Abstract
Protozoa detection and classification from freshwaters and microscopic imaging are critical components in environmental monitoring, parasitology, science, biological processes, and scientific research. Bacterial and parasitic contamination of water plays an important role in society health. Conventional methods often rely on manual identification, resulting in time-consuming analyses and limited scalability. In this study, we propose a real-time protozoa detection framework using the YOLOv4 algorithm, a state-of-the-art deep learning model known for its exceptional speed and accuracy. Our dataset consists of objects of the protozoa species, such as Bdelloid Rotifera, Stylonychia Pustulata, Paramecium, Hypotrich Ciliate, Colpoda, Lepocinclis Acus, and Clathrulina Elegans, which are in freshwaters and have different shapes, sizes, and movements. One of the major properties of our work is to create a dataset by forming different cultures from various water sources like rainwater and puddles. Our network architecture is carefully tailored to optimize the detection of protozoa, ensuring precise localization and classification of individual organisms. To validate our approach, extensive experiments are conducted using real-world microscopic image datasets. The results demonstrate that the YOLOv4-based model achieves outstanding detection accuracy and significantly outperforms traditional methods in terms of speed and precision. The real-time capabilities of our framework enable rapid analysis of large-scale datasets, making it highly suitable for dynamic environments and time-sensitive applications. Furthermore, we introduce a user-friendly interface that allows researchers and environmental professionals to effortlessly deploy our YOLOv4-based protozoa detection tool. We conducted f1-score 0.95, precision 0.92, sensitivity 0.98, and mAP 0.9752 as evaluating metrics. The proposed model achieved 97% accuracy. After reaching high efficiency, a desktop application was developed to provide testing of the model. The proposed framework’s speed and accuracy have significant implications for various fields, ranging from a support tool for paramesiology/parasitology studies to water quality assessments, offering a powerful tool to enhance our understanding and preservation of ecosystems.
1. Introduction
The insufficiency of the world’s freshwater resources day by day and the increasing population requires more careful and meticulous use of water. Fresh water is used by humans in activities such as drinking water, animal husbandry, irrigation of agricultural lands, and aquaculture. Bacterial and parasitic contamination of water leads to the spread of waterborne infections.
Bacteria that infects humans from fresh water sources causes health concerns such as intestinal diseases, anemia, muscle pain, rotavirus-related diseases, hepatitis, edema, and diarrhea [1]. The detection of bacteria in the beverages we use in daily life is of critical importance for our health.
Today, object detection and classification processes are performed intensively from microscopic images. Automatic and semi-automatic studies are available. Detection, identification, and classification of bacteria are important both in terms of working time and in terms of workforce. Various methods are used to detect the presence of bacteria. Studies for the detection and diagnosis of protozoa are divided into three different groups. First, laboratory techniques are used. Secondly, image processing techniques are used for protozoa detection. Finally, studies are carried out using deep learning techniques.
Deep learning has made significant contributions to the healthcare field besides various areas like real-time vehicle detection [2], offering innovative solutions and improvements in various areas, such as medical imaging diagnosis and image segmentation, drug discovery and development, electronic health records, genomics and personalized medicine, robotic surgery, and so on. The following examples can be given to the studies conducted using deep learning in these health fields: DNA damages on comet assay images [3], classification of white blood cells [4], classification of dentinal tubule occlusions [5], lung cancer nodule detection [6]. The applications of deep learning in healthcare continue to evolve, promising advancements in diagnostics, treatment, and overall patient care. However, it is important to address challenges such as data privacy, interpretability, and regulatory considerations to ensure the responsible and ethical deployment of these technologies in healthcare settings. An example of these applications is a study conducted to detect and classify malaria parasites from blood smear images. In this study, a total of 1920 images belonging to three classes were used. Using the CNN model, 95.11% and 99.59% success rates were achieved, respectively [7]. In another study, B-Lymphoblast cells were classified in blood cell images. Using the C-NMC dataset containing 12,528 images, the model was created by training on the CNN model. They achieved 99.4% sensitivity, 96.7% specificity, 98.3% AUC, and 98.5% accuracy [8]. In another, artificial intelligence applications were developed to determine the severity of the COVID-19 infection. Classical machine learning and deep learning models were compared using data including clinical, demographic, laboratory, and serology parameters. In this study, the XGBoost algorithm gave the highest accuracy result of 97.6% [9]. A mobile application based on an efficient lightweight CNN model was developed for the classification of B-ALL cancer cells. 3242 images were decoded and resized for data preparation and data augmentation. In the study, CNN-based EfficientNet, MobileNetV2, and NASNet Mobile models were used for comparison. The MobileNetV2 model achieved the highest efficiency (100%) in the test data [10].
Microscopic images may have different characteristics according to the color and tone of the light, and the mobility of protozoa, the deformation of some organisms, and the pollutants in the water also affect the image state.
The first of these is laboratory techniques. One of them is the physical and chemical spectrum method with PCR [11,12,13,14]. In this technique, sample preparation, partitioning, PCR amplification, and detection steps were implemented. They analyzed some different implementations of dPCR for detection of protozoan objects in samples. In [15,16], electrochemical detection was carried out. They build a point-of-care device containing a couple of electrochemical biosensors, media layers, and some other parts. It is difficult to build portable systems with high sensitivity and resilience, even when used with complicated matrices. In [17], microfluidic impedance cytometry method was applied. Another was DNA and RNA methods based on molecular biology [18]. The conventional approach for diagnosing parasites involves microscopic examination. Despite being labor-intensive and demanding experienced interpreters for optimal results, the proposed method remains extensively utilized for diagnosing protozoa, particularly in resource-limited settings. Nevertheless, numerous laboratories face a shortage of examiners proficient in consistently detecting the existence and classification of intestinal protozoa. This deficiency contributes to the incapacity to precisely recognize protozoa, distinguish pathogenic from non-pathogenic species, and discern artifacts during microscopic assessments, ultimately compromising the sensitivity and specificity of intestinal protozoa diagnosis.
Secondly, image processing techniques were used to detect protozoan presence. In [19], their proposed method consisted of a pre-processing operation to activate images, dividing them into regions and applying edge detection for feature extraction, ANN classification of regions protozoa or non-protozoa, morphological operations for deciding the regions, and applying an active contour method to regions whose location could not be identified precisely. Thirty images were used. The suggested method’s weakness is that it disregards the filamentous structure. In [20], the parasites were segmented with Active Contour Without Edge. They performed morphological operations for removing noise and thresholding to segment an image into two areas. Their dataset contains 50 images, and each file size is 140 × 140 pixels. The accuracy of the work is 97.57% and the False Negative Rate is 12.04%. In [21], the authors proposed a method to identify malaria parasites from microscopic images. Their dataset contains 117 microscopic fields of 3136 × 2352 pixels. In the beginning, they applied an image noise removal operation using Laplacian filter. The adaptive histogram thresholding technique was used to segment thin and thick smear images. As well, 8-connectivity was used to label the segmented images in HSV color space. According to the author, while the manual parasite count was 576, the parasite count by their proposed method was 627.
Lastly, in the current studies, algorithms such as Classical CNN, Retina Net, R-CNN, Fast R-CNN, Faster R-CNN, You Look Only Once (YOLO), and single shot detector (SSD) are among the deep learning methods that stand out in the detection of objects in images with digital systems. In [22], a classical CNN model was used to detect epidemic pathogens vibrio cholera and plasmodium falciparum. The training dataset includes 400 images of both classes and 80 images for testing. A Tensorflow framework was conducted to apply the CNN. Their CNN model has 6 convolutional layers, ReLU and 2 × 2 MaxPooling and follows by fully connected layer. Finally, the classification accuracy of the system is 94% and the validation accuracy is 97%. Another CNN method was used to detect Intestinal Protozoa by Blain and others in [23]. In [24], a segmentation-driven RetinaNet system was based on region-based convolutional neural networks (RCNN). Due to the insufficient data, augmentation was made with image enhancement methods for 69 images and 117 samples for eight species. However, this caused inaccurate detections. The mAP for the average precision of segmentation hierarchical retina was 93%. They implemented random horizontal flip augmentation to prevent facing local minimums. In [25], as an object detection algorithm, single short detection (SSD) and Faster R-CNN methods were used. In the study, there were 643 images with 750 × 750 dimensions. A faster R-CNN method consists of forming region predictions, and then CNN is applied to classify object type and the location of bounding box. The method has a region of interest to determine features and to categorize objects. The network is built on ResNet50 and ResNet101. As a performance evaluation metric, mAP is 94.48% for ResNet101, with a 42.8 ms inference time. And single shot detector (SSD) is implemented for multiple object detection. SSD is selected due to quick extraction and mobile use. The network consolidates predictions from multiple feature maps with varying resolutions to naturally determine objects of different sizes. In contrast to methods demanding object proposals, SSD eliminates the need for generating proposals and subsequent pixel or feature recalibrations because it handles all calculations in a single network. As a performance evaluation metric, mAP is 83.97% for InceptionV2, with a 2.24 ms inference time.
Apart from these methods, in [26], a hybrid method was applied to detect and classify intestinal parasites. In their study, there were 15 different types of parasites located in images from different parasite database groups. The dataset contains 12.225 images separated into 40% for training, 30% for validation, and 30% for testing. The study consisted of two sections. One of them was manual feature extraction (DS1) and support vector classification. In the second part, VGG-16 based on deep neural network was used for image feature extraction and classification (DS2). The hybrid model consisted of DS1 and DS2. The efficiency was not at the desired level and the complexity needed to be simplified. In [27], the authors applied U-Net, a fully convolutional network model for leishmaniasis parasite segmentation. A total of 37 images belong to the leishmania dataset, with five classes used in this work. As a performance evaluation metric, the average precision value for five classes is 75.32%. In [24], CNN-based UNet and Unet++ were implemented with 954 microscopic images with 1536 × 1536 resolution for bacillus anthracis bacteria. They reached a recall of 98% and a precision of 87% for the whole test images.
Table 1 shows a summary of some articles published in the last decade regarding the detection of protozoa, cholera pathogen, malaria parasite, intestinal protozoa.

Table 1.
A summary of studies on the identification of bacteria on images.
Utilizing deep learning for microscopic parasite diagnosis has the potential to significantly enhance efficiency for medical professionals, leading to a decrease in instances of misdiagnosis, overlooked diagnoses, and inappropriate medication usage. Despite the advancements achieved in the application of deep learning to diagnose protozoan parasites, several problems still persist. The main problem is the limited number of labeled datasets publicly available. To overcome this problem, some methods are used to extend datasets [32]. But, in medical studies, real data should be used instead of artificial data. However, different image reproduction methods are used in cases where there are not enough images. The datasets on the internet resources were not sufficient for our study and were not sufficient in terms of image quality. Therefore, unlike other methods, in this study, completely real data were used without needing image reproduction methods. Furthermore, videos and images were obtained at different angles and zoom amounts using a microscope integrated with a digital camera in the medical biology laboratory of the Faculty of Medicine.
Microscopes and other imaging devices are widely used for the classification of protozoa in the methods in the literature [13,16,33]. However, there are no real-time and fully automatic protozoa detectors and classifiers. In this study, a real-time and fully automatic recognition system was designed with the deep learning-based YOLO algorithm to detect protozoa bacteria and determine their species. With our proposed method, real-time protozoa detection is achieved in a short time and with higher performance. In addition, this developed system can also be used as a support tool for education by providing resources for paramesiology/parasitology studies and for the Single-Celled Creatures course, which is a laboratory application in the Medical Biology Department of the Faculty of Medicine. It also helps with the self-education of the students. Furthermore, this fully automatic classifier can determine the presence of bacteria in the water, the species, and the density of the variety. To put it briefly, this study can significantly contribute to the literature in terms of minimizing time loss and increasing the accuracy of determining bacteria in fresh water.
When we look at the studies, it is noteworthy that the images used are not at a sufficient level. There is a lot of data used in some studies due to the use of data augmentation methods. These images are artificial, not original images. The background of medical research should be conducted powerfully and accurately.
2. Methodology
The purpose of this study is to automatically detect protozoan existence and classify its species among different types. The newly developed approach consists of the classification stage, applied by You Only Look Once (YOLO). This object detection framework serves as the key concept of our model.
2.1. YOLO (You Only Look Once)
A deep neural network known as YOLO (You Only Look Once) is utilized for the purpose of identifying and classifying objects within microscopic images of protozoa. Demonstrating a favorable sensitivity of 90.82% [34], this network has also been employed to recognize and segment skin lesions, such as melanoma, benign nevi, and seborrheic keratoses, present in dermoscopic images. Due to this versatility, YOLO is a suitable candidate for detecting entities that may be present in microscopic images, encompassing various sizes, mobilities, and types. In addition to the diverse range of movement exhibited by protozoa, there exists a variation in shapes and physical characteristics across different species. The system performs image classification based on input images, and the object detection process involves determining the object’s location with respect to a bounding box that surrounds it [35].
2.2. Detector
In the detection stage of the one-stage object detection algorithm of the YOLO algorithm, dense prediction takes place. The last prediction step consists of a vector with three data groups. Firstly, the bounding box is predicted with its coordinates, containing center, height, and width, the confidence score of the prediction, and the label. Figure 1 shows the illustration of YOLO determining the location of objects. It splits the picture into (S S) grids shown in Figure 1a and then assigns a confidence score and class probabilities to each grid cell to create B potential bounding boxes in Figure 1b.
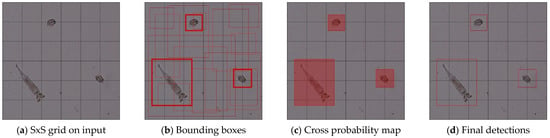
Figure 1.
Schematic presentation of YOLO algorithm.
The confidence is calculated by multiplying the probability of finding an object in the grid by the intersection over union (IOU) percentage, the intersection of the box in which the object is located and the predicted box, (see Figure 2), using the following formula:
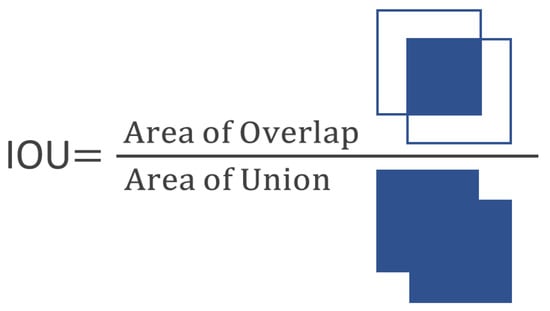
Figure 2.
Intersection over union (IOU), measurement for object detection performance.
The probability of the box containing an object is 70% when the probability of the object’s presence, denoted as P r(object), is 0.7. A confidence score of zero indicates the absence of an object in that cell. The confidence score is employed in the calculation of the mean Average Precision (mAP) at a specified threshold. If a grid cell predicts a 60% probability of containing a car (pr(Car) = 60%), there is a 60% likelihood that the cell indeed contains a car and a 40% likelihood that it does not [30]. The bounding box that is hovered over the image has five parameters in total: [x, y, w, h, confidence score]. Here, (x, y) represent the coordinates of the middle point of the bounding box, (w, h) the width and the height.
The standout characteristic of v4 lies in its ability to perform detections across three distinct scales. YOLO operates as a completely convolutional network, with its final output derived through the utilization of a 1 × 1 kernel on a feature map. In YOLOv4, the process of detection involves the application of 1 × 1 detection kernels on feature maps of varying sizes, strategically positioned at three different locations within the network [36].
The configuration of the detection kernel is 1 × 1 × (B × (5 + C)). In this context, B represents the quantity of bounding boxes that a cell within the feature map can predict. The value “5” pertains to the four attributes concerning bounding boxes and an additional object confidence score, while C denotes the total number of distinct classes. For YOLOv4 models trained on the COCO dataset, B corresponds to 3 and C is set at 80, resulting in a kernel dimension of 1 × 1 × 255 [37]. The resulting feature map generated through this kernel retains the same height and width as the previous feature map, while encompassing detection characteristics across its depth, as previously elaborated.
3. Experiment
In this section, we use our own dataset. To illustrate the success and adaptability of our system, we employ a variety of assessment indicators. For the training of the models, the Google Colaboratory environment, which provides especially high-level graphics card usage through the cloud platform offered by Google, was used. Google Colaboratory remote server computer was used for the training of the models with the video card model Tesla P100-PCIE-16GB. The related server is located in Taipei, Taiwan. We use Python (3.10) language in the Darknet framework to implement the algorithm.
3.1. Data Set Preparation
Living things need environments with suitable temperatures, humidity, pH, etc., to survive and reproduce. Microscopic organisms inhabit various aquatic environments, including ponds, lakes, streams, rivers, estuaries within ocean backwaters, and even, unexpectedly, rain puddles that have persisted for several days. Although single-celled organisms are very common in nature, their cultures must be prepared before they can be examined in laboratories. For this purpose, our collection jars were prepared completely clean and detergent free. Water samples were collected from rain puddles. Four different culture mixtures were prepared.
Culture cases were kept in a semi-dark environment for an average of 8–10 days, to allow for bacteria to appear. Then, samples from these different cultures were taken and examined under a microscope. After the bacteria started to form, video and images were obtained with a high-resolution camera. Sampling procedures were repeated at 1–2 day intervals to ensure the formation of different species, and the data set was enriched. The system for our study consisting of a microscope, camera and computer is shown in Figure 3.

Figure 3.
The examining protozoa with a camera.
Due to the continuous movement of many protozoa species in the images taken as video, three images were obtained randomly from the frame sets in each second. The data set was created by combining the images obtained from the videos and the images taken as photographs. The data set consists of 4653 color images with 1280 × 720 resolution related to 8 species. The training set is 3257 images, the validation set is 931 images, and the test set is 465 images. There are 11,252 protozoa objects in the whole data set consisting of 4653 images. Our data set includes Bdelloid Rotifera, Stylonychia Pustulata, Paramecium, Hypotrich Ciliate, Colpoda, Lepocinclis Acus, Clathrulina Elegans freshwater protozoa species shown on Figure 4.
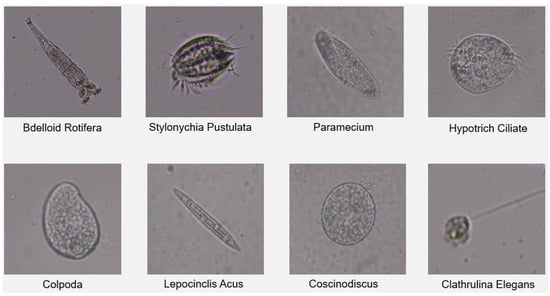
Figure 4.
Protozoan types in the dataset.
3.2. Labeling
MakeSense.ai was used to label images. This is an online tool for labelling images, and it makes the process of preparing a dataset much easier and faster [38]. The system creates YOLO format, VOC XML format, and a single CSV file.
The dataset consisting of 4653 images was uploaded to the MakeSense system, and protozoa species belonging to 8 classes were labeled. The labeling operation is handled in MakeSense system shown in Figure 5. While some images have a single object, some images contain 25 objects. After all images were tagged, labeled data was created in YOLO format. In YOLO format, the class ID, x and y coordinates of the midpoint of the rectangle, width and height values of the rectangle are normalized in each line.
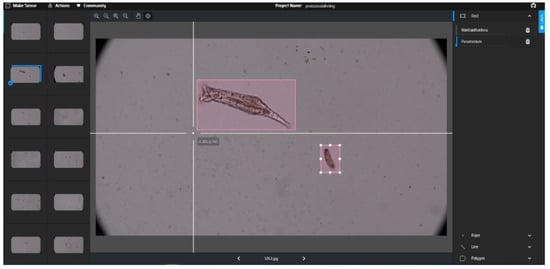
Figure 5.
Labeling with MakeSense.
3.3. Performance Matrix
The Tesla P100-PCIE-16GB was the main equipment used in the experiment.
To assess the experimental findings, we consider accuracy and recall rates as well as frames recognized per second (FPS). We compute the IoU of the detection and reference border boxes to determine the results as true or false. IoU is true when above and equal to 0.5 and false when less than 0.5.
The protozoa dataset has 8 categories, hence multi-class classification was performed. The metrics given in Equations (2)–(4) are calculated using indices, considering the values in the confusion matrix acquired in such classifications. These indices are True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN). TP is the true predicted class numbers among each category, whereas TN is the total number of objects that have been accurately categorized in all categories but the one that applies. TN is the number of incorrectly categorized objects from the relevant category. FP shows the total number of objects that were incorrectly categorized in all categories but the one that applies.
Accuracy is calculated as the ratio of the number of correctly predicted objects for each class to the total number of predicted objects. The average success rate is obtained by taking the average of all classes shown in Equation (2). Precision is the ratio of true positives to the sum of all positives shown in Equation (3). The positives are the sum of true and false predictions. Sensitivity is the ratio of correctly predicted class to actual assets. The average sensitivity is calculated as the average of the sensitivities of the classes shown in Equation (4).
3.4. Result and Discussion
In this study, real-time protozoa detection was realized by using YOLOv4 on Darknet-53 framework. The evaluation of the study is performed by average accuracy, average precision, average sensitivity, f1-score, and mAP metrics. The class-based average precision values were also obtained and are shown in Table 2. Class-8 has the highest precision rate among classes. The evaluation metrics of the system for YOLOv4 method are f1-score 95%, precision 92%, sensitivity 98%, and mean Average Precision (mAP) 97.52% values shown in Table 2.

Table 2.
Performance evaluation values of protozoa detection.
Table 3 shows class average precision values belonging to eight different classes. Average precision value of Class-3 (Colpoda) is 92.37% with 419 true positive and 58 false positive values. Colpoda protozoa has the lowest mAP value by YOLOv4 due to the similarity in shape with some other protozoan types. This is the lowest perception among all classes. Class-8 (Hypotrich Ciliate) achieved the highest mAP value among all classes.

Table 3.
Training results per class with YOLOv4.
YOLOv4 creates bounding boxes of detected objects. Figure 6 shows the IoU bounding box values of the protozoa classes. As seen in the figure, the places marked with red boxes show the Bdelloid Rotifera, blue boxes show the Paramecium, yellow boxes show the Stylonychia Pustulata, green boxes show the Clathrulina Elegans. These boxes show the highest confidence score among cross-probability maps. For example, Bdelloid Rotifera was detected on the image 96–98% of the time, even though the bacterium is constantly on the move and constantly changes its shape, as shown in Figure 6.
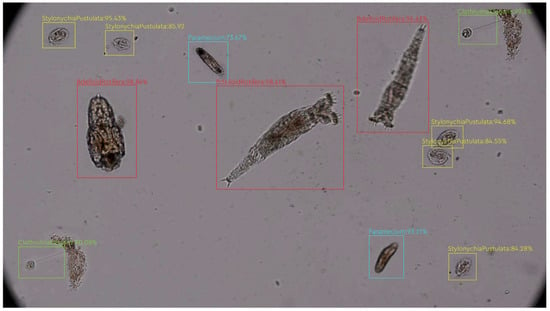
Figure 6.
Protozoa species detection test image.
Figure 7 displays the calculated mAP values of the valid data together with the loss function graph on the training iteration axis; as the model becomes more trained and learns to accept YOLO, the loss value drops. Typically, mAP climbs during the first few training iterations before falling as a result of the deep learning model overfitting after a given amount of training. Figure 7 shows the accuracy and loss rates after finishing training on valid bacterial classes data. YOLOv4 achieved an average accuracy of 97% and a loss value of 1.3792 with 0.5 IoU threshold value. The proposed YOLOv4 model misclassified 52 of 2063 objects out of 900 images in the test class. This result shows that the object detection accuracy of the YOLOv4 is quite high.
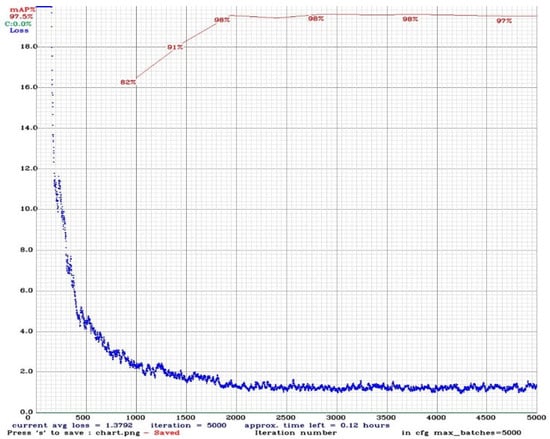
Figure 7.
mAP and loss graph.
Table 4 indicates the Faster R-CCN and SSD-based deep learning models, compared according to setting the batch size to 64. The optimal batch size was chosen from alternative values to reach the global optimum and the correct gradient value for our model. The most significant accuracy was achieved by YOLOv4 with a precision of 0.9752, FPS of 5. The FPS seems to be a bit low, but this can be increased with batch size and modifications to the model. But, in this case, there may be problems like global optimum and correct gradient value values. And, it may learn noise and it may get stuck at the local optimum and never reach the global optimum.

Table 4.
Performance evaluation values of protozoa detection.
4. Conclusions
The objective of this study is to create a technique for the automatic real-time recognition, segmentation, and classification of protozoa, with the potential for application to different species. According to the application results, successful results were obtained even though the protozoa of the same species in the images used were of different sizes and changed shape in motion. The detection of protozoa species in real time with deep learning, the application of the YOLO algorithm for the first time, and the creation of the data set by us add originality to this study. The developed system can be used in parasitology and paramesiology studies. Results such as the existence of single-celled organisms and the number of varieties can be obtained from the puddles. The system also gains importance in terms of providing information about the density of single-celled organisms and software support for researchers in the region. Finally, Karabük University Faculty of Medicine can use it as an educational support tool in terms of being a source for the single-celled organisms lesson, which is a medical biology application in term 1, and for students to train themselves. Lastly, the labeled dataset created specifically for this study will be shared later and will contribute to academic studies. In addition, more and different protozoa species will be added by expanding the data set.
5. Suggestions for the Future
This application is open for further development as the proposed method yields a high success rate. The dataset with eight protozoa species can be further expanded to provide researchers with an important field of study. In addition, the enriched dataset can be transformed into a medical biology application that can be used in medical schools. Methods can be explored to fine-tune the YOLOv4 model to enhance its performance under challenging imaging conditions, such as low light, high background noise, or varying magnifications. Developing an interactive user interface will allow users, especially domain experts, to annotate and correct the model’s predictions in real time. Collaboration can be handled with experts in automation to integrate the protozoa detection algorithm with automated sample processing systems. This could lead to a fully automated workflow for real-time protozoa detection in environmental samples.
Author Contributions
Conceptualization, İ.K.; data creation, İ.K. and M.K.T.; data validation, M.K.T.; methodology, İ.K. and M.K.T.; writing—original draft preparation, İ.K. and İ.R.K.; writing—review and editing, İ.K., M.K.T. and İ.R.K.; visualization, İ.K.; supervisor, İ.R.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data is not publicly available due to further studies.
Acknowledgments
This work was supported by Scientific Research Projects Unit of the Karabük University, Project Number: KBÜ-BAP-16/2-DR-102.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pandey, P.K.; Kass, P.H.; Soupir, M.L.; Biswas, S.; Singh, V.P. Contamination of Water Resources by Pathogenic Bacteria. AMB Express 2014, 4, 1–16. [Google Scholar] [CrossRef]
- Al Bayati, M.A.Z.; Çakmak, M. Real-Time Vehicle Detection for Surveillance of River Dredging Areas Using Convolutional Neural Networks. Int. J. Image Graph. Signal Process. 2023, 15, 17–28. [Google Scholar] [CrossRef]
- Atila, Ü.; Baydilli, Y.Y.; Sehirli, E.; Turan, M.K. Classification of DNA Damages on Segmented Comet Assay Images Using Convolutional Neural Network. Comput. Methods Programs Biomed. 2020, 186, 105192. [Google Scholar] [CrossRef] [PubMed]
- Baydilli, Y.Y.; Atila, Ü. Classification of White Blood Cells Using Capsule Networks. Comput. Med. Imaging Graph. 2020, 80, 101699. [Google Scholar] [CrossRef] [PubMed]
- Duru, A.; Karaş, İ.R.; Karayürek, F.; Gülses, A. A Deep Learning Approach for Classification of Dentinal Tubule Occlusions. Appl. Artif. Intell. 2022, 36, 2094446. [Google Scholar] [CrossRef]
- Ahmed, A.H.; Alwan, H.B.; Çakmak, M. Convolutional Neural Network-Based Lung Cancer Nodule Detection Based on Computer Tomography. In Lecture Notes in Networks and Systems; Springer: Singapore, 2023; Volume 572, pp. 89–102. [Google Scholar]
- Fasihfar, Z.; Rokhsati, H.; Sadeghsalehi, H.; Ghaderzadeh, M.; Gheisari, M. AI-Driven Malaria Diagnosis: Developing a Robust Model for Accurate Detection and Classification of Malaria Parasites. Iran. J. Blood Cancer 2023, 15, 112–124. [Google Scholar] [CrossRef]
- Ghaderzadeh, M.; Hosseini, A.; Asadi, F.; Abolghasemi, H.; Bashash, D.; Roshanpoor, A. Automated Detection Model in Classification of B-Lymphoblast Cells from Normal B-Lymphoid Precursors in Blood Smear Microscopic Images Based on the Majority Voting Technique. Sci. Program 2022, 2022, 4801671. [Google Scholar] [CrossRef]
- Ghaderzadeh, M.; Asadi, F.; Ramezan Ghorbani, N.; Almasi, S.; Taami, T. Toward Artificial Intelligence (AI) Applications in the Determination of COVID-19 Infection Severity: Considering AI as a Disease Control Strategy in Future Pandemics. Iran. J. Blood Cancer 2023, 15, 93–111. [Google Scholar] [CrossRef]
- Hosseini, A.; Eshraghi, M.A.; Taami, T.; Sadeghsalehi, H.; Hoseinzadeh, Z.; Ghaderzadeh, M.; Rafiee, M. A Mobile Application Based on Efficient Lightweight CNN Model for Classification of B-ALL Cancer from Non-Cancerous Cells: A Design and Implementation Study. Inform. Med. Unlocked 2023, 39, 101244. [Google Scholar] [CrossRef]
- Skotarczak, B. Methods for Parasitic Protozoans Detection in the Environmental Samples. Parasite 2009, 16, 183–190. [Google Scholar] [CrossRef]
- Maas, L.; Dorigo-Zetsma, J.W.; de Groot, C.J.; Bouter, S.; Plötz, F.B.; van Ewijk, B.E. Detection of Intestinal Protozoa in Paediatric Patients with Gastrointestinal Symptoms by Multiplex Real-Time PCR. Clin. Microbiol. Infect. 2014, 20, 545–550. [Google Scholar] [CrossRef]
- Le Calvez, T.; Trouilhé, M.C.; Humeau, P.; Moletta-Denat, M.; Frère, J.; Héchard, Y. Detection of Free-Living Amoebae by Using Multiplex Quantitative PCR. Mol. Cell. Probes 2012, 26, 116–120. [Google Scholar] [CrossRef]
- Baltrušis, P.; Höglund, J. Digital PCR: Modern Solution to Parasite Diagnostics and Population Trait Genetics. Parasit. Vectors 2023, 16, 143. [Google Scholar] [CrossRef]
- Houssin, T.; Bridle, H.; Senez, V. Electrochemical Detection. In Waterborne Pathogens; Academic Press: Cambridge, MA, USA, 2021; pp. 147–187. [Google Scholar] [CrossRef]
- da Silva, A.D.; Paschoalino, W.J.; Neto, R.C.; Kubota, L.T. Electrochemical Point-of-Care Devices for Monitoring Waterborne Pathogens: Protozoa, Bacteria, and Viruses—An Overview. Case Stud. Chem. Environ. Eng. 2022, 5, 100182. [Google Scholar] [CrossRef]
- McGrath, J.S.; Honrado, C.; Spencer, D.; Horton, B.; Bridle, H.L.; Morgan, H. Analysis of Parasitic Protozoa at the Single-Cell Level Using Microfluidic Impedance Cytometry. Sci. Rep. 2017, 7, 2601. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, P.; Tripathi, D.; Pandey, S.; Tapadar, S.; Bhattacharjee, A.; Das, D.; Palwan, E.; Rani, M.; Kumar, A. Molecular Biology Techniques for the Detection of Contaminants in Wastewater. In Wastewater Treatment: Cutting-Edge Molecular Tools, Techniques and Applied Aspects; Elsevier: Amsterdam, The Netherlands, 2021; pp. 217–235. [Google Scholar] [CrossRef]
- Boztoprak, H.; Özbay, Y. Detection of Protozoa in Wastewater Using ANN and Active Contour in Image Processing. Istanb. Univ. J. Electr. Electron. Eng. 2013, 13, 1661–1666. [Google Scholar]
- Abidin, S.R.; Salamah, U.; Nugroho, A.S. Segmentation of Malaria Parasite Candidates from Thick Blood Smear Microphotographs Image Using Active Contour without Edge. In Proceedings of the 2016 1st International Conference on Biomedical Engineering (IBIOMED), Yogyakarta, Indonesia, 5–6 October 2016; IEEE: New York, NY, USA, 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Dave, I.R.; Upla, K.P. Computer Aided Diagnosis of Malaria Disease for Thin and Thick Blood Smear Microscopic Images. In Proceedings of the 2017 4th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 2–3 February 2017; pp. 561–565. [Google Scholar] [CrossRef]
- Traore, B.B.; Kamsu-Foguem, B.; Tangara, F. Deep Convolution Neural Network for Image Recognition. Ecol. Inform. 2018, 48, 257–268. [Google Scholar] [CrossRef]
- Mathison, B.A.; Kohan, J.L.; Walker, J.F.; Smith, R.B.; Ardon, O.; Ardon, O.; Couturier, M.R.; Couturier, M.R. Detection of Intestinal Protozoa in Trichrome-Stained Stool Specimens by Use of a Deep Convolutional Neural Network. J. Clin. Microbiol. 2020, 58, e02053-19. [Google Scholar] [CrossRef]
- Pho, K.; Mohammed Amin, M.K.; Yoshitaka, A. Segmentation-Driven Hierarchical RetinaNet for Detecting Protozoa in Micrograph. Int. J. Semant. Comput. 2019, 13, 393–413. [Google Scholar] [CrossRef]
- Nakasi, R.; Mwebaze, E.; Zawedde, A.; Tusubira, J.; Akera, B.; Maiga, G. A New Approach for Microscopic Diagnosis of Malaria Parasites in Thick Blood Smears Using Pre-Trained Deep Learning Models. SN Appl. Sci. 2020, 2, 1255. [Google Scholar] [CrossRef]
- Osaku, D.; Cuba, C.F.; Suzuki, C.T.N.; Gomes, J.F.; Falcão, A.X. Automated Diagnosis of Intestinal Parasites: A New Hybrid Approach and Its Benefits. Comput. Biol. Med. 2020, 123, 103917. [Google Scholar] [CrossRef] [PubMed]
- Górriz, M.; Aparicio, A.; Raventós, B.; Vilaplana, V.; Sayrol, E.; López-Codina, D. Leishmaniasis Parasite Segmentation and Classification Using Deep Learning; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2018; Volume 10945, pp. 53–62. [Google Scholar] [CrossRef]
- Hoorali, F.; Khosravi, H.; Moradi, B. Automatic Bacillus Anthracis Bacteria Detection and Segmentation in Microscopic Images Using UNet++. J. Microbiol. Methods 2020, 177, 106056. [Google Scholar] [CrossRef] [PubMed]
- de Souza Oliveira, A.; Guimarães Fernandes Costa, M.; das Graças Vale Barbosa, M.; Ferreira Fernandes Costa Filho, C. A New Approach for Malaria Diagnosis in Thick Blood Smear Images. Biomed. Signal Process. Control 2022, 78, 103931. [Google Scholar] [CrossRef]
- Abdurahman, F.; Fante, K.A.; Aliy, M. Malaria Parasite Detection in Thick Blood Smear Microscopic Images Using Modified YOLOV3 and YOLOV4 Models. BMC Bioinform. 2021, 22, 112. [Google Scholar] [CrossRef]
- Nakasi, R.; Mwebaze, E.; Zawedde, A. Mobile-Aware Deep Learning Algorithms for Malaria Parasites and White Blood Cells Localization in Thick Blood Smears. Algorithms 2021, 14, 17. [Google Scholar] [CrossRef]
- Zhang, C.; Jiang, H.; Jiang, H.; Xi, H.; Chen, B.; Liu, Y.; Juhas, M.; Li, J.; Zhang, Y. Deep Learning for Microscopic Examination of Protozoan Parasites. Comput. Struct. Biotechnol. J. 2022, 20, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Althomali, R.H.; Abdu Musad Saleh, E.; Gupta, J.; Mohammed Baqir Al-Dhalimy, A.; Hjazi, A.; Hussien, B.M.; AL-Erjan, A.M.; Jalil, A.T.; Romero-Parra, R.M.; Barboza-Arenas, L.A. State-of-the-Art of Portable (Bio)Sensors Based on Smartphone, Lateral Flow and Microfluidics Systems in Protozoan Parasites Monitoring: A Review. Microchem. J. 2023, 191, 108804. [Google Scholar] [CrossRef]
- Ünver, H.M.; Ayan, E. Skin Lesion Segmentation in Dermoscopic Images with Combination of Yolo and Grabcut Algorithm. Diagnostics 2019, 9, 72. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. ArXiv 2020. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2014; Volume 8693. [Google Scholar] [CrossRef]
- Skalski, P. Make Sense. Available online: https://github.com/SkalskiP/make-sense/ (accessed on 18 December 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).