SemiPolypSeg: Leveraging Cross-Pseudo Supervision and Contrastive Learning for Semi-Supervised Polyp Segmentation
Abstract
1. Introduction
2. Related Works
2.1. Principal Models in Deep Learning Polyp Segmentation
2.2. Semi-Supervised Medical Image Segmentation Methods
2.2.1. Pseudo-Label-Based Methods
2.2.2. Consistency-Regularization-Based Methods
2.2.3. GAN-Based Methods
2.2.4. Contrastive-Learning-Based Methods
3. Materials and Methods
3.1. Outline of the SemiPolypSeg Framework
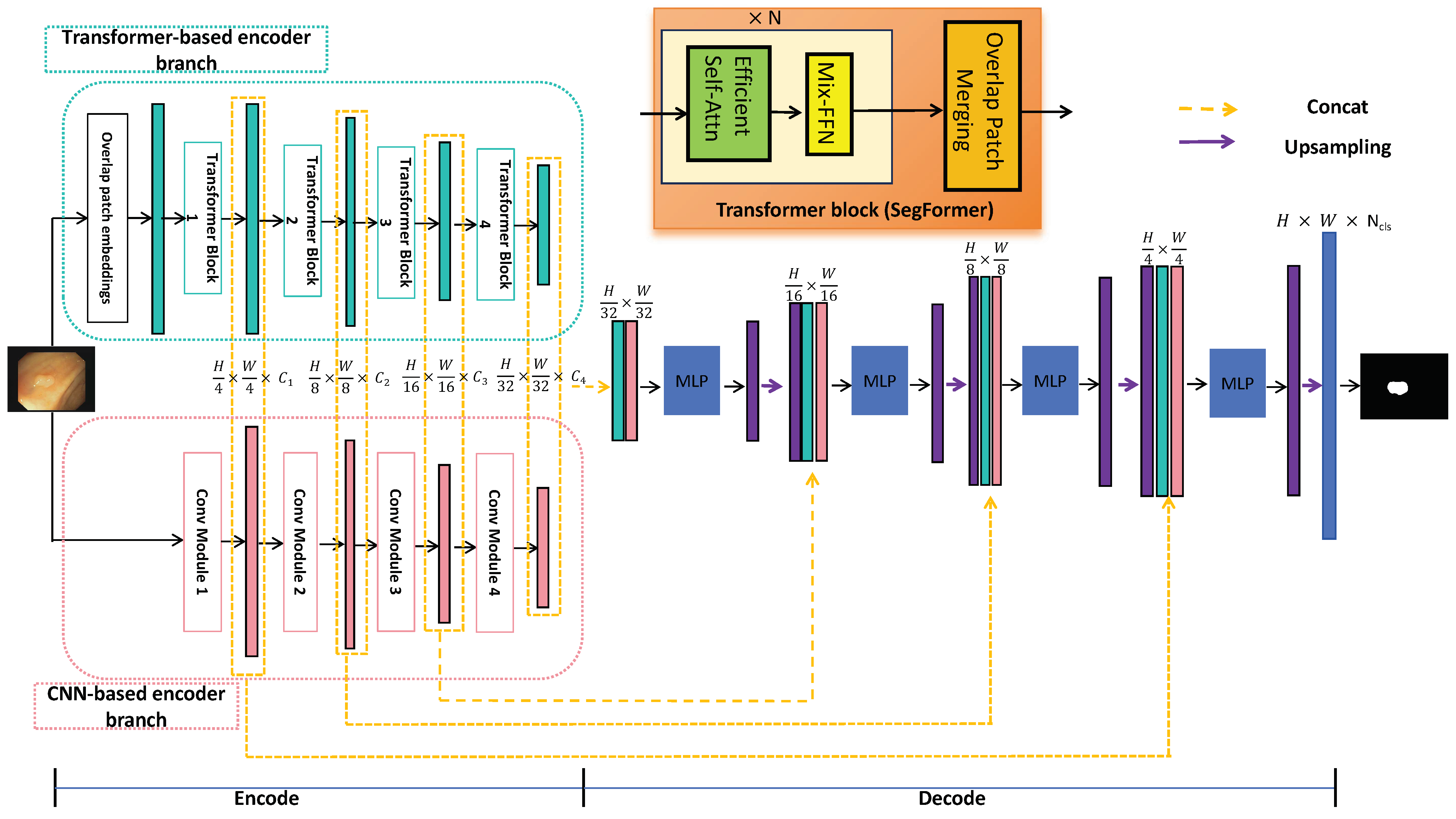
3.2. Hybrid Transformer–CNN Segmentation Networks
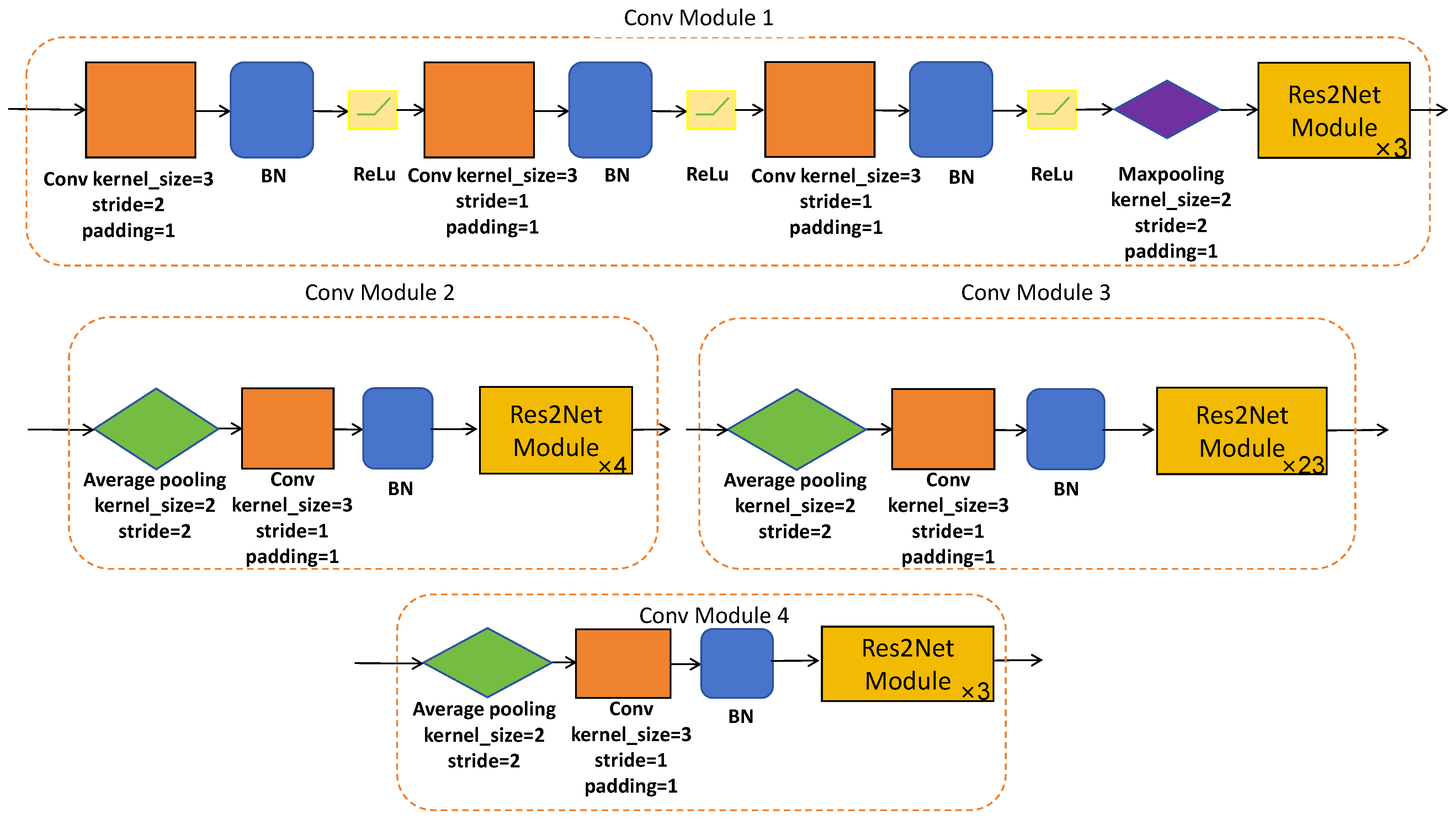
3.2.1. CNN-Based Encoder Branch
3.2.2. Transformer-Based Encoder Branch
3.2.3. ALL-MLP Decoder
3.3. Cross-Pseudo Supervision and Contrastive Learning
3.4. Loss Function
| Algorithm 1 SemiPolypSeg (training) |
| Input: Define the segmentation networks , projectors , batch size B, maximum epoch , labeled images , unlabeled images , and all datasets . Parameter: of Output: . Initialization: Initialize network and projector parameters and .
|
4. Results
4.1. Datasets and Evaluation Metrics
4.2. Implementation Details
4.3. Methods Comparison Analysis
4.4. Ablation Experiments
5. Discussion
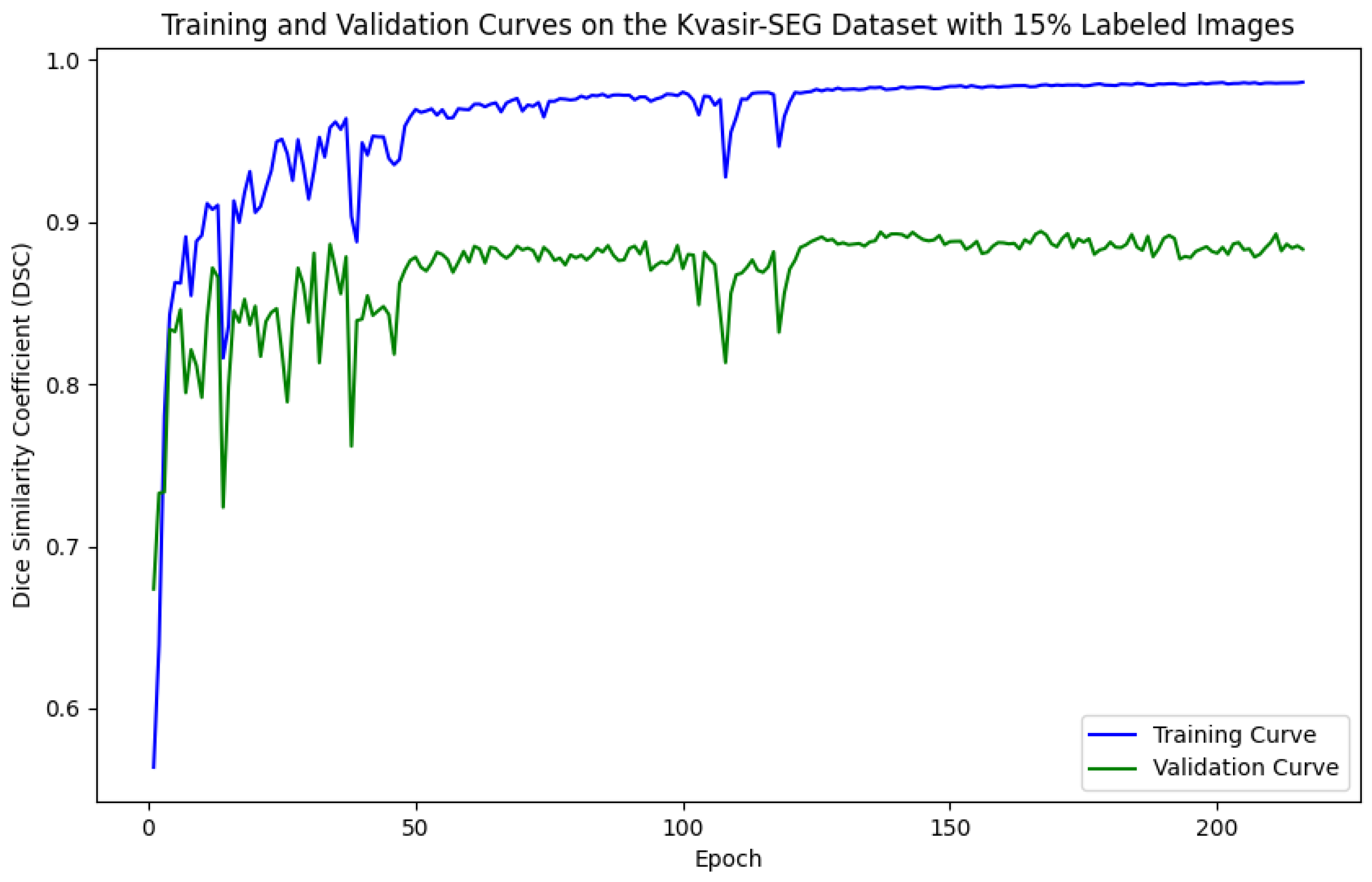
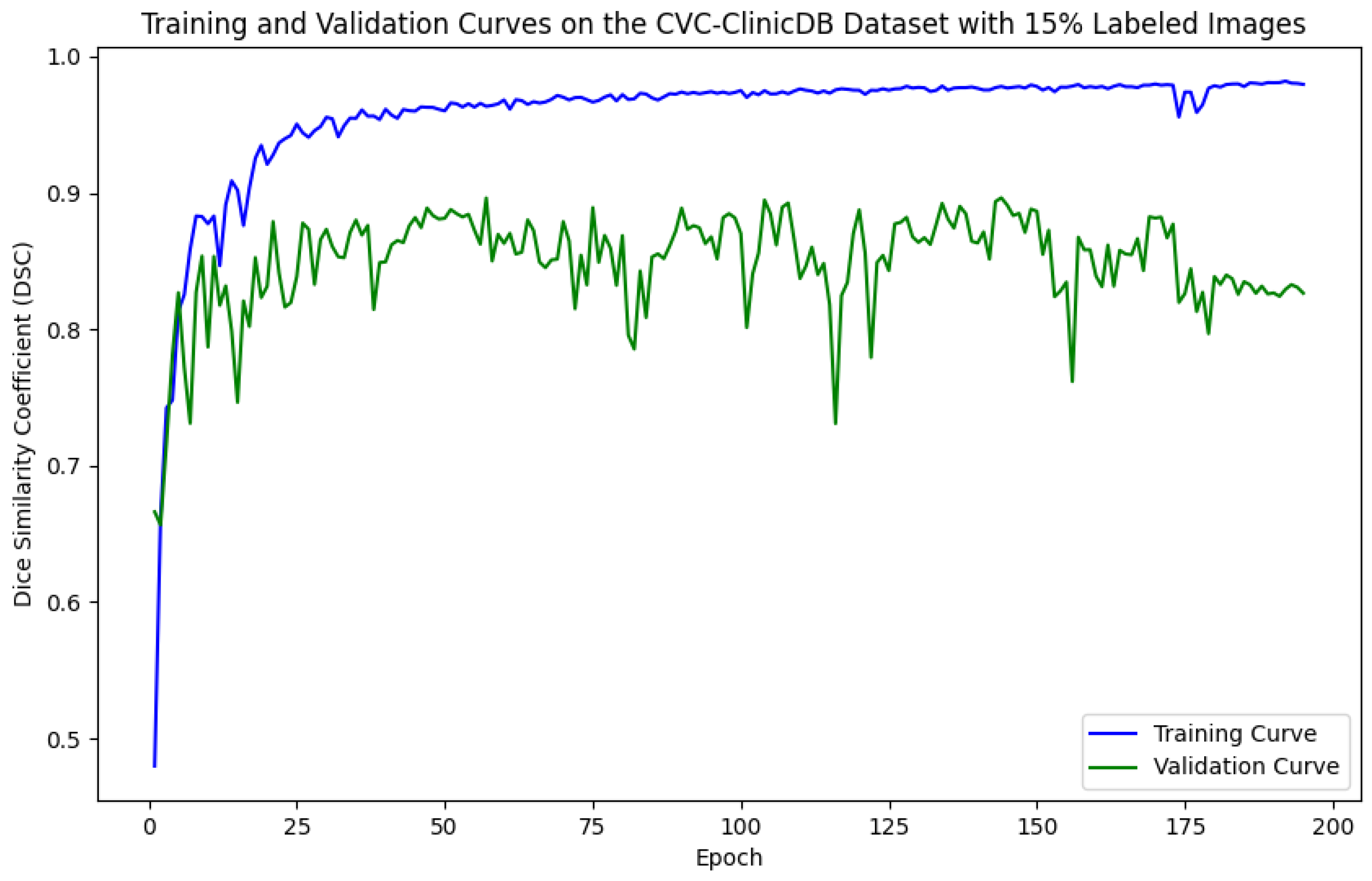
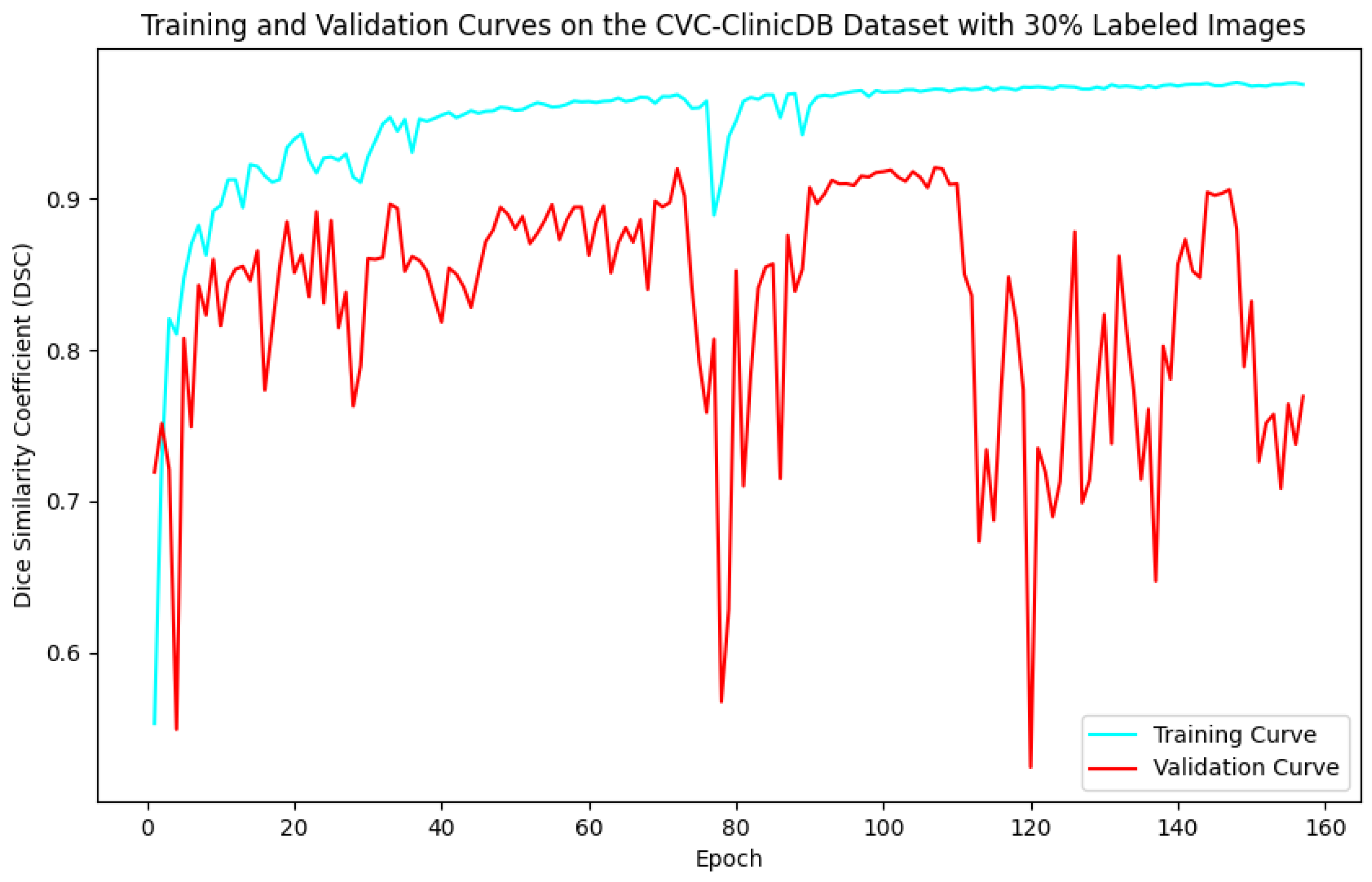
5.1. Evaluation of Training and Validation Performance
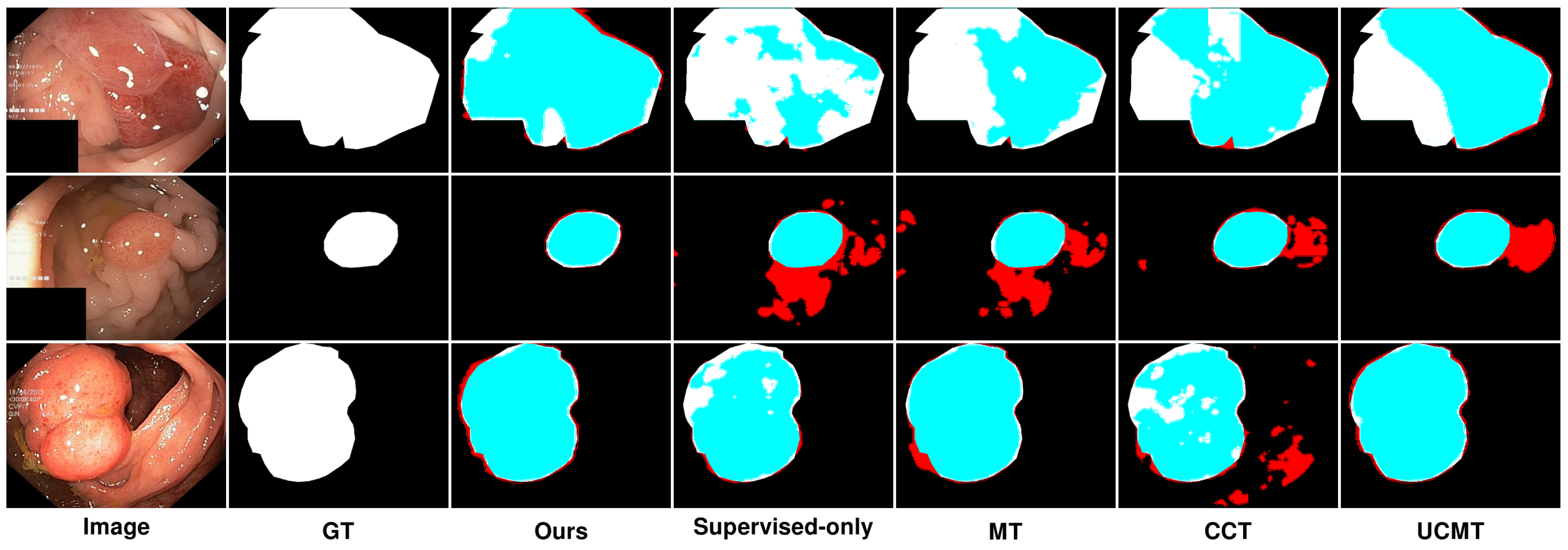
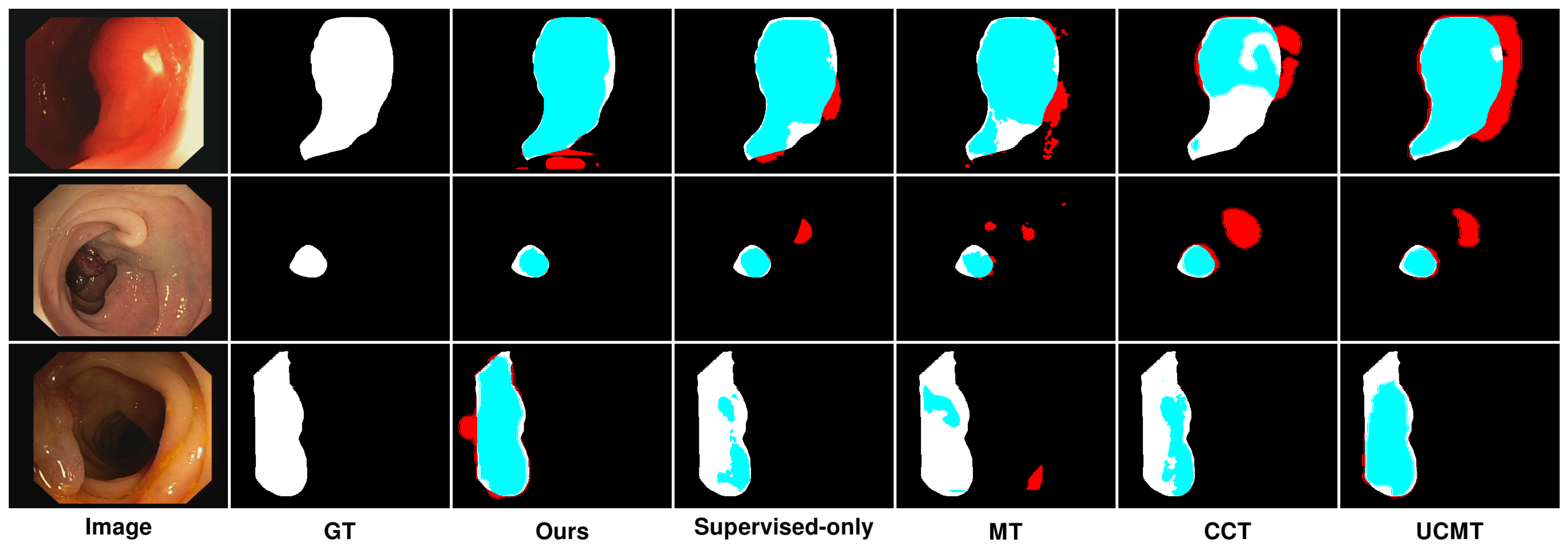
5.2. Qualitative Results Analysis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, S.; Sinha, S.; Jha, S.; Jagtap, B. Computer-aided automated diminutive colonic polyp detection in colonoscopy by using deep machine learning system; first indigenous algorithm developed in India. Indian J. Gastroenterol. 2023, 42, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yang, J.; Qi, Z. Unsupervised anomaly segmentation via deep feature reconstruction. Neurocomputing 2021, 424, 9–22. [Google Scholar] [CrossRef]
- Noor, S.; Waqas, M.; Saleem, M.I.; Minhas, H.N. Automatic object tracking and segmentation using unsupervised SiamMask. IEEE Access 2021, 9, 106550–106559. [Google Scholar] [CrossRef]
- Chen, X.; Yuan, Y.; Zeng, G.; Wang, J. Semi-supervised semantic segmentation with cross pseudo supervision. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 2613–2622. [Google Scholar]
- Alonso, I.; Sabater, A.; Ferstl, D.; Montesano, L.; Murillo, A.C. Semi-supervised semantic segmentation with pixel-level contrastive learning from a class-wise memory bank. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 8219–8228. [Google Scholar]
- Xiang, C.; Gan, V.J.; Guo, J.; Deng, L. Semi-supervised learning framework for crack segmentation based on contrastive learning and cross pseudo supervision. Measurement 2023, 217, 113091. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems 30; Neural Information Processing Systems Foundation, Inc. (NeurIPS): Long Beach, CA, USA, 2017; Volume 30. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Akbari, M.; Mohrekesh, M.; Nasr-Esfahani, E.; Soroushmehr, S.R.; Karimi, N.; Samavi, S.; Najarian, K. Polyp segmentation in colonoscopy images using fully convolutional network. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 69–72. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Zhang, Y.; Liu, H.; Hu, Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, Proceedings of the 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24; Springer: Cham, Switzerland, 2021; pp. 14–24. [Google Scholar]
- Sanderson, E.; Matuszewski, B.J. FCN-transformer feature fusion for polyp segmentation. In Proceedings of the Annual Conference on Medical Image Understanding and Analysis; Springer: Berlin/Heidelberg, Germany, 2022; pp. 892–907. [Google Scholar]
- Fan, D.P.; Zhou, T.; Ji, G.P.; Zhou, Y.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Inf-net: Automatic COVID-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging 2020, 39, 2626–2637. [Google Scholar] [CrossRef]
- Lyu, F.; Ye, M.; Carlsen, J.F.; Erleben, K.; Darkner, S.; Yuen, P.C. Pseudo-label guided image synthesis for semi-supervised covid-19 pneumonia infection segmentation. IEEE Trans. Med. Imaging 2022, 42, 797–809. [Google Scholar] [CrossRef]
- Shen, Z.; Cao, P.; Yang, H.; Liu, X.; Yang, J.; Zaiane, O.R. Co-training with high-confidence pseudo labels for semi-supervised medical image segmentation. arXiv 2023, arXiv:2301.04465. [Google Scholar]
- Tarvainen, A.; Valpola, H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. In Advances in Neural Information Processing Systems 30; Neural Information Processing Systems Foundation, Inc. (NeurIPS): Long Beach, CA, USA, 2017; Volume 30. [Google Scholar]
- Ouali, Y.; Hudelot, C.; Tami, M. Semi-supervised semantic segmentation with cross-consistency training. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 12674–12684. [Google Scholar]
- Wu, Y.; Xu, M.; Ge, Z.; Cai, J.; Zhang, L. Semi-supervised left atrium segmentation with mutual consistency training. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, Proceedings of the 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II 24; Springer: Cham, Switzerland, 2021; pp. 297–306. [Google Scholar]
- Li, D.; Yang, J.; Kreis, K.; Torralba, A.; Fidler, S. Semantic segmentation with generative models: Semi-supervised learning and strong out-of-domain generalization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 8300–8311. [Google Scholar]
- Tan, Y.; Wu, W.; Tan, L.; Peng, H.; Qin, J. Semi-supervised medical image segmentation based on generative adversarial network. J. New Media 2022, 4, 155. [Google Scholar] [CrossRef]
- Li, G.; Wang, J.; Tan, Y.; Shen, L.; Jiao, D.; Zhang, Q. Semi-supervised medical image segmentation based on GAN with the pyramid attention mechanism and transfer learning. Multimed. Tools Appl. 2024, 83, 17811–17832. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, D.; Xu, X.; Shi, Y. Semi-supervised contrastive learning for label-efficient medical image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, Proceedings of the 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II 24; Springer: Cham, Switzerland, 2021; pp. 481–490. [Google Scholar]
- Liu, Q.; Gu, X.; Henderson, P.; Deligianni, F. Multi-Scale Cross Contrastive Learning for Semi-Supervised Medical Image Segmentation. arXiv 2023, arXiv:2306.14293. [Google Scholar]
- Chen, Y.; Chen, F.; Huang, C. Combining contrastive learning and shape awareness for semi-supervised medical image segmentation. Expert Syst. Appl. 2024, 242, 122567. [Google Scholar] [CrossRef]
- Shen, Y.; Lu, Y.; Jia, X.; Bai, F.; Meng, M.Q.H. Task-relevant feature replenishment for cross-centre polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2022; pp. 599–608. [Google Scholar]
- Wei, J.; Hu, Y.; Li, G.; Cui, S.; Kevin Zhou, S.; Li, Z. BoxPolyp: Boost generalized polyp segmentation using extra coarse bounding box annotations. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2022; pp. 67–77. [Google Scholar]
- Yang, C.; Guo, X.; Chen, Z.; Yuan, Y. Source free domain adaptation for medical image segmentation with fourier style mining. Med. Image Anal. 2022, 79, 102457. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, L.; Lu, H. Automatic polyp segmentation via multi-scale subtraction network. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. Springer: Cham, Switzerland, 2021; pp. 120–130. [Google Scholar]
- Tomar, N.K.; Jha, D.; Riegler, M.A.; Johansen, H.D.; Johansen, D.; Rittscher, J.; Halvorsen, P.; Ali, S. Fanet: A feedback attention network for improved biomedical image segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2022, 34, 9375–9388. [Google Scholar]
- Li, Q.; Yang, G.; Chen, Z.; Huang, B.; Chen, L.; Xu, D.; Zhou, X.; Zhong, S.; Zhang, H.; Wang, T. Colorectal polyp segmentation using a fully convolutional neural network. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, Biomedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 14–16 October 2017; pp. 1–5. [Google Scholar]
- Brandao, P.; Mazomenos, E.; Ciuti, G.; Caliò, R.; Bianchi, F.; Menciassi, A.; Dario, P.; Koulaouzidis, A.; Arezzo, A.; Stoyanov, D. Fully convolutional neural networks for polyp segmentation in colonoscopy. In Proceedings of the Medical Imaging 2017: Computer-Aided Diagnosis; Spie: San Diego, CA, USA, 2017; Volume 10134, pp. 101–107. [Google Scholar]
- Srivastava, A.; Jha, D.; Chanda, S.; Pal, U.; Johansen, H.D.; Johansen, D.; Riegler, M.A.; Ali, S.; Halvorsen, P. MSRF-Net: A multi-scale residual fusion network for biomedical image segmentation. IEEE J. Biomed. Health Inform. 2021, 26, 2252–2263. [Google Scholar] [CrossRef]
- Song, P.; Li, J.; Fan, H. Attention based multi-scale parallel network for polyp segmentation. Comput. Biol. Med. 2022, 146, 105476. [Google Scholar] [CrossRef]
- Patel, K.; Bur, A.M.; Wang, G. Enhanced u-net: A feature enhancement network for polyp segmentation. In Proceedings of the 2021 18th Conference on Robots and Vision (CRV), Burnaby, BC, Canada, 26–28 May 2021; pp. 181–188. [Google Scholar]
- Nisa, S.Q.; Ismail, A.R. Dual U-Net with Resnet Encoder for Segmentation of Medical Images. Int. J. Adv. Comput. Sci. Appl. 2022, 13, 537–542. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Qin, Y.; Xia, H.; Song, S. RT-Net: Region-enhanced attention transformer network for polyp segmentation. Neural Process. Lett. 2023, 55, 11975–11991. [Google Scholar] [CrossRef]
- Liu, R.; Duan, S.; Xu, L.; Liu, L.; Li, J.; Zou, Y. A fuzzy transformer fusion network (FuzzyTransNet) for medical image segmentation: The case of rectal polyps and skin lesions. Appl. Sci. 2023, 13, 9121. [Google Scholar] [CrossRef]
- Liu, Z.; Hu, H.; Lin, Y.; Yao, Z.; Xie, Z.; Wei, Y.; Ning, J.; Cao, Y.; Zhang, Z.; Dong, L.; et al. Swin transformer v2: Scaling up capacity and resolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 12009–12019. [Google Scholar]
- Nanni, L.; Fantozzi, C.; Loreggia, A.; Lumini, A. Ensembles of convolutional neural networks and transformers for polyp segmentation. Sensors 2023, 23, 4688. [Google Scholar] [CrossRef] [PubMed]
- Tolstikhin, I.O.; Houlsby, N.; Kolesnikov, A.; Beyer, L.; Zhai, X.; Unterthiner, T.; Yung, J.; Steiner, A.; Keysers, D.; Uszkoreit, J.; et al. Mlp-mixer: An all-mlp architecture for vision. Adv. Neural Inf. Process. Syst. 2021, 34, 24261–24272. [Google Scholar]
- Lian, D.; Yu, Z.; Sun, X.; Gao, S. As-mlp: An axial shifted mlp architecture for vision. arXiv 2021, arXiv:2107.08391. [Google Scholar]
- Yu, T.; Li, X.; Cai, Y.; Sun, M.; Li, P. S2-mlp: Spatial-shift mlp architecture for vision. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 297–306. [Google Scholar]
- Touvron, H.; Bojanowski, P.; Caron, M.; Cord, M.; El-Nouby, A.; Grave, E.; Izacard, G.; Joulin, A.; Synnaeve, G.; Verbeek, J.; et al. Resmlp: Feedforward networks for image classification with data-efficient training. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 5314–5321. [Google Scholar] [CrossRef] [PubMed]
- Valanarasu, J.M.J.; Patel, V.M. Unext: Mlp-based rapid medical image segmentation network. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2022; pp. 23–33. [Google Scholar]
- Amini, M.R.; Feofanov, V.; Pauletto, L.; Hadjadj, L.; Devijver, E.; Maximov, Y. Self-training: A survey. arXiv 2022, arXiv:2202.12040. [Google Scholar]
- Ning, X.; Wang, X.; Xu, S.; Cai, W.; Zhang, L.; Yu, L.; Li, W. A review of research on co-training. Concurr. Comput. Pract. Exp. 2023, 35, e6276. [Google Scholar] [CrossRef]
- Peng, J.; Estrada, G.; Pedersoli, M.; Desrosiers, C. Deep co-training for semi-supervised image segmentation. Pattern Recognit. 2020, 107, 107269. [Google Scholar] [CrossRef]
- Li, X.; Yu, L.; Chen, H.; Fu, C.W.; Xing, L.; Heng, P.A. Transformation-consistent self-ensembling model for semisupervised medical image segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 523–534. [Google Scholar] [CrossRef]
- Xie, Z.; Tu, E.; Zheng, H.; Gu, Y.; Yang, J. Semi-supervised skin lesion segmentation with learning model confidence. In Proceedings of the ICASSP 2021–2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1135–1139. [Google Scholar]
- Luo, X.; Chen, J.; Song, T.; Wang, G. Semi-supervised medical image segmentation through dual-task consistency. Proc. Aaai Conf. Artif. Intell. 2021, 35, 8801–8809. [Google Scholar] [CrossRef]
- Basak, H.; Bhattacharya, R.; Hussain, R.; Chatterjee, A. An exceedingly simple consistency regularization method for semi-supervised medical image segmentation. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March2022; pp. 1–4. [Google Scholar]
- Cho, H.; Han, Y.; Kim, W.H. Anti-adversarial Consistency Regularization for Data Augmentation: Applications to Robust Medical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2023; pp. 555–566. [Google Scholar]
- Wang, Y.; Xiao, B.; Bi, X.; Li, W.; Gao, X. Mcf: Mutual correction framework for semi-supervised medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 15651–15660. [Google Scholar]
- Wu, C.; Zhang, W.; Han, J.; Wang, H. Multi-Consistency Training for Semi-Supervised Medical Image Segmentation. J. Shanghai Jiaotong Univ. (Sci.) 2024, 29, 1–15. [Google Scholar] [CrossRef]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. In Proceedings of the 37th International Conference on Machine Learning, Online, 13–18 July 2020; pp. 1597–1607. [Google Scholar]
- Park, T.; Efros, A.A.; Zhang, R.; Zhu, J.Y. Contrastive learning for unpaired image-to-image translation. In Proceedings of the Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020; Proceedings, Part IX 16. Springer: Cham, Switzerland, 2020; pp. 319–345. [Google Scholar]
- Chen, P.; Liu, S.; Zhao, H.; Jia, J. Gridmask data augmentation. arXiv 2020, arXiv:2001.04086. [Google Scholar]
- Gao, S.H.; Cheng, M.M.; Zhao, K.; Zhang, X.Y.; Yang, M.H.; Torr, P. Res2net: A new multi-scale backbone architecture. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; De Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In Proceedings of the MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, Republic of Korea, 5–8 January 2020; Proceedings, Part II 26. Springer: Cham, Switzerland, 2020; pp. 451–462. [Google Scholar]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. Resunet++: An advanced architecture for medical image segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; pp. 225–2255. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]



| Related Studies | Methods | Typical Works | Key Characteristics |
|---|---|---|---|
| Principal model architectures | CNN-based methods | [10,11,12], etc. | Pros: Strong local feature extraction capabilities. Cons: Limited ability to capture global dependencies, which may lead to loss of feature information. |
| Transformer-based methods | [13,14], etc. | Pros: Excellent global modeling capabilities, able to capture long-range dependencies. Cons: High computational complexity, longer training times. | |
| Hybrid network methods | [15,16], etc. | Pros: Combines the strengths of CNNs and transformers, capturing both local features and global dependencies. Cons: Complex structure, challenging to design effective fusion modules. | |
| Our method | Current study | Innovative hybrid architecture that combines transformer, CNN, and MLP, which captures both local features and global dependencies and enhances the feature fusion workflow. | |
| Semi-supervised learning | Pseudo-label-based methods | [17,18,19], etc. | Pros: Effective even with limited labeled data, iterative refinement of predictions. Cons: Initial pseudo-label quality is crucial; poor labels can degrade performance. |
| Consistency-regularization-based methods | [20,21,22], etc. | Pros: Ensures model consistency, effective use of unlabeled data. Cons: Requires multiple forward passes, increasing computational demands. | |
| GAN-based methods | [23,24,25], etc. | Pros: Captures complex patterns, improves generalization across domains. Cons: High training complexity, stability issues, and significant computational resources required. | |
| Contrastive-learning-based methods | [26,27,28], etc. | Pros: Learns robust feature representations, effective with large amounts of unlabeled data. Cons: Requires careful selection of contrastive pairs; balancing contrastive loss can be complex. | |
| Our method | Current study | Innovative semi-supervised framework combining CPS with patch-wise contrastive learning, enhancing feature representation and segmentation accuracy. |
| Datasets | Number of Images | Size | Train | Valid | Test |
|---|---|---|---|---|---|
| Kvasir-SEG [65] | 1000 | Variable | 800 | 100 | 100 |
| CV-ClinicDB [66] | 612 | 384 × 288 | 488 | 62 | 62 |
| Lr | Weight Decay | ||
|---|---|---|---|
| 0.0001 | 0.9 | 0.999 | 0.0001 |
| Method | Labeled/Unlabeled | DSC (%) |
|---|---|---|
| Fully supervised (HTCSNet) | 800/0 | 90.95 |
| Baseline (supervised-only) | 120/0 | 86.33 |
| MT (DeepLabv3+) [20] | 120/680 | 87.44 |
| CCT (DeepLabv3+) [21] | 120/680 | 81.14 |
| CMT (DeepLabv3+) [19] | 120/680 | 88.08 |
| UCMT (DeepLabv3+) [19] | 120/680 | 88.68 |
| SemiPolypSeg (DeepLabv3+) (ours) | 120/680 | 89.03 |
| SemiPolypSeg (HTCSNet) (ours) | 120/680 | 89.68 |
| Baseline (supervised-only) | 240/0 | 87.45 |
| MT (DeepLabv3+) [20] | 240/560 | 88.72 |
| CCT (DeepLabv3+) [21] | 240/560 | 84.67 |
| CMT (DeepLabv3+) [19] | 240/560 | 88.61 |
| UCMT (DeepLabv3+) [19] | 240/560 | 89.06 |
| SemiPolypSeg (DeepLabv3+) (ours) | 240/560 | 89.91 |
| SemiPolypSeg (HTCSNet) (ours) | 240/560 | 90.62 |
| Method | Labeled/Unlabeled | DSC (%) |
|---|---|---|
| Fully supervised (HTCSNet) | 488/0 | 90.90 |
| Baseline (supervised-only) | 74/0 | 85.16 |
| MT (DeepLabv3+) [20] | 74/414 | 84.19 |
| CCT (DeepLabv3+) [21] | 74/414 | 74.20 |
| CMT (DeepLabv3+) [19] | 74/414 | 85.88 |
| UCMT (DeepLabv3+) [19] | 74/414 | 87.30 |
| SemiPolypSeg (DeepLabv3+) (ours) | 74/414 | 89.47 |
| SemiPolypSeg (HTCSNet) (ours) | 74/414 | 89.72 |
| Baselinen(supervised-only) | 148/0 | 86.38 |
| MT (DeepLabv3+) [20] | 148/340 | 84.40 |
| CCT (DeepLabv3+) [21] | 148/340 | 78.46 |
| CMT (DeepLabv3+) [19] | 148/340 | 86.83 |
| UCMT (DeepLabv3+) [19] | 148/340 | 87.51 |
| SemiPolypSeg (DeepLabv3+) (ours) | 148/340 | 89.61 |
| SemiPolypSeg (HTCSNet) (ours) | 148/340 | 90.06 |
| Network | Loss-sup | Loss-sim | Loss-cps | DSC |
|---|---|---|---|---|
| HTCSNet (baseline) | ✓ | 85.16 | ||
| HTCSNet | ✓ | ✓ | 85.29 | |
| HTCSNet | ✓ | ✓ | 88.10 | |
| HTCSNet (w/o CNN) | ✓ | ✓ | ✓ | 84.24 |
| HTCSNet (w/o Transformer) | ✓ | ✓ | ✓ | 85.73 |
| HTCSNet | ✓ | ✓ | ✓ | 89.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, P.; Liu, G.; Liu, H. SemiPolypSeg: Leveraging Cross-Pseudo Supervision and Contrastive Learning for Semi-Supervised Polyp Segmentation. Appl. Sci. 2024, 14, 7852. https://doi.org/10.3390/app14177852
Guo P, Liu G, Liu H. SemiPolypSeg: Leveraging Cross-Pseudo Supervision and Contrastive Learning for Semi-Supervised Polyp Segmentation. Applied Sciences. 2024; 14(17):7852. https://doi.org/10.3390/app14177852
Chicago/Turabian StyleGuo, Ping, Guoping Liu, and Huan Liu. 2024. "SemiPolypSeg: Leveraging Cross-Pseudo Supervision and Contrastive Learning for Semi-Supervised Polyp Segmentation" Applied Sciences 14, no. 17: 7852. https://doi.org/10.3390/app14177852
APA StyleGuo, P., Liu, G., & Liu, H. (2024). SemiPolypSeg: Leveraging Cross-Pseudo Supervision and Contrastive Learning for Semi-Supervised Polyp Segmentation. Applied Sciences, 14(17), 7852. https://doi.org/10.3390/app14177852






