Epileptic Seizure Detection in Neonatal EEG Using a Multi-Band Graph Neural Network Model
Abstract
1. Introduction
2. Database
3. Methods
3.1. Preprocessing
3.2. Graph Generation
3.2.1. Decomposition
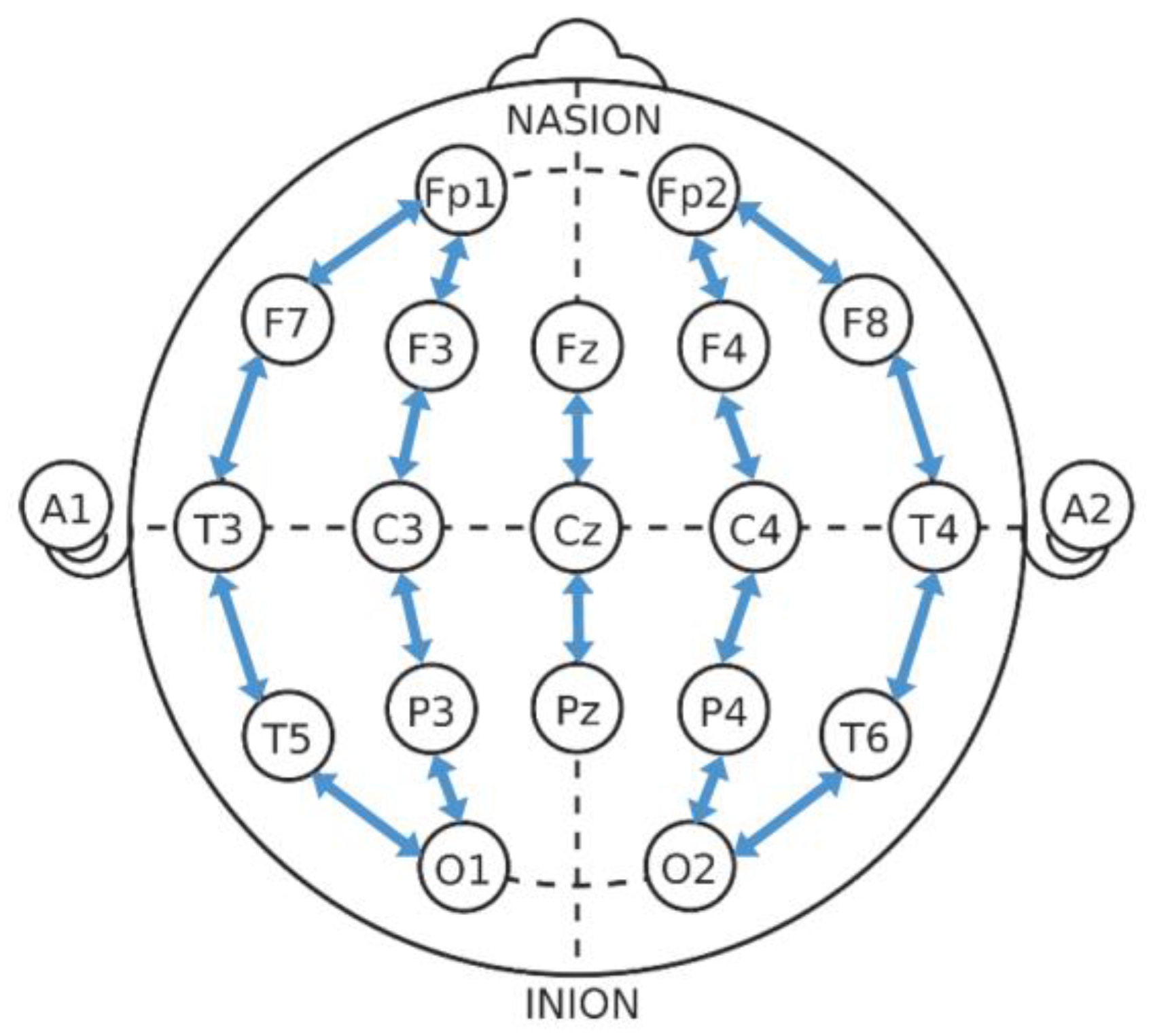
3.2.2. Graph Edge Generation
3.2.3. Graph Node Generation
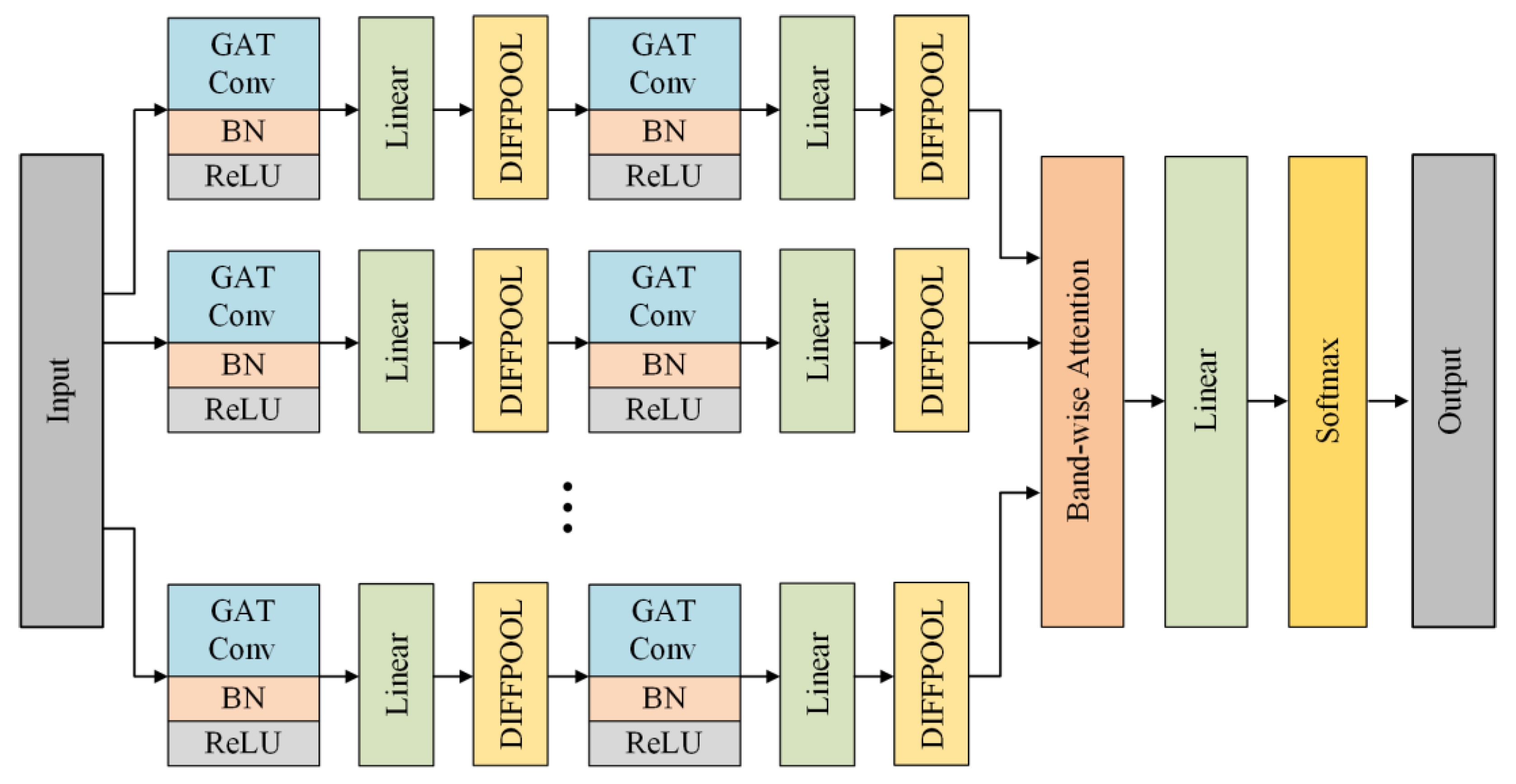
3.3. Multi-Band Graph Neural Network
3.3.1. Graph Convolution
3.3.2. Graph Pooling
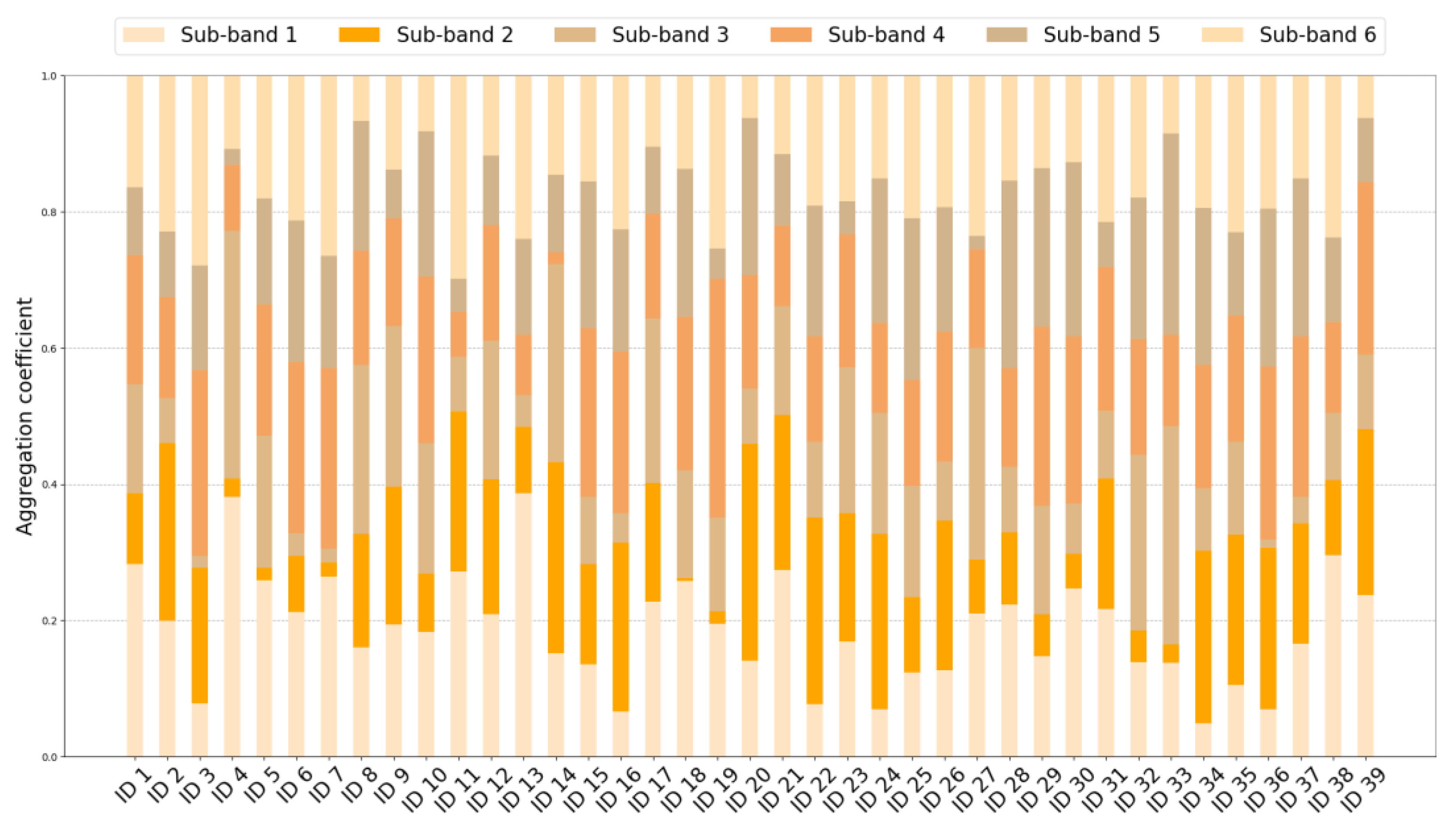
3.3.3. Graph Aggregation
4. Experimental Results
- Area under the receiver operating characteristic curve (AUC): The receiver operating characteristic (ROC) curve is a plot of sensitivity versus (1-specificity) and the AUC illustrates the performance of a binary classifier with varied decision threshold. The AUC values range from “0” to “1”, and an ideal classifier will have an AUC equal to “1”;
- Positive predictive value (PPV): The PPV is the probability that epochs with positive labels are truly seizure epochs. In this paper, the PPV illustrates the ability of a classifier to capture the real seizure epochs;
- Negative predictive value (NPV): The NPV is the probability that epochs with negative labels are truly non-seizure epochs. In this paper, the NPV illustrates the ability of a classifier to avoid false alarms.
5. Discussion
| Author | Subject | Method | Validation | AUC (%) |
|---|---|---|---|---|
| Tapani et al. [9] | 79 | SVM | Leave-one-out cross-validation | 95.7 |
| O’Shea et al. [15] | 79 | 2D CNN | Leave-one-out cross-validation | 95.6 |
| Caliskan et al. [39] | 39 | Pretrained DCNN | Training (50%) and test (50%) | 99.2 |
| Tanveer et al. [16] | 39 | Ensemble 2D CNN | 10-fold cross-validation | 99.3 |
| Abbas et al. [41] | 14 | t-test and ROC analysis | - | 77.9 |
| Raeisi et al. [19] | 39 | GCNN | Leave-one-out cross-validation | 94.7 |
| Diykh et al. [42] | 39 | Ensemble classifiers | Leave-one-out cross-validation | 94.0 |
| Zhou et al. [40] | 15 | LMA-EEGNet | 5-fold cross-validation | 98.6 |
| Nelson et al. [20] | - | SSGNN | K-fold cross-validation | 97.5 |
| Proposed method | 39 | MBGNN | 10-fold cross-validation | 99.1 |
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bye, A.M.E.; Flanagan, D. Spatial and Temporal Characteristics of Neonatal Seizures. Epilepsia 1995, 36, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Wietstock, S.O.; Bonifacio, S.L.; Sullivan, J.E.; Nash, K.B.; Glass, H.C. Continuous Video Electroencephalographic (EEG) Monitoring for Electrographic Seizure Diagnosis in Neonates: A Single-Center Study. J. Child Neurol. 2016, 31, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Hahn, J.S.; Heldt, G.P.; Coen, R.W. Detection of Neonatal Seizures through Computerized EEG Analysis. Electroencephalogr. Clin. Neurophysiol. 1992, 82, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Gotman, J.; Flanagan, D.; Zhang, J.; Rosenblatt, B. Automatic Seizure Detection in the Newborn: Methods and Initial Evaluation. Electroencephalogr. Clin. Neurophysiol. 1997, 103, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Navakatikyan, M.A.; Colditz, P.B.; Burke, C.J.; Inder, T.E.; Richmond, J.; Williams, C.E. Seizure Detection Algorithm for Neonates Based on Wave-Sequence Analysis. Clin. Neurophysiol. 2006, 117, 1190–1203. [Google Scholar] [CrossRef]
- Deburchgraeve, W.; Cherian, P.J.; De Vos, M.; Swarte, R.M.; Blok, J.H.; Visser, G.H.; Govaert, P.; Van Huffel, S. Automated Neonatal Seizure Detection Mimicking a Human Observer Reading EEG. Clin. Neurophysiol. 2008, 119, 2447–2454. [Google Scholar] [CrossRef]
- Thomas, E.M.; Temko, A.; Lightbody, G.; Marnane, W.P.; Boylan, G.B. Gaussian Mixture Models for Classification of Neonatal Seizures Using EEG. Physiol. Meas. 2010, 31, 1047–1064. [Google Scholar] [CrossRef]
- Temko, A.; Thomas, E.; Marnane, W.; Lightbody, G.; Boylan, G. EEG-Based Neonatal Seizure Detection with Support Vector Machines. Clin. Neurophysiol. 2011, 122, 464–473. [Google Scholar] [CrossRef]
- Tapani, K.T.; Vanhatalo, S.; Stevenson, N.J. Time-Varying EEG Correlations Improve Automated Neonatal Seizure Detection. Int. J. Neural Syst. 2019, 29, 1850030. [Google Scholar] [CrossRef]
- Zhou, M.; Duan, N.; Liu, S.; Shum, H.-Y. Progress in Neural NLP: Modeling, Learning, and Reasoning. Engineering 2020, 6, 275–290. [Google Scholar] [CrossRef]
- Nazir, S.; Dickson, D.M.; Akram, M.U. Survey of Explainable Artificial Intelligence Techniques for Biomedical Imaging with Deep Neural Networks. Comput. Biol. Med. 2023, 156, 106668. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.Y.; Abu-Salih, B.; Qaddoura, R.; Al-Zoubi, A.M.; Palade, V.; Pham, D.-S.; Ser, J.D.; Muhammad, K. Deep Neural Networks in the Cloud: Review, Applications, Challenges and Research Directions. Neurocomputing 2023, 545, 126327. [Google Scholar] [CrossRef]
- Islam, M.M.; Nooruddin, S.; Karray, F.; Muhammad, G. Human Activity Recognition Using Tools of Convolutional Neural Networks: A State of the Art Review, Data Sets, Challenges, and Future Prospects. Comput. Biol. Med. 2022, 149, 106060. [Google Scholar] [CrossRef]
- Ansari, A.H.; Cherian, P.J.; Caicedo, A.; Naulaers, G.; Vos, M.D.; Huffel, S.V. Neonatal Seizure Detection Using Deep Convolutional Neural Networks. Int. J. Neural Syst. 2019, 29, 1850011. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, A.; Lightbody, G.; Boylan, G.; Temko, A. Neonatal Seizure Detection from Raw Multi-Channel EEG Using a Fully Convolutional Architecture. Neural Netw. 2020, 123, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Tanveer, M.A.; Khan, M.J.; Sajid, H.; Naseer, N. Convolutional Neural Networks Ensemble Model for Neonatal Seizure Detection. J. Neurosci. Methods 2021, 358, 109197. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, C.; Zhang, G.; Wang, Y.; Chen, X.; Jia, W.; Yuan, Q.; Xu, F.; Zheng, Y. EEG-Based Seizure Detection Using Linear Graph Convolution Network with Focal Loss. Comput. Methods Programs Biomed. 2021, 208, 106277. [Google Scholar] [CrossRef]
- Zeng, D.; Huang, K.; Xu, C.; Shen, H.; Chen, Z. Hierarchy Graph Convolution Network and Tree Classification for Epileptic Detection on Electroencephalography Signals. IEEE Trans. Cogn. Dev. Syst. 2021, 13, 955–968. [Google Scholar] [CrossRef]
- Raeisi, K.; Khazaei, M.; Croce, P.; Tamburro, G.; Comani, S.; Zappasodi, F. A Graph Convolutional Neural Network for the Automated Detection of Seizures in the Neonatal EEG. Comput. Methods Programs Biomed. 2022, 222, 106950. [Google Scholar] [CrossRef]
- Nelson, M.; Rajendran, S.; Khalaf, O.I.; Hamam, H.; Nelson, M.; Rajendran, S.; Khalaf, O.I.; Hamam, H. Deep-Learning-Based Intelligent Neonatal Seizure Identification Using Spatial and Spectral GNN Optimized with the Aquila Algorithm. MATH 2024, 9, 19645–19669. [Google Scholar] [CrossRef]
- Stevenson, N.J.; Tapani, K.; Lauronen, L.; Vanhatalo, S. A Dataset of Neonatal EEG Recordings with Seizure Annotations. Sci. Data 2019, 6, 190039. [Google Scholar] [CrossRef]
- Song, M.; Li, L.; Guo, J.; Liu, T.; Li, S.; Wang, Y.; Qurat Ul Ain; Wang, J. A New Method for Muscular Visual Fatigue Detection Using Electrooculogram. Biomed. Signal Process. Control 2020, 58, 101865. [Google Scholar] [CrossRef]
- Safder, S.N.-U.-H.; Akram, M.U.; Dar, M.N.; Khan, A.A.; Khawaja, S.G.; Subhani, A.R.; Niazi, I.K.; Gul, S. Analysis of EEG Signals Using Deep Learning to Highlight Effects of Vibration-Based Therapy on Brain. Biomed. Signal Process. Control 2023, 83, 104605. [Google Scholar] [CrossRef]
- Faust, O.; Acharya, U.R.; Adeli, H.; Adeli, A. Wavelet-Based EEG Processing for Computer-Aided Seizure Detection and Epilepsy Diagnosis. Seizure 2015, 26, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Light, G.A.; Hsu, J.L.; Hsieh, M.H.; Meyer-Gomes, K.; Sprock, J.; Swerdlow, N.R.; Braff, D.L. Gamma Band Oscillations Reveal Neural Network Cortical Coherence Dysfunction in Schizophrenia Patients. Biol. Psychiatry 2006, 60, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Uhlhaas, P.J.; Singer, W. Neural Synchrony in Brain Disorders: Relevance for Cognitive Dysfunctions and Pathophysiology. Neuron 2006, 52, 155–168. [Google Scholar] [CrossRef]
- Huang, N.E.; Shen, Z.; Long, S.R.; Wu, M.C.; Shih, H.H.; Zheng, Q.; Yen, N.C.; Tung, C.C.; Liu, H.H. The empirical mode decomposition and the Hilbert spectrum for nonlinear and non-stationary time series analysis. Proc. R. Soc. London. Ser. Math. Phys. Eng. Sci. 1998, 454, 903–995. [Google Scholar] [CrossRef]
- Batista, G.E.A.P.A.; Keogh, E.J.; Tataw, O.M.; de Souza, V.M.A. CID: An Efficient Complexity-Invariant Distance for Time Series. Data Min. Knowl. Disc. 2014, 28, 634–669. [Google Scholar] [CrossRef]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph Attention Networks. arXiv 2018, arXiv:1710.10903. [Google Scholar] [CrossRef]
- Hechtlinger, Y.; Chakravarti, P.; Qin, J. A Generalization of Convolutional Neural Networks to Graph-Structured Data. arXiv 2017, arXiv:1704.08165. [Google Scholar] [CrossRef]
- Hamilton, W.L.; Ying, R.; Leskovec, J. Inductive Representation Learning on Large Graphs. arXiv 2017, arXiv:1706.02216. [Google Scholar] [CrossRef]
- Ying, R.; You, J.; Morris, C.; Ren, X.; Hamilton, W.L.; Leskovec, J. Hierarchical Graph Representation Learning with Differentiable Pooling. Adv. Neural Inf. Process. Syst. 2018, 31. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Fluss, R.; Faraggi, D.; Reiser, B. Estimation of the Youden Index and Its Associated Cutoff Point. Biom. J. 2005, 47, 458–472. [Google Scholar] [CrossRef]
- Shi, S. Visualizing Data Using GTSNE. arXiv 2021, arXiv:2108.01301. [Google Scholar] [CrossRef]
- Usman, S.M.; Usman, M.; Fong, S. Epileptic Seizures Prediction Using Machine Learning Methods. Comput. Math. Methods Med. 2017, 9074759. [Google Scholar] [CrossRef]
- Emami, A.; Kunii, N.; Matsuo, T.; Shinozaki, T.; Kawai, K.; Takahashi, H. Seizure Detection by Convolutional Neural Network-Based Analysis of Scalp Electroencephalography Plot Images. NeuroImage Clin. 2019, 22, 101684. [Google Scholar] [CrossRef]
- Açıkoğlu, M.; Tuncer, S.A. Incorporating Feature Selection Methods into a Machine Learning-Based Neonatal Seizure Diagnosis. Med. Hypotheses 2020, 135, 109464. [Google Scholar] [CrossRef]
- Caliskan, A.; Rencuzogullari, S. Transfer Learning to Detect Neonatal Seizure from Electroencephalography Signals. Neural Comput. Applic. 2021, 33, 12087–12101. [Google Scholar] [CrossRef]
- Zhou, W.; Zheng, W.; Feng, Y.; Li, X. LMA-EEGNet: A Lightweight Multi-Attention Network for Neonatal Seizure Detection Using EEG Signals. Electronics 2024, 13, 2354. [Google Scholar] [CrossRef]
- Abbas, A.K.; Azemi, G.; Ravanshadi, S.; Omidvarnia, A. An EEG-Based Methodology for the Estimation of Functional Brain Connectivity Networks: Application to the Analysis of Newborn EEG Seizure. Biomed. Signal Process. Control 2021, 63, 102229. [Google Scholar] [CrossRef]
- Diykh, M.; Miften, F.S.; Abdulla, S.; Deo, R.C.; Siuly, S.; Green, J.H.; Oudahb, A.Y. Texture Analysis Based Graph Approach for Automatic Detection of Neonatal Seizure from Multi-Channel EEG Signals. Measurement 2022, 190, 110731. [Google Scholar] [CrossRef]



| ID | TL 1 (s) | SL 2 (s) | CSL 3 (s) | ||
|---|---|---|---|---|---|
| Expert 1 | Expert 2 | Expert 3 | |||
| 1 | 6993 | 1602 | 3141 | 997 | 745 |
| 2 | 3425 | 925 | 1161 | 987 | 850 |
| 3 | 3841 | 3325 | 3189 | 3184 | 3136 |
| 4 | 3654 | 625 | 970 | 608 | 605 |
| 5 | 3550 | 882 | 1041 | 862 | 862 |
| 6 | 7488 | 99 | 127 | 40 | 40 |
| 7 | 15,416 | 1271 | 1421 | 1346 | 1235 |
| 8 | 3726 | 2142 | 2413 | 2314 | 2084 |
| 9 | 6898 | 1471 | 573 | 1308 | 425 |
| 10 | 5941 | 620 | 4247 | 1355 | 242 |
| 11 | 5493 | 171 | 583 | 60 | 20 |
| 12 | 9006 | 2204 | 2679 | 2182 | 2089 |
| 13 | 3987 | 751 | 1794 | 1054 | 639 |
| 14 | 5707 | 43 | 42 | 39 | 39 |
| 15 | 3844 | 697 | 1379 | 640 | 311 |
| 16 | 6709 | 284 | 798 | 73 | 43 |
| 17 | 3529 | 182 | 181 | 168 | 167 |
| 18 | 6491 | 455 | 452 | 452 | 452 |
| 19 | 5082 | 492 | 532 | 451 | 451 |
| 20 | 6095 | 2983 | 4505 | 2864 | 2681 |
| 21 | 4629 | 2259 | 2517 | 2229 | 2177 |
| 22 | 5837 | 601 | 627 | 513 | 79 |
| 23 | 9684 | 8081 | 9450 | 8542 | 7856 |
| 24 | 3360 | 341 | 397 | 335 | 315 |
| 25 | 3606 | 211 | 312 | 201 | 200 |
| 26 | 9850 | 930 | 898 | 902 | 823 |
| 27 | 4701 | 385 | 320 | 491 | 291 |
| 28 | 3920 | 119 | 100 | 86 | 86 |
| 29 | 5852 | 382 | 390 | 380 | 380 |
| 30 | 3900 | 373 | 1041 | 2040 | 344 |
| 31 | 11,350 | 1738 | 1878 | 1548 | 1548 |
| 32 | 4900 | 1424 | 1648 | 1576 | 1342 |
| 33 | 3973 | 2268 | 3455 | 2716 | 2250 |
| 34 | 3730 | 868 | 1140 | 1047 | 809 |
| 35 | 3955 | 920 | 925 | 918 | 918 |
| 36 | 3821 | 477 | 600 | 429 | 375 |
| 37 | 4193 | 258 | 462 | 520 | 256 |
| 38 | 4971 | 2216 | 2513 | 2011 | 1915 |
| 39 | 3297 | 198 | 475 | 242 | 179 |
| Layer | Parameters | Sizes | |
|---|---|---|---|
| Input | Node: 6 × 19 × 11 | - | |
| Edge: 6 × 19 × 19 | |||
| Six Channels | GAT Convolution 1 | Hidden Channels: 32 | Conv Weight: 32 × 11 |
| Conv Bias: 32 | |||
| Batch Normalization 1 | Batch Size: 32 | Batch Norm Weight: 32 | |
| Batch Norm Bias: 32 | |||
| Linear 1 | Node Number: 6 | Assignment Weight: 6 × 32 | |
| DIFFPOOL 1 | Assignment Bias: 6 | ||
| GAT Convolution 2 | Hidden Channels: 32 | Conv Weight: 32 × 32 | |
| Conv Bias: 32 | |||
| Batch Normalization 2 | Batch Size: 32 | Batch Norm Weight: 32 | |
| Batch Norm Bias: 32 | |||
| Linear 2 | Node Number: 1 | Assignment Weight: 1 × 32 | |
| DIFFPOOL 2 | Assignment Bias: 1 | ||
| Attention Layer | - | Attention Weight: 1 × 6 | |
| Attention Bias: 1 | |||
| Linear 3 | Hidden Channels: 2 | Linear Weight: 2 × 32 Linear Bias: 2 | |
| Softmax | - | - | |
| ID | AUC(%) | PPV(%) | NPV(%) |
|---|---|---|---|
| 1 | 99.10 ± 0.13 | 94.90 ± 1.22 | 96.09 ± 0.90 |
| 2 | 99.07 ± 0.17 | 95.69 ± 1.10 | 95.26 ± 1.00 |
| 3 | 96.78 ± 0.32 | 90.07 ± 1.49 | 90.72 ± 1.88 |
| 4 | 99.19 ± 0.15 | 94.80 ± 1.11 | 96.83 ± 1.01 |
| 5 | 99.81 ± 0.05 | 98.01 ± 0.48 | 98.01 ± 0.57 |
| 6 | 99.99 ± 0.01 | 94.38 ± 0.81 | 99.90 ± 0.10 |
| 7 | 99.78 ± 0.06 | 97.54 ± 0.55 | 98.24 ± 0.18 |
| 8 | 99.12 ± 0.11 | 96.76 ± 0.79 | 94.00 ± 0.68 |
| 9 | 99.64 ± 0.06 | 98.09 ± 0.53 | 97.10 ± 0.58 |
| 10 | 99.83 ± 0.04 | 97.95 ± 0.67 | 98.05 ± 0.36 |
| 11 | 99.84 ± 0.24 | 85.34 ± 3.42 | 98.80 ± 1.78 |
| 12 | 99.36 ± 0.06 | 94.94 ± 0.72 | 97.11 ± 0.68 |
| 13 | 94.16 ± 0.53 | 81.58 ± 1.53 | 91.28 ± 0.91 |
| 14 | 99.99 ± 0.01 | 93.78 ± 1.13 | 99.94 ± 0.05 |
| 15 | 1.00 ± 0.00 | 99.33 ± 0.03 | 99.78 ± 0.12 |
| 16 | 99.91 ± 0.06 | 94.12 ± 0.94 | 99.25 ± 0.43 |
| 17 | 99.84 ± 0.04 | 97.63 ± 1.19 | 97.70 ± 1.03 |
| 18 | 1.00 ± 0.00 | 99.55 ± 0.02 | 1.00 ± 0.00 |
| 19 | 99.99 ± 0.01 | 99.33 ± 0.23 | 99.66 ± 0.11 |
| 20 | 95.26 ± 0.30 | 86.36 ± 2.25 | 89.76 ± 2.11 |
| 21 | 99.81 ± 0.03 | 98.19 ± 0.44 | 97.82 ± 0.63 |
| 22 | 99.85 ± 0.03 | 97.11 ± 0.45 | 97.76 ± 0.65 |
| 23 | 93.24 ± 0.37 | 87.68 ± 1.43 | 83.72 ± 1.74 |
| 24 | 99.91 ± 0.03 | 99.05 ± 0.33 | 98.31 ± 0.65 |
| 25 | 99.95 ± 0.03 | 98.53 ± 0.46 | 99.20 ± 0.29 |
| 26 | 99.70 ± 0.09 | 97.27 ± 0.67 | 98.59 ± 0.56 |
| 27 | 99.85 ± 0.04 | 97.62 ± 0.66 | 98.45 ± 0.36 |
| 28 | 99.83 ± 0.06 | 96.25 ± 1.23 | 98.78 ± 0.36 |
| 29 | 1.00 ± 0.00 | 99.45 ± 0.02 | 99.70 ± 0.08 |
| 30 | 99.03 ± 0.20 | 92.75 ± 0.53 | 96.50 ± 0.74 |
| 31 | 99.55 ± 0.07 | 95.98 ± 0.52 | 97.43 ± 0.21 |
| 32 | 99.72 ± 0.07 | 98.06 ± 0.40 | 97.71 ± 0.51 |
| 33 | 99.20 ± 0.11 | 94.69 ± 0.81 | 97.26 ± 0.97 |
| 34 | 99.32 ± 0.16 | 95.77 ± 0.65 | 96.92 ± 0.49 |
| 35 | 98.77 ± 0.12 | 95.86 ± 0.69 | 93.22 ± 0.45 |
| 36 | 98.80 ± 0.15 | 95.69 ± 1.40 | 93.01 ± 1.23 |
| 37 | 99.25 ± 0.18 | 97.66 ± 0.95 | 94.33 ± 0.82 |
| 38 | 99.04 ± 0.11 | 93.48 ± 0.57 | 96.32 ± 0.32 |
| 39 | 99.68 ± 0.09 | 97.20 ± 1.17 | 97.26 ± 0.61 |
| Avg. | 99.11 ± 1.55 | 95.34 ± 4.18 | 96.66 ± 3.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; Zhao, M. Epileptic Seizure Detection in Neonatal EEG Using a Multi-Band Graph Neural Network Model. Appl. Sci. 2024, 14, 9712. https://doi.org/10.3390/app14219712
Tang L, Zhao M. Epileptic Seizure Detection in Neonatal EEG Using a Multi-Band Graph Neural Network Model. Applied Sciences. 2024; 14(21):9712. https://doi.org/10.3390/app14219712
Chicago/Turabian StyleTang, Lihan, and Menglian Zhao. 2024. "Epileptic Seizure Detection in Neonatal EEG Using a Multi-Band Graph Neural Network Model" Applied Sciences 14, no. 21: 9712. https://doi.org/10.3390/app14219712
APA StyleTang, L., & Zhao, M. (2024). Epileptic Seizure Detection in Neonatal EEG Using a Multi-Band Graph Neural Network Model. Applied Sciences, 14(21), 9712. https://doi.org/10.3390/app14219712






