Efficient Diagnosis of Autism Spectrum Disorder Using Optimized Machine Learning Models Based on Structural MRI
Abstract
1. Introduction
- Can the proposed FS methods improve the accuracy of ML models in ASD classification?
- Which of the proposed optimized models performs the best in predicting ASD in terms of accuracy on the two public datasets?
- Does combining personal features data with sMRI yield better results in ASD classification compared to using only sMRI data?
2. Related Work
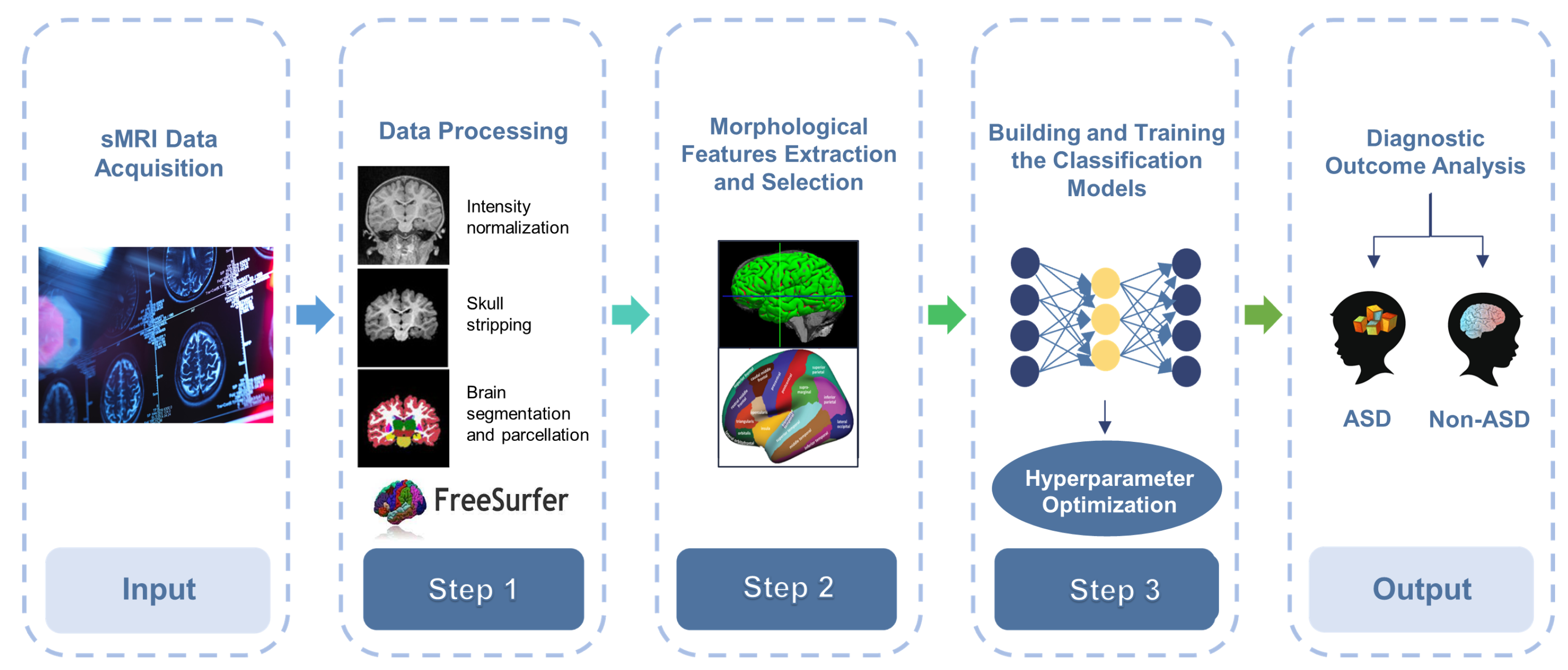
3. Methods and Materials
3.1. Data Acquisition
3.2. Pre-Processing
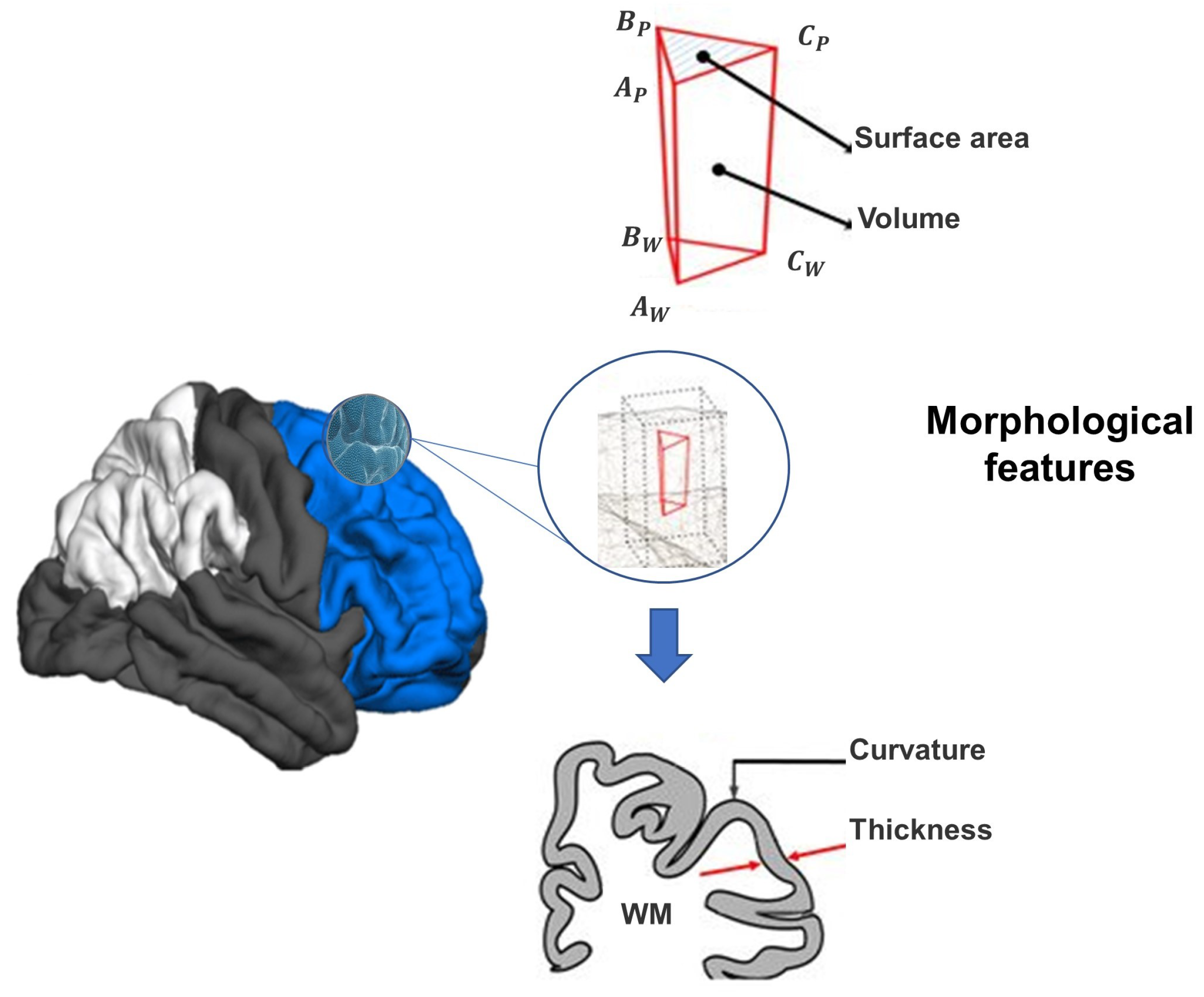
3.2.1. sMRI Pre-Processing and Features Extraction
3.2.2. Data Cleaning and Integration
3.2.3. Data Transformation
3.3. Data Splitting
3.4. Feature Selection
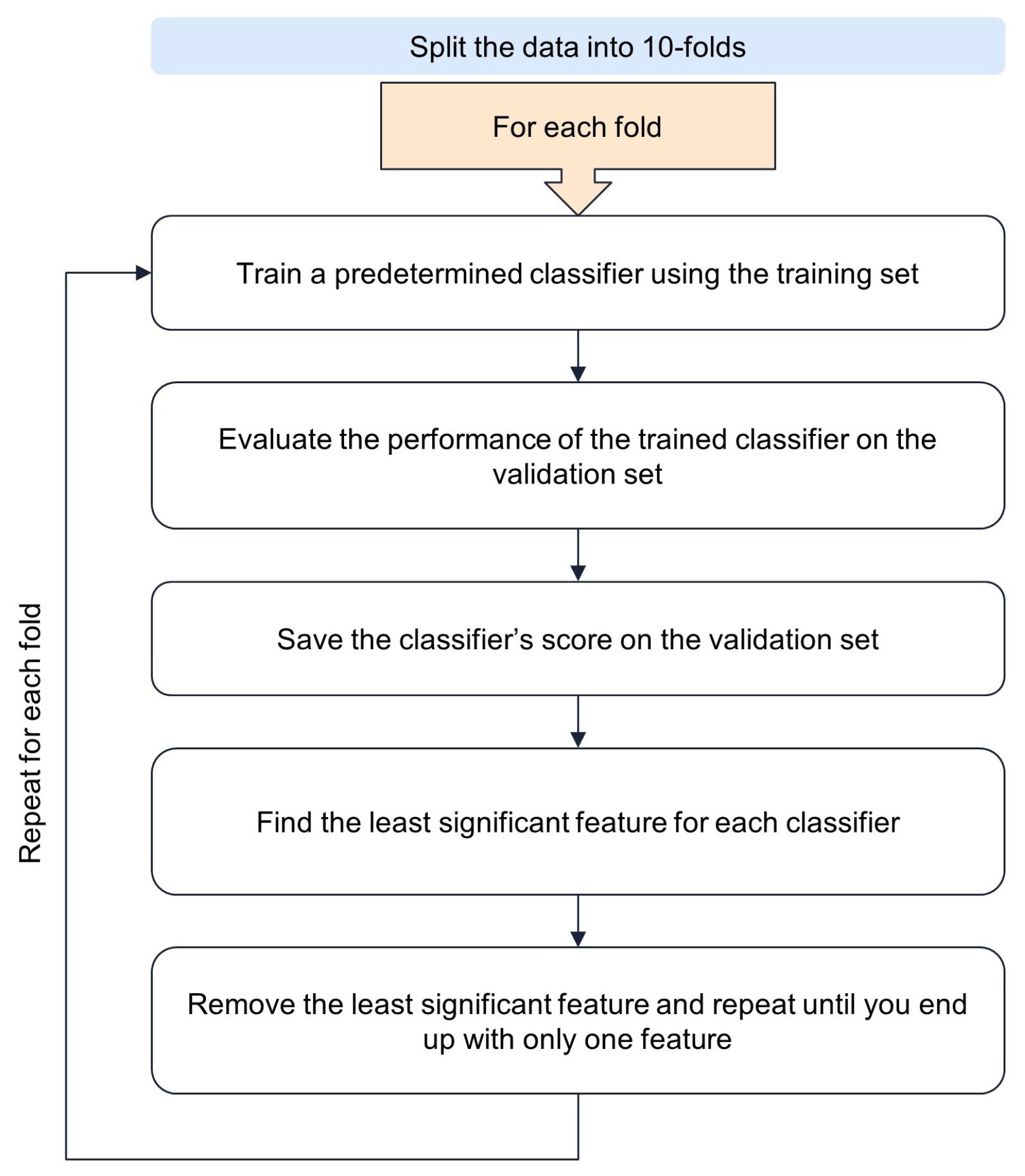
3.4.1. Recursive Feature Elimination with Cross-Validation (RFECV)
3.4.2. Boruta
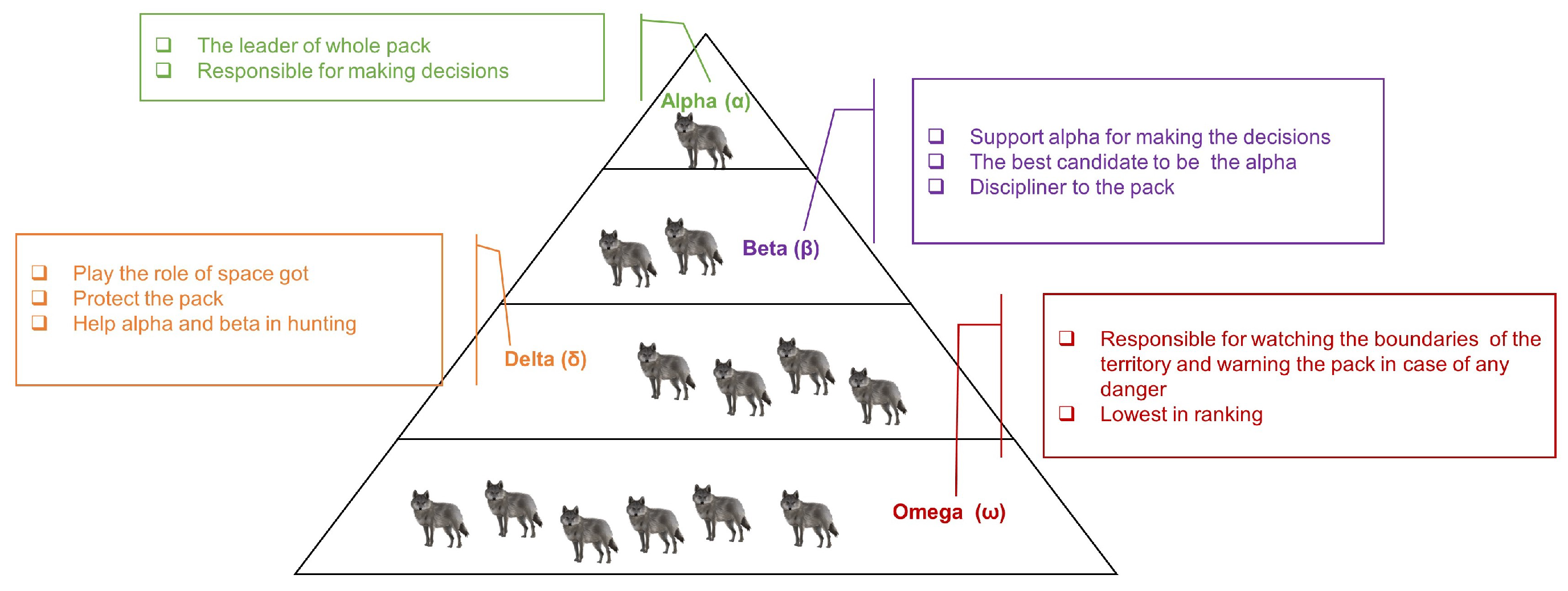
3.4.3. Grey Wolf Optimizer (GWO)-Based Algorithm
- Initialize the positions of the wolves randomly within the search space.
- Prey Encircling: update the positions of the wolves based on the alpha wolf’s position, aiming to encircle the prey. The position of each wolf is updated using the following equation: , where is the position of the ith wolf, is the position of the alpha wolf, is a random vector, and is a coefficient vector.
- Prey Attacking: update the positions of the wolves to attack the prey. The position of each wolf is updated using the following equation: , where is the position of the prey.
- Searching: update the positions of the wolves to search for the prey. The position of each wolf is updated using the following equation: , where is a random vector, and is a coefficient vector.
- Boundary Checking: ensure that the updated positions of the wolves remain within the defined search space.
- Fitness Evaluation: evaluate the fitness of each wolf based on the problem-specific fitness function.
- Select the three wolves with the best fitness values as the alpha, beta, and delta wolves, respectively.
- Update the positions of the omega wolves based on the positions of the alpha, beta, and delta wolves.
- Termination: repeat steps 2–8 until a termination criterion is met (e.g., a maximum number of iterations or a desired fitness value is reached).
3.5. Models Development and Training
3.5.1. Machine Learning (ML) Algorithms
- Support vector machine (SVM):SVM finds an optimal decision boundary (hyperplane) in a high-dimensional space to separate different classes. It maximizes the margin between classes and can handle non-linear classification using kernel methods. SVM is suitable for handling high-dimensional data such as sMRI, but training time can be long [5].
- Naïve Bayes (NB):NB is based on Bayes’ theorem and assumes that features are independent and contribute independently to the final prediction. Gaussian NB, which follows the normal distribution, is used in this work. NB is known for its simplicity and efficiency, and it is particularly useful when dealing with large datasets [52,53].
- Decision Tree (DT):DT is a flowchart-like tree structure that represents a series of decisions and their outcomes. Starting from a root node, it uses a top-down greedy search to produce a DT with decision and leaf nodes without backtracking over the space of possible branches. The DT branches describe the dataset’s features. The final classification is made at the leaf nodes of the tree. DTs are intuitive and easy to interpret, but they can be prone to overfitting [54].
- Random Forest (RF):RF is an ensemble learning method that combines multiple weak learners (e.g., DTs) to create a robust model. Each tree is trained on a random subset of the original dataset using a bagging technique. The final prediction is determined by majority voting. RF improves performance and reduces overfitting compared to a single DT [17].
- Extreme Gradient Boosting (XGB):XGB is an optimized version of the GBC algorithm. It is designed for speed and performance, utilizing three techniques: implementation of sparse-aware that handles missing data values automatically; utilization of a block structure to facilitate the parallelization of tree construction; and continuous training to further enhance the model’s performance that has been fitted to new data [14,55].
- Category Boosting (CatBoost):CatBoost is a gradient boosting approach that specifically works well with categorical features. It creates a set of DTs with identical splitting criteria throughout the entire level. Each successive tree is produced with a lower loss than the previous one. CatBoost is well-balanced and less sensitive to overfitting [55].
- Multilayer perceptron (MLP):MLP is an artificial neural network with input, output, and hidden layers. The input layer receives the data represented by a digital vector to be processed. Classification is handled by the output layer. A number of hidden layers make up the MLP’s true computational engine. MLP processes input data through linear and nonlinear transformations, transmitting the data forward. The network learns from feedback on prediction errors through the backpropagation learning process, adjusting weights for improved predictions [56].
3.5.2. Hyperparameter Optimization
- Random Search:Random search explores a specified number of random hyperparameter combinations. It is faster than grid search but does not guarantee finding the optimal combination. Here, the RandomizedSearchCV function from sklearn was used with 20 iterations to find the best hyperparameters [58].
- GWO-based hyperparameters tuning:GWO is a nature-inspired algorithm. It is faster and more likely to find the best solution compared to random search. The GWO algorithm was used for hyperparameter tuning with the NatureInspiredSearchCV function from the sklearn_nature_inspired_algorithms library. Parameters such as the model name, population size and maximum stagnating generation were set for the optimization process [57,59]. Here, the population size is set to 50 and the max stagnating generation is 20.
3.6. Model Testing and Performance Evaluation
4. Results and Discussion
4.1. Result Analysis
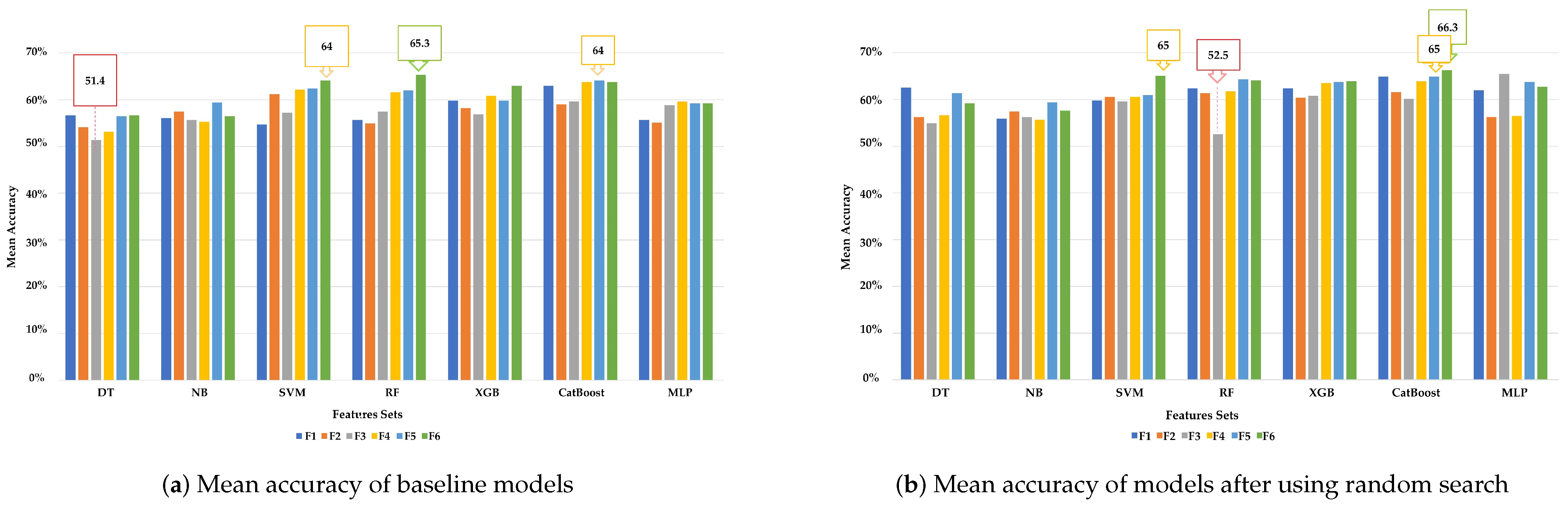
4.1.1. Experiment 1 Results
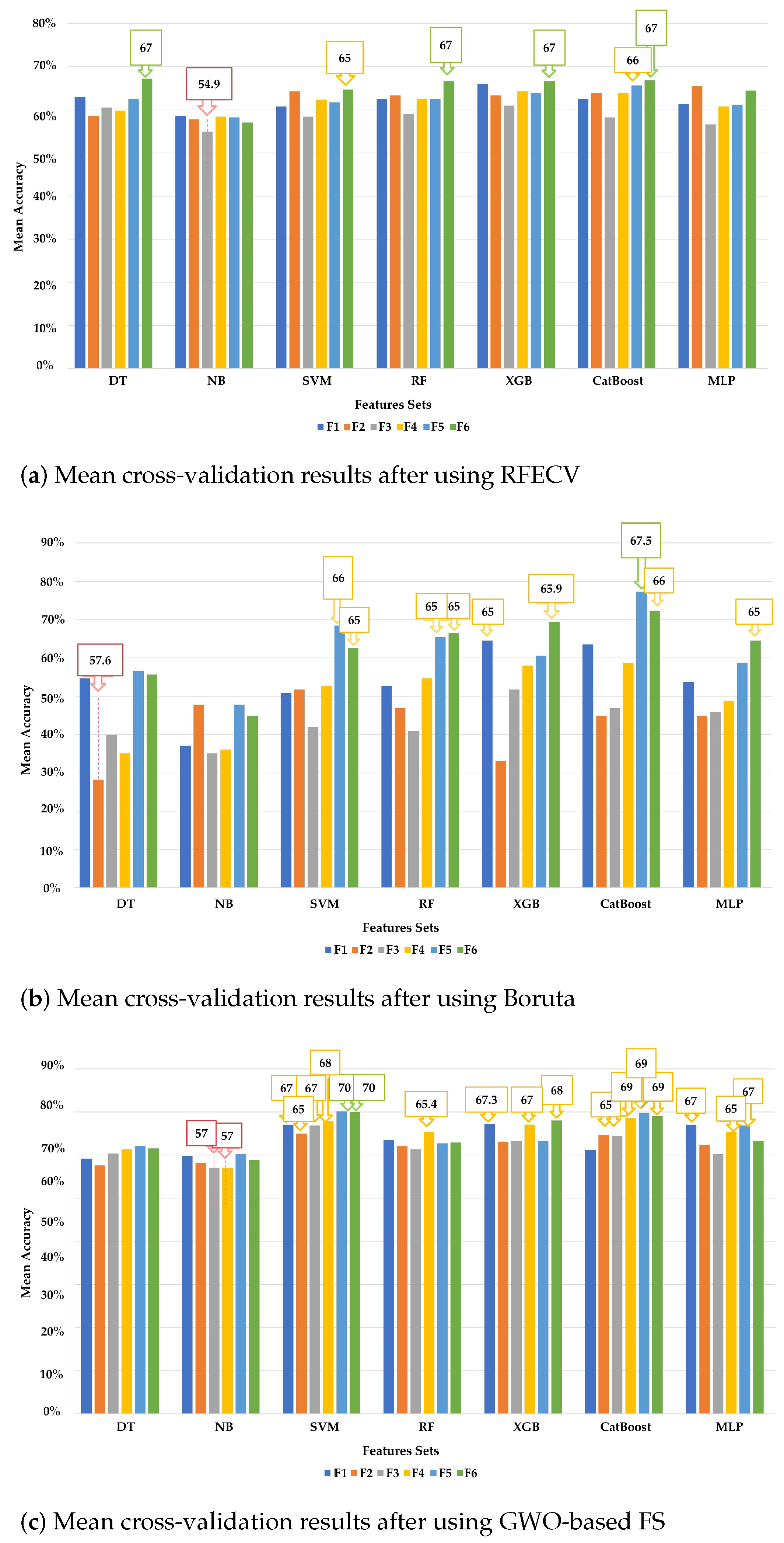
4.1.2. Experiment 2 Results
4.1.3. Experiment 3 Results
4.2. Discussion
Comparison with Previous Methods
4.3. Limitations
- This work is limited to ASD and non-ASD (TD) classification tasks, and the accuracy and classification of ASD subgroups are open questions.
- Several algorithms have been evaluated for classifying ASD based on age, sex, and brain morphological features; behavioral features or clinical test results that could be informative are not included.
- Limited availability of sMRI images for children in the studied age group poses challenges for effective ML model training and increases the risk of overfitting.
- The findings are limited to the age group of 5–10 years, and applying them to different age groups may impact accuracy due to age-related brain differences. The complexity of selecting stable and discriminating biomarkers between age groups further contributes to these limitations.
- Relying on specific brain segmentation methods such as our method of segmentation according to the DK atlas, may lead to biases and limitations. It should be emphasized that our findings can be replicated using different data or atlases.
- The reported accuracy may be insufficient for clinical use due to data variability, heterogeneity, and limitations of multi-site datasets used. However, we followed ML best practices to the best of our knowledge. As data heterogeneity increases, the training, validation, and testing folds used to evaluate a model’s performance can diverge significantly. This divergence can lead to poor performance in fold testing, ultimately reducing the cross-validated estimated generalized predictive performance of the model.
- An inherent limitation is the cross-sectional design chosen, which limits understanding of potential changes over time. Longitudinal designs can provide a more nuanced understanding of ASD.
4.4. Future Works
- Gather comprehensive and diverse datasets to account for the heterogeneity of ASD data and develop a robust, generalizable, and more clinically useful model.
- Conduct longitudinal studies to understand the developmental trajectory of ASD-related brain changes, identify predictive biomarkers, and improve early detection and intervention strategies.
- Investigate multimodal imaging-based classification by combining sMRI data with other methods (fMRI, EEG, genetic data) to provide a more comprehensive understanding of the neurobiological mechanisms underlying ASD, reveal additional biomarkers and improve the accuracy of diagnosis [28].
- Perform fine-grained analysis of subtypes or phenotypic variations within the ASD spectrum to identify unique biomarkers and enable personalized diagnostic approaches and targeted interventions.
- Validate ML-based diagnostic systems in real-world clinical settings to assess their feasibility, acceptability, and clinical utility.
- DL methods differ from traditional ML models as they eliminate the need for manual feature extraction and minimize information loss. However, training DL networks and uncovering intricate patterns requires extensive datasets. In our research, we intend to investigate DL models using large datasets, employing techniques such as data augmentation, generative adversarial networks, and transfer learning [5,27].
- Integrate Explainable AI (XAI) techniques to improve the interpretability of DL algorithms in the diagnostic process, enhancing their clinical utility and gaining trust and acceptance from clinicians and stakeholders. XAI explores the decision-making process, provides explanations for system behavior, and offers insights into future performance [67].
- Use T7 scans to obtain accurate and clinically useful biomarkers for ASD diagnoses.
- Explore the potential of ML techniques for early classification of infants at risk for ASD, addressing challenges associated with processing and interpreting MRI images in pediatric brains.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Su, J.Y. Effects of in Utero Exposure to CASPR2 Autoantibodies on Neurodevelopment and Autism Spectrum Disorder. Master’s Thesis, Donald and Barbara Zucker School of Medicine at Hofstra/Northwell, Hempstead, NY, USA, 2023. [Google Scholar]
- Autism Spectrum Disorders and Other Developmental Disorders: From Raising Awareness to Building Capacity. Available online: https://apps.who.int/ (accessed on 25 March 2023).
- Autism Rates by Country. 2022. Available online: https://worldpopulationreview.com/country-rankings/autism-rates-by-country (accessed on 1 March 2023).
- Anderson, D.; Lord, C.; Risi, S.; DiLavore, P.; Shulman, C.; Thurm, A.; Pickles, A. Diagnostic and statistical manual of mental disorders. In The Linguistic And Cognitive Effects Of Bilingualism On Children With Autism Spectrum Disorders; American Psychiatric Association: Washington, DC, USA, 2017; Volume 21, p. 175. [Google Scholar]
- Bahathiq, R.; Banjar, H.; Bamaga, A.; Jarraya, S. Machine learning for autism spectrum disorder diagnosis using structural magnetic resonance imaging: Promising but challenging. Front. Neuroinf. 2022, 16, 949926. [Google Scholar] [CrossRef] [PubMed]
- Mostapha, M. Learning from Complex Neuroimaging Datasets. Ph.D. Thesis, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA, 2020. [Google Scholar]
- Li, G.; Chen, M.; Li, G.; Wu, D.; Lian, C.; Sun, Q.; Shen, D.; Wang, L. A longitudinal MRI study of amygdala and hippocampal subfields for infants with risk of autism. Graph Learn. Med. Imaging 2019, 11849, 164–171. [Google Scholar]
- Ali, M.; ElNakieb, Y.; Elnakib, A.; Shalaby, A.; Mahmoud, A.; Ghazal, M.; Yousaf, J.; Abu Khalifeh, H.; Casanova, M.; Barnes, G.; et al. The Role of Structure MRI in Diagnosing Autism. Diagnostics 2022, 12, 165. [Google Scholar] [CrossRef] [PubMed]
- Rojas, D.; Peterson, E.; Winterrowd, E.; Reite, M.; Rogers, S.; Tregellas, J. Regional gray matter volumetric changes in autism associated with social and repetitive behavior symptoms. BMC Psychiatry 2006, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Ye, H.; Heidari, A.; Zheng, L.; Hu, Z.; Chen, H.; Turabieh, H.; Mafarja, M.; Wu, P. Analysis of COVID-19 severity from the perspective of coagulation index using evolutionary machine learning with enhanced brain storm optimization. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 4874–4887. [Google Scholar] [CrossRef]
- Muhammed Niyas, K.P.; Thiyagarajan, P. Feature selection using efficient fusion of Fisher Score and greedy searching for Alzheimer’s classification. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 4993–5006. [Google Scholar]
- Morris, C.; Rekik, I. Autism spectrum disorder diagnosis using sparse graph embedding of morphological brain networks. In Graphs in Biomedical Image Analysis, Computational Anatomy and Imaging Genetics; Springer: Cham, Switzerland, 2017; pp. 12–20. [Google Scholar]
- Soussia, M.; Rekik, I. Unsupervised manifold learning using high-order morphological brain networks derived from T1-w MRI for autism diagnosis. Front. Neuroinform. 2018, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Fang, H.; Wu, J.; Xiao, C.; Xiao, T.; Qian, L.; Liang, F.; Xiao, Z.; Chu, K.; Ke, X. Diagnostic model generated by MRI-derived brain features in toddlers with autism spectrum disorder. Autism Res. 2017, 10, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Yassin, W.; Nakatani, H.; Zhu, Y.; Kojima, M.; Owada, K.; Kuwabara, H.; Gonoi, W.; Aoki, Y.; Takao, H.; Natsubori, T.; et al. Machine-learning classification using neuroimaging data in schizophrenia, autism, ultra-high risk and first-episode psychosis. Transl. Psychiatry 2020, 10, 278. [Google Scholar] [CrossRef]
- Katuwal, G. Machine Learning Based Autism Detection Using Brain Imaging. Ph.D. Thesis, Rochester Institute of Technology, Rochester, NY, USA, 2017. [Google Scholar]
- Xu, M.; Calhoun, V.; Jiang, R.; Yan, W.; Sui, J. Brain imaging-based machine learning in autism spectrum disorder: Methods and applications. J. Neurosci. Methods 2021, 361, 109271. [Google Scholar] [CrossRef]
- Demirhan, A. The effect of feature selection on multivariate pattern analysis of structural brain MR images. Phys. Medica 2018, 47, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.; Barnes, G.; Nitzken, M.; Switala, A.; Shalaby, A.; Hosseini-Asl, E.; Casanova, M.; Keynton, R.; Khalil, A.; El-Baz, A. A new deep-learning approach for early detection of shape variations in autism using structural mri. In Proceedings of the 2017 IEEE International Conference On Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 1057–1061. [Google Scholar]
- Squarcina, L.; Nosari, G.; Marin, R.; Castellani, U.; Bellani, M.; Bonivento, C.; Fabbro, F.; Molteni, M.; Brambilla, P. Automatic classification of autism spectrum disorder in children using cortical thickness and support vector machine. Brain Behav. 2021, 11, e2238. [Google Scholar] [CrossRef] [PubMed]
- Bilgen, I.; Guvercin, G.; Rekik, I. Machine learning methods for brain network classification: Application to autism diagnosis using cortical morphological networks. J. Neurosci. Methods 2020, 343, 108799. [Google Scholar] [CrossRef] [PubMed]
- Eill, A.; Jahedi, A.; Gao, Y.; Kohli, J.; Fong, C.; Solders, S.; Carper, R.; Valafar, F.; Bailey, B.; Müller, R. Functional connectivities are more informative than anatomical variables in diagnostic classification of autism. Brain Connect. 2019, 9, 604–612. [Google Scholar] [CrossRef]
- Katuwal, G.; Cahill, N.; Baum, S.; Michael, A. The predictive power of structural MRI in Autism diagnosis. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015, 2015, 4270–4273. [Google Scholar] [PubMed]
- Gorriz, J.; Ramıirez, J.; Segovia, F.; Martinez, F.; Lai, M.; Lombardo, M.; Baron-Cohen, S.; Consortium, M.; Suckling, J. A machine learning approach to reveal the neurophenotypes of autisms. Int. J. Neural Syst. 2019, 29, 1850058. [Google Scholar] [CrossRef] [PubMed]
- Akhavan Aghdam, M.; Sharifi, A.; Pedram, M. Combination of rs-fMRI and sMRI data to discriminate autism spectrum disorders in young children using deep belief network. J. Digit. Imaging 2018, 31, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Ke, F.; Choi, S.; Kang, Y.H.; Cheon, K.-A.; Lee, S.W. Exploring the structural and strategic bases of autism spectrum disorders with deep learning. IEEE Access 2020, 8, 153341–153352. [Google Scholar] [CrossRef]
- Eslami, T.; Almuqhim, F.; Raiker, J.; Saeed, F. Machine Learning Methods for Diagnosing Autism Spectrum Disorder and Attention-Deficit/Hyperactivity Disorder Using Functional and Structural MRI: A Survey. Front. Neuroinform. 2021, 14, 62. [Google Scholar] [CrossRef]
- Itani, S.; Thanou, D. Combining anatomical and functional networks for neuropathology identification: A case study on autism spectrum disorder. Med. Image Anal. 2021, 69, 101986. [Google Scholar] [CrossRef]
- Chen, C.; Keown, C.; Jahedi, A.; Nair, A.; Pflieger, M.; Bailey, B.; Müller, R. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage Clin. 2015, 8, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Alsuliman, M.; Al-Baity, H. Efficient Diagnosis of Autism with Optimized Machine Learning Models: An Experimental Analysis on Genetic and Personal Characteristic Datasets. Appl. Sci. 2022, 12, 3812. [Google Scholar] [CrossRef]
- Ahmed, H.; Soliman, H.; Elmogy, M. Early detection of Alzheimer’s disease using single nucleotide polymorphisms analysis based on gradient boosting tree. Comput. Biol. Med. 2022, 146, 105622. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, A.; Yan, C.; Li, Q.; Denio, E.; Castellanos, F.; Alaerts, K.; Anderson, J.; Assaf, M.; Bookheimer, S.; Dapretto, M.; et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry. 2014, 19, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, A.; O’connor, D.; Chen, B.; Alaerts, K.; Anderson, J.; Assaf, M.; Balsters, J.; Baxter, L.; Beggiato, A.; Bernaerts, S.; et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci. Data 2017, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Autism Brain Imaging Data Exchange! Available online: https://fcon_1000.projects.nitrc.org/indi/abide/ (accessed on 25 October 2023).
- Cyberduck: Libre Server and Cloud Storage Browser for Mac and Windows with Support for FTP, SFTP, WebDAV, Amazon S3, OpenStack Swift, Backblaze B2, Microsoft Azure & Onedrive, Google Drive and Dropbox. Available online: https://cyberduck.io/ (accessed on 25 October 2023).
- O’Connor, D.; Clark, D.; Milham, M.; Craddock, R. Sharing data in the cloud. GigaScience 2016, 5. [Google Scholar] [CrossRef]
- Brain Imaging Data Structure. Available online: https://bids.neuroimaging.io/ (accessed on 25 March 2023).
- NITRC: Mricrogl. Available online: https://www.nitrc.org/projects/mricrogl/ (accessed on 25 March 2023).
- Book, G.; Stevens, M.; Assaf, M.; Glahn, D.; Pearlson, G. Neuroimaging data sharing on the neuroinformatics database platform. Neuroimage 2016, 124, 1089–1092. [Google Scholar] [CrossRef] [PubMed]
- Fischl, B. FreeSurfer. Neuroimage 2012, 62, 774–781. [Google Scholar] [CrossRef]
- Khodatars, M.; Shoeibi, A.; Ghassemi, N.; Jafari, M.; Khadem, A.; Sadeghi, D.; Moridian, P.; Hussain, S.; Alizadehsani, R.; Zare, A.; et al. Deep Learning for Neuroimaging-based Diagnosis and Rehabilitation of Autism Spectrum Disorder: A Review. arXiv 2020, arXiv:2007.01285. [Google Scholar] [CrossRef]
- Faraji, R.; Ganji, Z.; Alreza, Z.; Akbari-Lalimi, H.; Zare, H. Volume-based and Surface-Based Methods in Autism compared with Healthy Controls; Are Freesurfer and CAT12 in Agreement? Preprints. 2022. Available online: https://www.researchsquare.com/article/rs-1840707/v1 (accessed on 25 March 2023).
- Bas-Hoogendam, J.; Steenbergen, H.; Tissier, R.; Houwing-Duistermaat, J.; Westenberg, P.; Wee, N. Subcortical brain volumes, cortical thickness and cortical surface area in families genetically enriched for social anxiety disorder—A multiplex multigenerational neuroimaging study. EBioMedicine 2018, 36, 410–428. [Google Scholar] [CrossRef]
- Bisong, E. Introduction to Scikit-learn. In Building Machine Learning and Deep Learning Models on Google Cloud Platform; Apress: Berkeley, CA, USA, 2019; pp. 215–229. [Google Scholar]
- Shankar, K.; Lakshmanaprabu, S.; Khanna, A.; Tanwar, S.; Rodrigues, J.; Roy, N. Alzheimer detection using Group Grey Wolf Optimization based features with convolutional classifier. Comput. Electr. Eng. 2019, 77, 230–243. [Google Scholar]
- Wang, X.; Li, J. Detecting communities by the core-vertex and intimate degree in complex networks. Phys. A 2013, 392, 2555–2563. [Google Scholar] [CrossRef]
- Ali, M.; Elnakieb, Y.; Shalaby, A.; Mahmoud, A.; Switala, A.; Ghazal, M.; Khelifi, A.; Fraiwan, L.; Barnes, G.; El-Baz, A. Autism classification using smri: A recursive features selection based on sampling from multi-level high dimensional spaces. In Proceedings of the 2021 IEEE 18th International Symposium On Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 267–270. [Google Scholar]
- Scikit-Learn-Contrib Python Implementations of the Boruta AllRelevant Feature Selection Method. Available online: https://github.com/scikit-learn-contrib/boruta_py (accessed on 25 March 2023).
- Kursa, M.; Jankowski, A.; Rudnicki, W. Boruta–a system for feature selection. Fundam. Inf. 2010, 101, 271–285. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.; Lewis, A. Grey wolf optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- NiaPy’s Documentation. Available online: https://niapy.org/en/stable/_modules/index.html (accessed on 12 July 2022).
- Tang, R.; Zhang, X. CART decision tree combined with Boruta feature selection for medical data classification. In Proceedings of the 2020 5th IEEE International Conference on Big Data Analytics (ICBDA), Xiamen, China, 8–11 May 2020; pp. 80–84. [Google Scholar]
- Khan, A.; Zubair, S. Development of a three tiered cognitive hybrid machine learning algorithm for effective diagnosis of Alzheimer’s disease. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 8000–8018. [Google Scholar] [CrossRef]
- Ogwo, O. Medical Data Classification Using Binary Brain Storm Optimization. Master’s Thesis, Texas A&M University-Corpus Christi, Corpus Christi, TX, USA, 2019. [Google Scholar]
- Mahapatra, S.; Sahu, S. ANOVA-PSO based feature selection and gradient boosting machine classifier for improved protein-protein interaction prediction. Proteins 2021, 90, 443–454. [Google Scholar] [CrossRef]
- Mellema, C.; Treacher, A.; Nguyen, K.; Montillo, A. Multiple deep learning architectures achieve superior performance diagnosing autism spectrum disorder using features previously extracted from structural and functional mri. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1891–1895. [Google Scholar]
- Vrbančič, G.; Pečnik, Š.; Podgorelec, V. Identification of COVID-19 X-ray images using CNN with optimized tuning of transfer learning. In Proceedings of the 2020 International Conference On Innovations In Intelligent Systems And Applications (INISTA), Novi Sad, Serbia, 24–26 August 2020; pp. 1–8. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Nugroho, A.; Suhartanto, H. Hyper-parameter tuning based on random search for densenet optimization. In Proceedings of the 2020 7th International Conference on Information Technology, Computer, and Electrical Engineering (ICITACEE), Semarang, Indonesia, 24–25 September 2020; pp. 96–99. [Google Scholar]
- Class NatureInspiredSearchCV—Sklearn Nature Inspired Algorithms Documentation. Available online: https://sklearn-nature-inspired-algorithms.readthedocs.io/en/latest/introduction/nature-inspired-search-cv.html (accessed on 25 March 2023).
- McCormick, K.; Salcedo, J. SPSS Statistics for Data Analysis and Visualization; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Jiao, Y.; Chen, R.; Ke, X.; Chu, K.; Lu, Z.; Herskovits, E. Predictive models of autism spectrum disorder based on brain regional cortical thickness. Neuroimage 2010, 50, 589–599. [Google Scholar] [CrossRef]
- Nordahl, C.; Dierker, D.; Mostafavi, I.; Schumann, C.; Rivera, S.; Amaral, D.; Van Essen, D. Cortical folding abnormalities in autism revealed by surface-based morphometry. J. Neurosci. 2007, 27, 11725–11735. [Google Scholar] [CrossRef]
- Hong, S.; Hyung, B.; Paquola, C.; Bernhardt, B. The superficial white matter in autism and its role in connectivity anomalies and symptom severity. Cereb. Cortex 2019, 29, 4415–4425. [Google Scholar] [CrossRef]
- Ecker, C.; Ginestet, C.; Feng, Y.; Johnston, P.; Lombardo, M.; Lai, M.; Suckling, J.; Palaniyappan, L.; Daly, E.; Murphy, C.; et al. Brain surface anatomy in adults with autism: The relationship between surface area, cortical thickness, and autistic symptoms. JAMA Psychiatry 2013, 70, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kim, S.; Lee, J.; Lee, W. Brain Age Prediction: A Comparison between Machine Learning Models Using Brain Morphometric Data. Sensors 2022, 22, 8077. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Ye, Q.; Xia, J. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf. Fusion 2022, 77, 29–52. [Google Scholar] [CrossRef] [PubMed]
- Shattuck, D.; Leahy, R. BrainSuite: An automated cortical surface identification tool. Med. Image Anal. 2002, 6, 129–142. [Google Scholar] [CrossRef]
- Bloch, L.; Friedrich, C. Comparison of Automated Volume Extraction with FreeSurfer and FastSurfer for Early Alzheimer’s Disease Detection with Machine Learning. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 113–118. [Google Scholar]







| Feature Selection | Feature Selection Method | Modality | Ref., Year | Biomarker | Dataset | Subjects | Age | Preprocessing Tools | Classifiers | Validation | Best ACC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ML | |||||||||||
| Without FS | - | sMRI + personal and behavioral features (PBC) | [16], 2017 | GM, WM, CSF and total intracranial volume | ABIDE I | ASD = 114, TD = 108 | 6–13 years | FreeSurfer | RF, XGB | 10-fold CV | Highest ACC by RF: 60% |
| - | sMRI | [14], 2017 | Volume, CT, Cortical surface | private data | ASD = 46, Development delay = 39 | 18 to 37 months | FreeSurfer | RF, NB, SVM | 5-fold CV | CT + RF: 80.9 ± 1.5 | |
| - | sMRI + fMRI | [28] 2021 | Graph signals | ABIDE I pre-processed | ASD = 201, TD = 251 | 6–18 years old | GBC, SVM | DT | LOOCV | ACC: 67.7 | |
| Supervised sample FS: Filter | Variable importance measures in RF | fMRI, sMRI and DWI | [22], 2019 | ROI-based FC and various anatomic features | Private data | ASD = 46, TD = 47 | 13.6 ± 2.8 years | FreeSurfer, FSL and AFNI | RF | - | Highest ACC: RF combining the top 19 variables: 92.5 |
| 1st: SelectKBest Algorithm, 2nd: Minimum Redundancy Maximum Relevance | sMRI | [21], 2020 | Cortical MBN | ABIDE I | ASD = 100, TD = 100 | Unknown | FreeSurfer | LR, SVM, DT, LDA, KNN, QDA, RF, AdaBoost, GBC, XGB | - | GBC 1st: 70% | |
| statistical test | GM and WM | [24], 2019 | sMRI | MRC AIMS collected data | ASD = 60, TD = 60 | 18–49 years | SPM 12 | SVM | Groups of CV | Highest ACC: 86% | |
| Supervised sample FS: wrapper | greedy forward-feature selection | T1-sMRI | [20], 2021 | Regional CT | Private data | ASD = 40, TD = 36 | 9.5 ± 3.4 years | FreeSurfer | SVM | LOOCV | 84.2% |
| Unsupervised FS | sparse graph embedding | T1w-sMRI | [12], 2017 | Morphological brain connectivity using a set of cortical attributes | ABIDE I | ASD = 59, TD = 43 | Unknown | FreeSurfer | SVM | LOOCV | 61.76% |
| Unsupervised Dimensionality Reduction | PCA | sMRI | [15], 2020 | CT, SA and sub-cortical features | Private data | Schizophrenia = 64, ASD = 36, TD = 106 | Schizophrenia = 14–60, ASD = 20–44, TD = 16–60 years | FreeSurfer and Enhancing Neuroimaging Genetics | SVM, DT, LR, KNN, RF, AdaBoost | 10-fold CV | Highest Acc: multi-class classification LR + CT = 69, ASD vs. TD binary classification => 70 |
| DL | |||||||||||
| Supervised sample FS: Filter | ReliefF and mRMR | CT | [18], 2018 | sMRI | 5 datasets | ASD = 325, TD = 325 | 17.8 ± 7.4 years | FreeSurfer | NN, SVM, KNN | 5-fold CV | 62% |
| Supervised sample FS: Wrapper | RFECV | sMRI | [8], 2022 | set of morphological features | ABIDE I | ASD = 530, TD = 573 | 6–64 years | FreeSurfer | LASSO, RF, SVM, and NN | 4-fold CV | NN Avg ACC = 71% |
| Unsupervised Dimensionality Reduction | PCA | sMRI and rs-fMRI | [25], 2018 | Regional based mean time series+ GM+ WM | ABIDE I and ABIDE II | ASD = 116, TD = 69 | 5–10 Years | SPM 8 | DBN of depth 3 + LR | 10-fold CV | 65.56% |
| Dataset | ASD% | Male% | Age (years) | Total Participants |
|---|---|---|---|---|
| ABIDE I | 47.7 | 82.7 | 6.4–10.9 | 220 |
| ABIDE II | 44.7 | 69.4 | 5.1–10.9 | 418 |
| KAU | 57.6 | 66.7 | 5.4–10.8 | 33 |
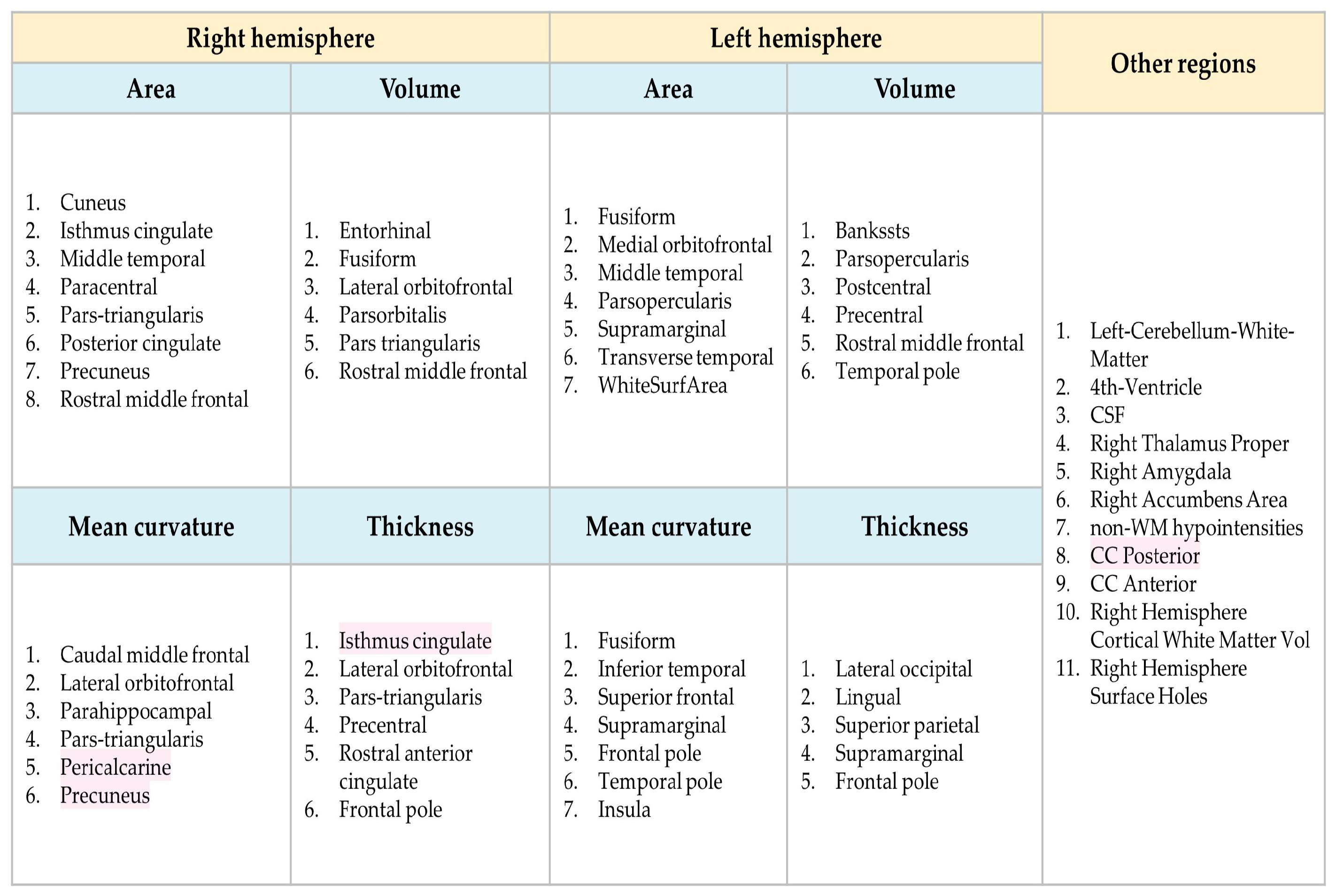
| Cortical Regions | |||||
|---|---|---|---|---|---|
| #Label | Label Name | Name | #Label | Label Name | Name |
| 1 | lh bankssts | Banks of superior temporal sulcus | 35 | rh bankssts | Banks of superior temporal sulcus |
| 2 | lh caudal anteriorcingulate | Caudal anterior cingulate cortex | 36 | rh caudalanteriorcingulate | Caudal anterior cingulate cortex |
| 3 | lh caudal middlefrontal | Caudal middle frontal gyrus | 37 | rh caudal middlefrontal | Caudal middle frontal gyrus |
| 4 | lh cuneus | Cuneus | 38 | rh cuneus | Cuneus |
| 5 | lh entorhinal | Entorhinal cortex | 39 | rh entorhinal | Entorhinal cortex |
| 6 | lh fusiform | Fusiform gyrus | 40 | rh fusiform | Fusiform gyrus |
| 7 | lh inferiorparietal | Inferior parietal lobule | 41 | rh inferiorparietal | Inferior parietal lobule |
| 8 | lh inferiortemporal | Inferior temporal gyrus | 42 | rh inferiortemporal | Inferior temporal gyrus |
| 9 | lh lateraloccipital | Lateral occipital gyrus | 43 | rh lateraloccipital | Lateral occipital gyrus |
| 10 | lh caudal lateralorbitofrontal | Lateral orbitofrontal gyrus | 44 | rh caudal lateralorbitofrontal | Lateral orbitofrontal gyrus |
| 11 | lh lingual | Lingual gyrus | 45 | rh lingual | Lingual gyrus |
| 12 | lh caudal medialorbitofrontal | Medial orbitofrontal gyrus | 46 | rh caudal medialorbitofrontal | Medial orbitofrontal gyrus |
| 13 | lh middletemporal | Medial temporal gyrus | 47 | rh middletemporal | Medial temporal gyrus |
| 14 | lh parahippocampal | Parahippocampal gyrus | 48 | rh parahippocampal | Parahippocampal gyrus |
| 15 | lh paracentral | Paracentral gyrus | 49 | rh paracentral | Paracentral gyrus |
| 16 | lh parsopercularis | Pars opercularis | 50 | rh parsopercularis | Pars opercularis |
| 17 | lh parsorbitalis | Pars orbitalis | 51 | rh parsorbitalis | Pars orbitalis |
| 18 | lh parstriangularis | Pars triangularis | 52 | rh parstriangularis | Pars triangularis |
| 19 | lh pericalcarine | Pericalcarine gyrus | 53 | rh pericalcarine | Pericalcarine gyrus |
| 20 | lh postcentral | Postcentral gyrus | 54 | rh postcentral | Postcentral gyrus |
| 21 | lh posterior cingulate | Posterior cingulate cortex | 55 | rh posteriorcingulate | Posterior cingulate cortex |
| 22 | lh precentral | Precentral gyrus | 56 | rh precentral | Precentral gyrus |
| 23 | lh precuneus | Precuneus | 57 | rh precuneus | Precuneus |
| 24 | lh rostral anteriorcingulate | Rostral anterior cingulate cortex | 58 | rh rostral anteriorcingulate | Rostral anterior cingulate cortex |
| 25 | lh rostral middlefrontal | Rostral middle frontal gyrus | 59 | rh rostral middlefrontal | Rostral middle frontal gyrus |
| 26 | lh superiorfrontal | Superior frontal gyrus | 60 | rh superiorfrontal | Superior frontal gyrus |
| 27 | lh superiorparietal | Superior parietal gyrus | 61 | rh superiorparietal | Superior parietal gyrus |
| 28 | lh superiortemporal | Superior temporal gyrus | 62 | rh superiortemporal | Superior temporal gyrus |
| 29 | lh supramarginal | Supramarginal gyrus | 63 | rh supramarginal | Supramarginal gyrus |
| 30 | lh frontalpole | Frontal pole | 64 | rh frontalpole | Frontal pole |
| 31 | lh temporalpole | Temporal pole | 65 | rh temporalpole | Temporal pole |
| 32 | lh transversetemporal | Transverse temporal gyrus | 66 | rh transversetemporal | Transverse temporal gyrus |
| 33 | lh insula | Insula | 67 | rh insula | Insula |
| 34 | lh isthmuscingulate | Isthmus cingulate | 68 | rh isthmuscingulate | Isthmus cingulate |
| Sub-Cortical regions | |||||
| 69 | Left Thalamus Proper | Thalamus | 85 | Right-Caudate | Caudate nucleus |
| 70 | Left-Hippocampus | Hippocampus | 86 | Right-Amygdala | Amygdala |
| 71 | Left-Caudate | Caudate nucleus | 87 | Right Accumbens area | Nucleus Accumbens |
| 72 | Left-Amygdala | Amygdala | 88 | Right-Lateral-Ventricle | Lateral ventricles |
| 73 | Left-Accumbens-area | Nucleus Accumbens | 89 | Right-Inf-Lat-Vent | |
| 74 | Left-Lateral-Ventricle | Lateral ventricles | 90 | Right Cerebellum White Matter | Cerebellum-White-Matter |
| 75 | Left-Inf-Lat-Vent | 91 | Surfaces holes | ||
| 76 | Left-Cerebellum-White-Matter | Cerebellum-White-Matter | 92 | Right-Putamen | Putamen |
| 77 | Left-Putamen | Putamen | 94 | Right-choroid-plexus | Choroid-plexus |
| 78 | Left-Pallidum | Pallidum | 95 | Right-VentralDC | Ventral Diencephalon |
| 79 | Left-choroid-plexus | Choroid-plexus | 96 | Right-vessel | Vessel |
| 80 | Left-VentralDC | Ventral Diencephalon | 97 | 3rd-Ventricle | Ventricle |
| 81 | Left-vessel | Vessel | 98 | 4th-Ventricle | |
| 82 | Right-Thalamus-Proper | Thalamus | 99 | 5th-Ventricle | |
| 83 | Right-Hippocampus | Hippocampus | |||
| Model Name | Hyperparameter Name | Definition | Hyperparameter Value Range |
|---|---|---|---|
| Support Vector Machine (SVM) | C value | The penalty parameter | (0.1, 0.8, 0.9, 1, 1.1, 1.2, 1.3, 1.4, 0.25, 0.5, 0.75, 10, 100) |
| Kernel | Defining the algorithm | linear, rbf, poly, and sigmoid | |
| Degree | The degree of the polynomial kernel function (‘poly’) | 1, 2, 3, 4, 5, 6 | |
| Gamma | Kernel coefficient | scale, auto | |
| Decision Tree (DT) | Criterion | The function to measure the quality of a split | Gini and entropy |
| Max_depth | The maximum depth of the tree | None, 2, 4, 6, 8, 10, or 12 | |
| Random forest (RF) | N_estimators | Number of estimators | 10, 20, 50, 64, 100, 140, 200, and 256 |
| Min_samples split | Minimal sample count necessary to split an internal node | 1, 2, 3, 6 | |
| Min_samples_leaf | Minimum amount of samples at the tree’s leaves | 1, 6, and 10 | |
| Max_features | The number of features to consider for the best split | sqrt, log2, None | |
| Criterion of trees | The function to measure the quality of a split | Gini and entropy | |
| Max depth | The maximum depth of the tree | None, 2, 4, 5, 6, 7, 8, 16, or 30 | |
| Naïve Bayes (NB) | Smoothing | Laplace smoothing technique helps tackle the problem of zero probability | (, , , , , , , , ) |
| Extreme Gradient Boosting (XGB) | Max depth | Maximum depth of the individual estimators/trees | 3, 4, 5, 6, 8, 10, 12, 15 |
| Gamma | Minimum loss reduction required to partition a leaf node of the tree | 0, 0.1, 0.2, 0.3, 0.4 | |
| Colsample by tree | subsample ratio of columns when constructing each tree | 0.3, 0.4, 0.5, 0.7, 1 | |
| Learning rate | Step size shrinkage used in update to prevents overfitting | 0.05, 0.10, 0.15, 0.20, 0.25, 0.3 | |
| Min child weight | Minimum sum of instance weight needed in a child | 1, 3, 5, 7 | |
| Category Boosting (CatBoost) | Depth | Depth of the tree | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 |
| Learning rate | The rate at which the model weights are updated after working through each batch of training examples | 0.01, 0.02, 0.03, 0.04, 0.009 | |
| Iteration | The maximum number of trees that can be built | 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 250, 500 | |
| Multi-layer Perceptron (MLP) | Hidden layer sizes | Number of hidden layers | (50,50,50), (50,100,50), (100), (10,30,10), (100, 3), (3,3), and (20) |
| Activation | Activation function for the hidden layer | ‘identity’, ‘logistic’, ‘tanh’, or ‘relu’. | |
| Solver | The solver for weight optimization | ‘lbfgs’, ‘sgd’, or ‘adam’ | |
| Alpha | Strength of the L2 regularization term | 0.0001, , and 0.05 | |
| Learning rate | Learning rate schedule for weight updates | ‘constant’, ‘invscaling’, ‘adaptive’ |
| Experiment 1.1: Baseline Models | ||||||||
|---|---|---|---|---|---|---|---|---|
| Model | Feature Set/Number of Features | Model Performance on Training Dataset | Prediction Performance on Testing Dataset | |||||
| Mean Accuracy | ABIDE I + ABIDE II | KAU | ||||||
| Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | |||
| CatBoost | F1: cortical region’s volumes/66 | 62.94 | 58.81 | 57.81 | 56.76 | 63.64 | 63.64 | 61.84 |
| SVM | F2: volumes of sub-cortical regions/68 | 61.18 | 61.94 | 60.94 | 60.46 | 69.7 | 69.7 | 67.11 |
| CatBoost | F3: surface area of sub-cortical regions/70 | 59.61 | 62.72 | 61.72 | 60.39 | 54.55 | 54.55 | 53.01 |
| CatBoost | F4: cortical thickness of sub-cortical regions/70 | 63.73 | 61.16 | 60.16 | 58.03 | 72.73 | 72.73 | 67.86 |
| CatBoost | F5: mean curvatures of sub-cortical regions/68 | 64.12 | 69.75 | 68.75 | 67.47 | 63.64 | 63.64 | 62.78 |
| RF | F6: all features/342 | 65.29 | 65.84 | 62.5 | 61.56 | 48.48 | 48.48 | 44.92 |
| Experiment 1.2 : Models with Random Search | ||||||||
|---|---|---|---|---|---|---|---|---|
| Model | Feature Set /Number of Features | Model Performance on Training Dataset | Prediction Performance on Testing Dataset | |||||
| Mean Accuracy | ABIDE I + ABIDE II | KAU | ||||||
| Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | |||
| CatBoost | F1: cortical region’s volumes/66 | 64.91 | 59.59 | 58.59 | 57.68 | 78.79 | 78.79 | 78.76 |
| CatBoost | F2: volumes of sub-cortical regions/68 | 61.57 | 64.28 | 63.28 | 61.99 | 72.73 | 72.73 | 71.62 |
| XGB | F3: surface area of sub-cortical regions/70 | 60.78 | 59.59 | 58.59 | 58.68 | 57.58 | 57.58 | 55.64 |
| XGB | F4: cortical thickness of sub-cortical regions/70 | 63.53 | 60.38 | 59.38 | 58.95 | 66.67 | 66.67 | 63.53 |
| CatBoost | 63.92 | 62.72 | 61.72 | 58.91 | 84.85 | 84.85 | 82.14 | |
| CatBoost | F5: mean curvatures of sub-cortical regions/68 | 64.91 | 69.75 | 68.75 | 66.72 | 72.73 | 72.73 | 73.50 |
| CatBoost | F6: all features/342 | 66.27 | 65.84 | 64.84 | 63.09 | 51.5 | 48.49 | 44.92 |
| Model | Feature Set/Number of Features | Model Performance on Training Dataset | Prediction Performance on Testing Dataset | |||||
|---|---|---|---|---|---|---|---|---|
| Mean Accuracy | ABIDE I + ABIDE II | KAU | ||||||
| Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | |||
| Experiment 2.1: models with random search and recursive feature elimination with cross-validation (RFECV) | ||||||||
| XGB | F1: cortical region’s volumes/12 | 66.08 | 59.59 | 58.59 | 57.43 | 63.64 | 63.64 | 63.72 |
| MLP | F2: volumes of sub-cortical regions/17 | 65.49 | 64.28 | 63.28 | 61.74 | 69.7 | 69.7 | 67.11 |
| DT | F3: surface area of sub-cortical regions/16 | 60.59 | 58.81 | 57.81 | 59.01 | 66.67 | 66.67 | 63.53 |
| XGB | 60.98 | 55.69 | 54.69 | 53.05 | 57.58 | 57.58 | 57.52 | |
| XGB | F4: cortical thickness of sub-cortical regions/19 | 64.31 | 61.94 | 60.94 | 58.67 | 66.67 | 66.67 | 62.59 |
| CatBoost | F5: mean curvatures of sub-cortical regions/29 | 65.69 | 65.84 | 64.84 | 62.84 | 69.7 | 69.7 | 68.05 |
| DT | F6: all features/342 | 67.24 | 61.16 | 60.16 | 59.03 | 45.45 | 45.45 | 41.35 |
| Experiment 2.2: models with random search and Boruta | ||||||||
| XGB | F1: cortical region’s volumes/6 | 64.90 | 54.91 | 53.91 | 51.88 | 60.61 | 60.61 | 63.91 |
| CatBoost | 64.71 | 61.94 | 60.94 | 60.21 | 66.67 | 66.67 | 60.71 | |
| SVM | F2: volumes of sub-cortical regions/4 | 62.35 | 65.84 | 64.84 | 63.84 | 66.67 | 66.67 | 67.29 |
| XGB | F3: surface area of sub-cortical regions/3 | 62.35 | 56.47 | 56.48 | 54.98 | 48.48 | 48.48 | 48.68 |
| XGB | F4: cortical thickness of sub-cortical regions / 4 | 63.53 | 58.81 | 57.81 | 57.3 | 57.58 | 57.58 | 54.7 |
| CatBoost | 63.73 | 58.81 | 57.81 | 56.91 | 66.67 | 66.67 | 61.65 | |
| CatBoost | F5: mean curvatures of sub-cortical regions/4 | 67.45 | 62.72 | 61.72 | 60.14 | 72.73 | 72.73 | 72.56 |
| NB | F6: all features/8 | 70.13 | 65.84 | 64.84 | 60.01 | 60.61 | 60.61 | 55.45 |
| SVM | 70.08 | 60.38 | 59.38 | 56.86 | 54.55 | 54.55 | 52.08 | |
| RF | 70.05 | 58.03 | 57.03 | 55.09 | 66.67 | 51.52 | 52.26 | |
| Experiment 2.3: models with random search and grey wolf-based optimizer (GWO) | ||||||||
| SVM | F1: cortical region’s volumes/16 | 67.06 | 61.94 | 60.94 | 59.46 | 54.55 | 54.55 | 49.25 |
| XGB | 67.25 | 55.69 | 54.69 | 53.80 | 60.61 | 60.61 | 58.27 | |
| MLP | 67.06 | 59.59 | 58.59 | 57.18 | 60.61 | 60.61 | 56.39 | |
| SVM | F2: volumes of sub-cortical regions/19 | 65 | 60.38 | 59.38 | 57.86 | 57.86 | 57.58 | 56.58 |
| SVM | F3: surface area of sub-cortical regions/14 | 66.86 | 55.69 | 54.69 | 53.30 | 54.55 | 54.55 | 51.13 |
| XGB | F4: cortical thickness of sub-cortical regions/19 | 63.53 | 60.38 | 59.38 | 58.95 | 66.67 | 66.67 | 63.53 |
| CatBoost | 68.63 | 61.16 | 60.16 | 58.03 | 66.67 | 66.67 | 62.59 | |
| SVM | F5: mean curvatures of sub-cortical regions/11 | 70 | 63.5 | 62.5 | 60.31 | 69.7 | 69.7 | 67.12 |
| SVM | F6: all features/62 | 70 | 65.84 | 64.84 | 63.34 | 57.58 | 57.58 | 54.69 |
| Experiment 3: Models with GWO-Based Hyperparameter Tuning and Feature Selection Algorithms + Age and Gender | ||||||||
|---|---|---|---|---|---|---|---|---|
| Model | Feature Set/Number of Features | Model Performance on Training Dataset | Prediction Performance on Testing Dataset | |||||
| Mean Accuracy | ABIDE I + ABIDE II | KAU | ||||||
| Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | |||
| SVM | 70.19 | 63.5 | 62.5 | 60.31 | 69.67 | 69.67 | 67.12 | |
| XGB | F5: mean curvatures of sub-cortical regions/13 | 67.06 | 58.03 | 57.03 | 55.33 | 78.79 | 78.79 | 75 |
| CatBoost | 69.80 | 62.72 | 61.72 | 60.14 | 63.64 | 69.7 | 66.17 | |
| SVM | F6: all features/62 | 71 | 64.28 | 63.28 | 61.74 | 57.58 | 57.58 | 53.76 |
| XGB | 68.63 | 59.59 | 58.59 | 57.18 | 45.45 | 45.45 | 43.23 | |
| CatBoost | 69.02 | 62.72 | 61.72 | 60.14 | 52.51 | 51.52 | 50.38 | |
| Ref., Year | Modality | Biomarker | Feature Selection Method | Dataset | Subjects | Age | Preprocessing Tools | Classifiers | Validation | Best ACC | Number of Final Features | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [25], 2018 | sMRI + rs-fMRI | Regional based mean time series + GM + WM | Unsupervised Dimensionality Reduction | ABIDE I + ABIDE II | ASD = 116, TD = 69 | 5–10 Years | SPM 8 | Deep Belief Network of depth 3 + LR | 10-fold CV | 65.56% | 348 | |
| [26], 2020 | sMRI | 3D volumetric data | - | ABIDE I + ABIDE II | ASD = 946, TD = 1046 | 8–40 Years old | SPM 8 | 2D/3D CNN, 2D/3D STN, RNN, class activation mapping, recurrent attention model | 10-fold CV | Highest ACC by 3D CNN + 3D STN: 60% | - | |
| Our study | sMRI + Age and gender data | Cortical regions thickness, volume, mean curvature, surface area + subcortical regions volumes | RFECV, Boruta, GWO-based algorithm | ABIDE I + ABIDE II | ASD:311, TD:360 | 5–10 years old | FreeSurfer | NB, DT, RF, SVM, CatBoost, XGB, and MLP | 10-fold CV | Highest ACC by GWO-SVM:71% | GWO-SVM: 62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahathiq, R.A.; Banjar, H.; Jarraya, S.K.; Bamaga, A.K.; Almoallim, R. Efficient Diagnosis of Autism Spectrum Disorder Using Optimized Machine Learning Models Based on Structural MRI. Appl. Sci. 2024, 14, 473. https://doi.org/10.3390/app14020473
Bahathiq RA, Banjar H, Jarraya SK, Bamaga AK, Almoallim R. Efficient Diagnosis of Autism Spectrum Disorder Using Optimized Machine Learning Models Based on Structural MRI. Applied Sciences. 2024; 14(2):473. https://doi.org/10.3390/app14020473
Chicago/Turabian StyleBahathiq, Reem Ahmed, Haneen Banjar, Salma Kammoun Jarraya, Ahmed K. Bamaga, and Rahaf Almoallim. 2024. "Efficient Diagnosis of Autism Spectrum Disorder Using Optimized Machine Learning Models Based on Structural MRI" Applied Sciences 14, no. 2: 473. https://doi.org/10.3390/app14020473
APA StyleBahathiq, R. A., Banjar, H., Jarraya, S. K., Bamaga, A. K., & Almoallim, R. (2024). Efficient Diagnosis of Autism Spectrum Disorder Using Optimized Machine Learning Models Based on Structural MRI. Applied Sciences, 14(2), 473. https://doi.org/10.3390/app14020473







