology and KG in a design-centric case study in Vienna, located on the Nordbahnhof Freie Mitte site. More details on the case study and on the development of the respective network configuration are presented in a dedicated study [
20]. The main result of the demonstrator is to showcase the expressiveness and capture of the higher-level requirements defined in
Section 4.2 by using a network configuration in the CAD environment (cf.
Figure 3). Local species around the site are curated and provided by the Vienna municipality, and in addition, they can be obtained from GBIF API (cf.
Figure 11)—albeit with the common data quality issues encountered in the common citizen science approaches. The case study can be replicated to any other case study in a different location, provided that the local data are available. In this demonstration experiment, the widely used McNeel Rhinoceros 3D and the Grasshopper 3D platform were used. This platform consists of a 3D CAD environment (Rhinoceros 3D) that is interactively linked with a domain-specific node-based programming interface (Grasshopper 3D). Dedicated ETL workflows can be implemented within this node-based programming interface, using both native Grasshopper components and custom functionalities, e.g., as Python scripts encapsulated using the native
Grasshopper Python Script component. Since the Grasshopper 3D platform does not contain any components that can interact with ontologies or the GraphDB platform, the required functionalities were implemented as Python scripts in the Grasshopper environment. This implementation was based on the GraphDB REST API specification and the underlying RDF4J SPARQL endpoint.
The pipeline (cf.
Figure 12) that we describe in the demonstration section consists of various steps, starting with cleaning and mapping the designer network created by the end-users (designers) within the Rhinoceros 3D CAD environment. The designer network consists of nodes (cf.
Figure 3) represented both visually in the 3D CAD environment and mapped to the JSON-LD schema (
https://www.w3.org/TR/json-ld11/ (accessed on 17 March 2025)) within the Grasshopper platform. The implemented JSON-LD serialisation prepares the 3D CAD data to be pushed to GraphDB through the embedded RDF4J SPARQL endpoint. The implemented REST-based interface, consisting of the JSON-LD parser and GraphDB SPARQL endpoint, works bi-directionally, allowing the end-users to receive the data returned by the SPARQL endpoint, formatted in compliance with the native Grasshopper data typing and formatting. From the end-user perspective, the results of the SPARQL-based reasoning can be used in the Grasshopper-native ETL workflows. From the perspective of ontology engineering, data received from the Grasshopper platform can be validated and used for SPARQL-based reasoning. In the current implementation, before every run of the Grasshopper pipeline, the data were cleaned by calling
DROP GRAPH from the Grasshopper environment. This ensured that no stale data representing the past geometric condition of the 3D CAD environment were present in the knowledge graph. The rationale for choosing JSON-LD lies in the fact that JSON-LD is compatible with JSON, and the Grasshopper 3D platform features limited compatibility with JSON data streams through the jSwan Grasshopper plugin.
6.1. Mapping of Designer Network Data and Voxel Data Layers in EIM Ontology
The ECOLOPES Voxel Model contains environmental data describing the Viennese site of Nordbahnhof Freie Mitte. sunlightHoursSummer and sunlightHoursGrowingSeason properties map Voxel Data Layers to the respective attributes in the EIM Onology. For instance, the designer places an EcoNode, defines its subType as Plant, and sets dwc:vernacularName as “Silver fir”. The KG stores mappings between dwc:vernacularName (“Silver fir”) and dwc:scientificName (e.g., Abies alba), along with additional plant datasets and respective metadata. Consequently, sunlight requirements (minSunHours and maxSunHours) are retrieved for the given EcoNode.
6.2. Validation Process for EcoNodes and ArchiNodes
The presented functionality, showcasing the interaction between end-users, ECOLOPES Voxel Model, and knowledge graph utilises the GraphDB SPARQL endpoint to compare the
sunlightHoursGrowingSeason property retrieved from the ECOLOPES Voxel Model with the
minSunHours and
maxSunHours properties queried from the knowledge graph. This query returns a Boolean value (
?isValid) as the result of the comparison. In the designer networks workflow, the designer places an ArchiNode and defines its
subType as
Infrastructure or
Building. Similar validation and mapping processes are outlined for ArchiNodes, including the prey area that is predefined by the designer as a rectangular shape. The rule-based process is based on CONSTRUCT queries [
19] for the respective solar, proximity, and prey area constraints. In
Figure 13, one can see the query for the proximity constraint. The CONSTRUCT query is transformed into an INSERT query in order for the validation data to not only be computed but also materialised. The query takes into account multiple proximity constraints between different nodes
n in the range of
computational complexity. Variable ?res computes the
Euclidian distance between nodes using GraphDB’s math functions (
https://graphdb.ontotext.com/documentation/10.8/sparql-ext-functions-reference.html#mathematical-function-extensions (accessed on 17 March 2025)).
6.3. Preparing and Sending Network Data
At the beginning of the designer network workflow, the end-user inputs are read as data stored as key-value pairs, assigned to individual Rhinoceros 3D (ver. 7.3) objects representing network nodes. The 3D CAD environment provides the functionality to store key-value pairs as User Attributes attached to any native 3D CAD object, such as Rhino Point 3D. Moreover, the custom User Attributes are exposed in the Grasshopper 3D (ver. 7.3) interface and can be used by designers in the Grasshopper node-based programming interface. For this workflow, two custom Rhino commands,
PlaceArchiNode and
PlaceEcoNode (cf.
Figure 14), were implemented in Python (ver. 2.7), allowing the designers to place native Point 3D objects containing the required attributes and their values. The designers can construct networks in the 3D CAD environment, consisting of nodes, and the implemented Grasshopper pipeline converts and outputs data to JSON-LD, matching the ontology (TBox) schema. The developed Grasshopper components send the JSON-LD formatted Network Node data to the GraphDB (ver. 10.4) environment through the SPARQL endpoint.
6.4. Define Evaluation Constraints
The last part of the demonstrator pipeline enables the designer to select constraints to validate the
network configuration created in the 3D CAD environment. From within the Grasshopper interface, end-users can choose the Solar and Proximity constraints to be validated against, cf.
Figure 15, for visualisation of the validation, which is stored in GraphDB. The final output is a Boolean answer that is computed in GraphDB by the
AND (“join”) intersection of constraints, which mimics using the
MIN operator in the SPARQL query. The results of this validation are returned to the Grasshopper pipeline and visualised in the Rhinoceros 3D CAD interface. This mechanism provides visual feedback to the end-user, indicating which nodes in the designer networks do not fulfil the constraints inferred from the ontologies using SPARQL-based reasoning.
7. Conclusions and Future Work
This paper presents an ontology-driven approach to integrate diverse datasets—ranging from public APIs and endpoints and internal and local sources to CAD environments—into a unified RDF-based knowledge graph, facilitating the design of sites as well as fostering ecological building design. By leveraging OBDM, the approach balances virtualisation and materialisation, ensuring efficient data integration while supporting advanced queries, updates, and reasoning. The ECOLOPES ontologies and knowledge graph bridge the life sciences, geography, and geometric data, enabling designers to address the ecological constraints of solar radiation, proximity, and prey areas through interactive workflows. The application of OBDM has been demonstrated in the case of computing the permissible positions on a site where one can place a plant species (EcoNode) based on solar radiation and the plant’s requirements. Furthermore, this was extended to a case study comprising a network of Archi- and EcoNodes, where additional requirement constraints are applied, namely proximity and prey areas (cf.
Section 4.2). Through the application of a network configuration of nodes and the respective expressivity of the higher-level constraints, the case study highlighted the ontology’s practical application in designing habitats that promote species attractiveness and urban biodiversity.
The advantage of the proposed approach is that it is flexible, as different constraints have already been implemented and can be plugged in using Grasshopper components. Nevertheless, the addition of other constraints is possible to the extent that their implementation is provided using the SPARQL update queries. The latter limits the expressivity of the constraints subject to the expressivity of the SPARQL language [
19,
48] and its extensions for expressing those constraints. The next step of joining the constraints can be simply carried out using the implementation provided in this paper via the off-the-shelf Grasshopper components. The adoption of the workflow by the community is made possible due to the encapsulation of the complexity in Grasshopper components, through which different constraints can be enabled or disabled.
The problem setting is limited to the translational process of gathering the requirements from the design brief and creating the network of nodes. This translation (or mapping) is made by the designer subjectively. The automatic generation of nodes, which is out of the scope of this work, could possibly employ Generative AI in conjunction with the proposed ontologies and knowledge graph.
It is worth emphasising that the validation of the approach from the ecological point of view is left for future work. Furthermore, we plan to use SHACL to define constraints, as well as use a bespoke rule language (e.g., Datalog or SWRL (
https://www.w3.org/submissions/SWRL/ (accessed on 17 March 2025))) instead of SPARQL queries in order to adhere to Semantic Web standards. More importantly, we plan to extend the problem setting towards a generative process (geometric articulation) in ontology-aided generative computational design by employing more sophisticated approaches akin to answer set programming [
49,
50].