A Medical Image Segmentation Network with Multi-Scale and Dual-Branch Attention
Abstract
:1. Introduction
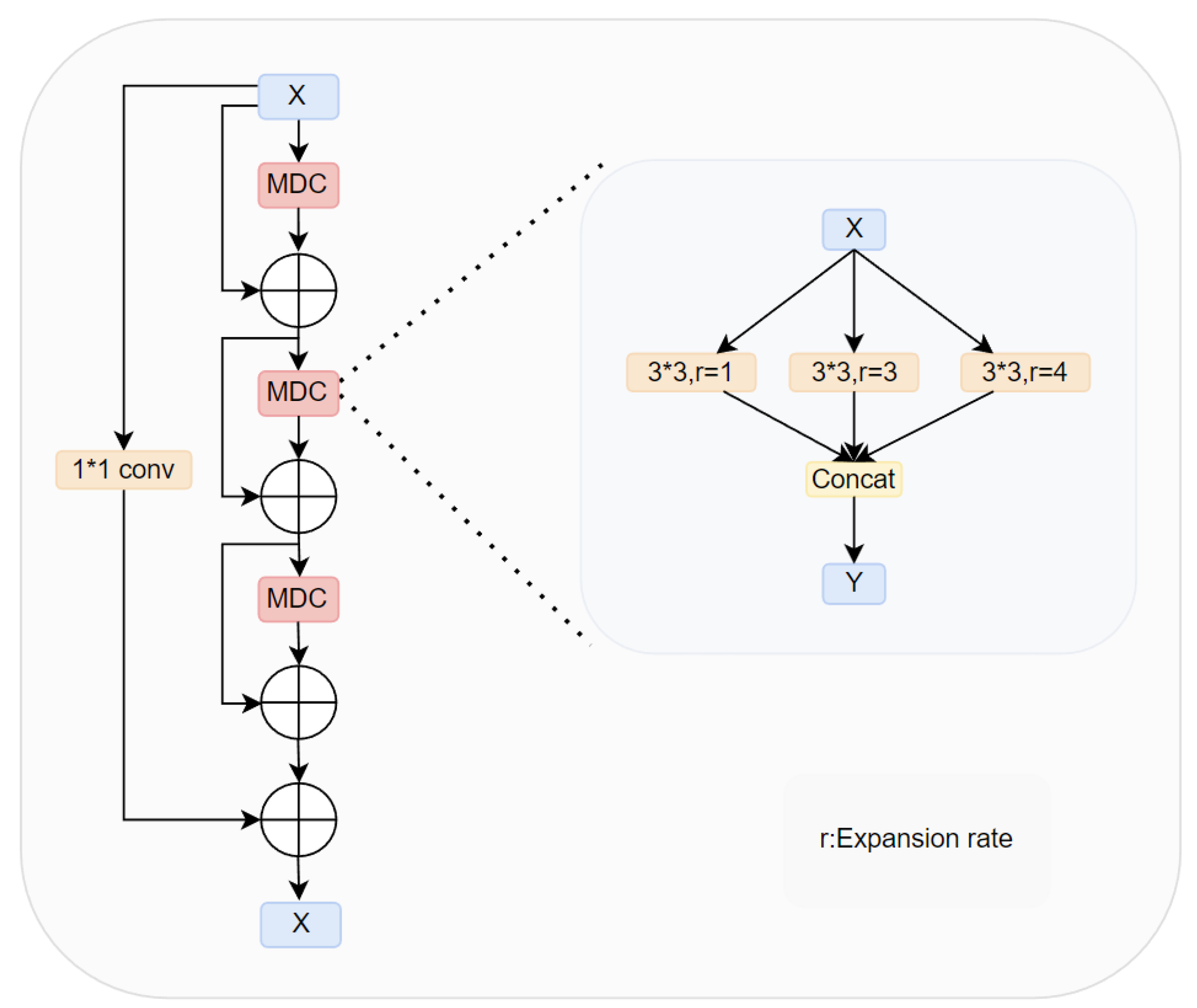
- We proposed a novel module MRD, which introduced convolution operations with different dilation rates during upsampling and extracted features through three parallel branches. This module enables the simultaneous capture of global and local semantic information, enhancing the multi-scale feature representation capability of the network. Additionally, the extracted semantic information is fused through dense connections to reduce the risk of information loss, thereby enhancing the feature representation ability and improving segmentation accuracy. The MRD effectively addresses the shortcomings of traditional methods in feature extraction and fusion, significantly enhancing the overall performance and segmentation results of the network.
- We propose a novel module PDA designed to process feature information in parallel from both channel and spatial dimensions. This module receives shallow semantic features from the encoder and multi-scale features from the decoder, effectively integrating features at different scales, emphasizing crucial global and local semantic information in each receptive field, suppressing noise, and enhancing the ability to segment target boundaries. The PDA allows for better integration of feature information and precise calculation of attention weights for the feature maps, thereby enhancing the overall performance of the network.
- To assess the robustness and generalization of our network, we conduct experimental analyses on four distinct medical image datasets: 2018 Data Science Bowl [15], ISIC 2018 [16,17], CVC-ClinicDB [18], and colon cancer slices dataset(images provided with authorization from the collaborating hospital). The assessment findings underscore notable advancements attained with our proposed model in contrast to medical image segmentation methods such as UNet and UNet++, thereby providing additional affirmation of the effectiveness of our approach.
2. Related Work
2.1. Medical Image Segmentation
2.2. Image Serialization
2.3. Attention Mechanism
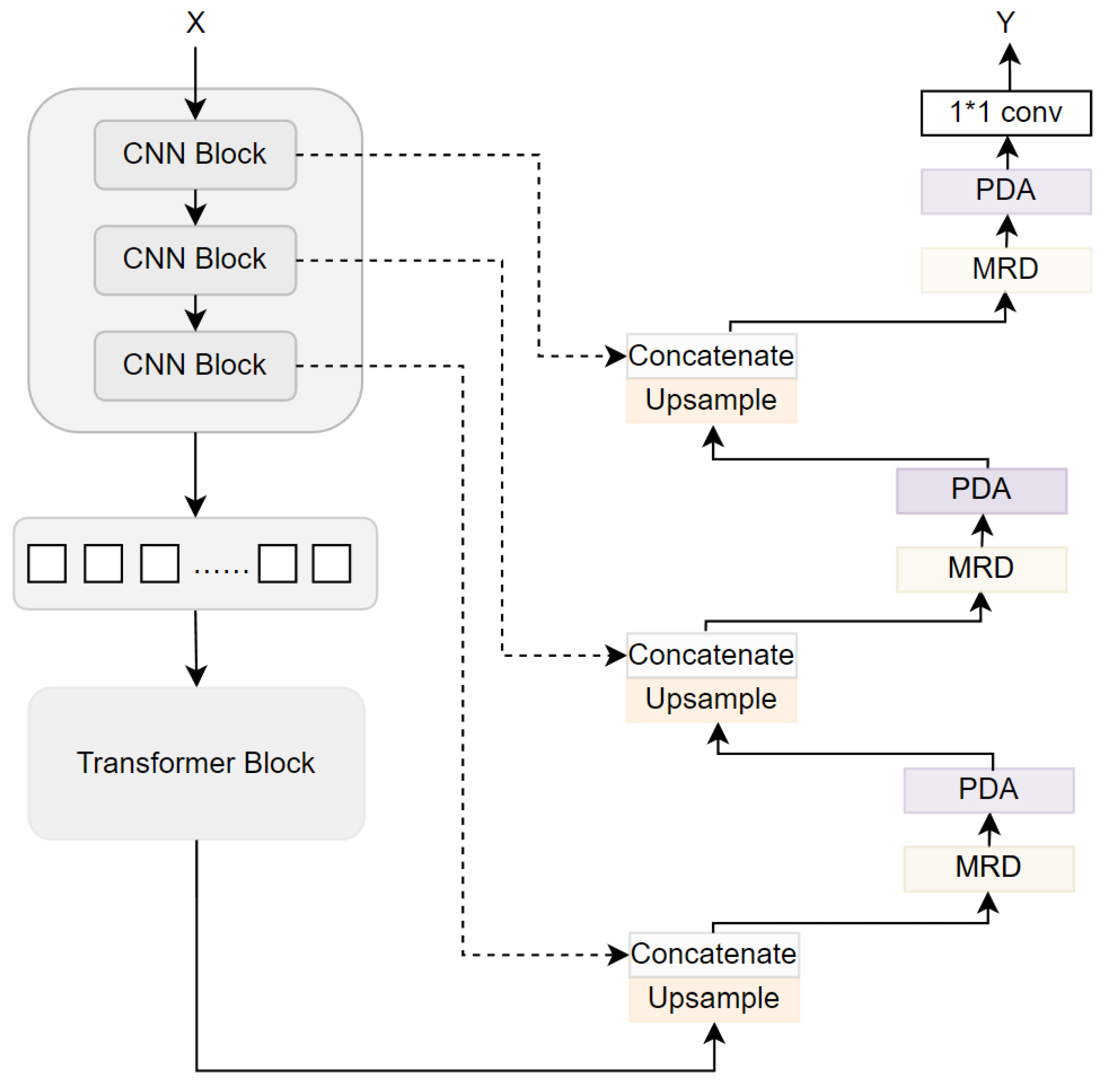
3. Method
3.1. Multi-Receptive Field Densely Connected Module
3.2. Parallel Dual-Branch Attention Module
4. Experiments and Results
4.1. Datasets and Evaluation Metric
4.2. Implementation Details
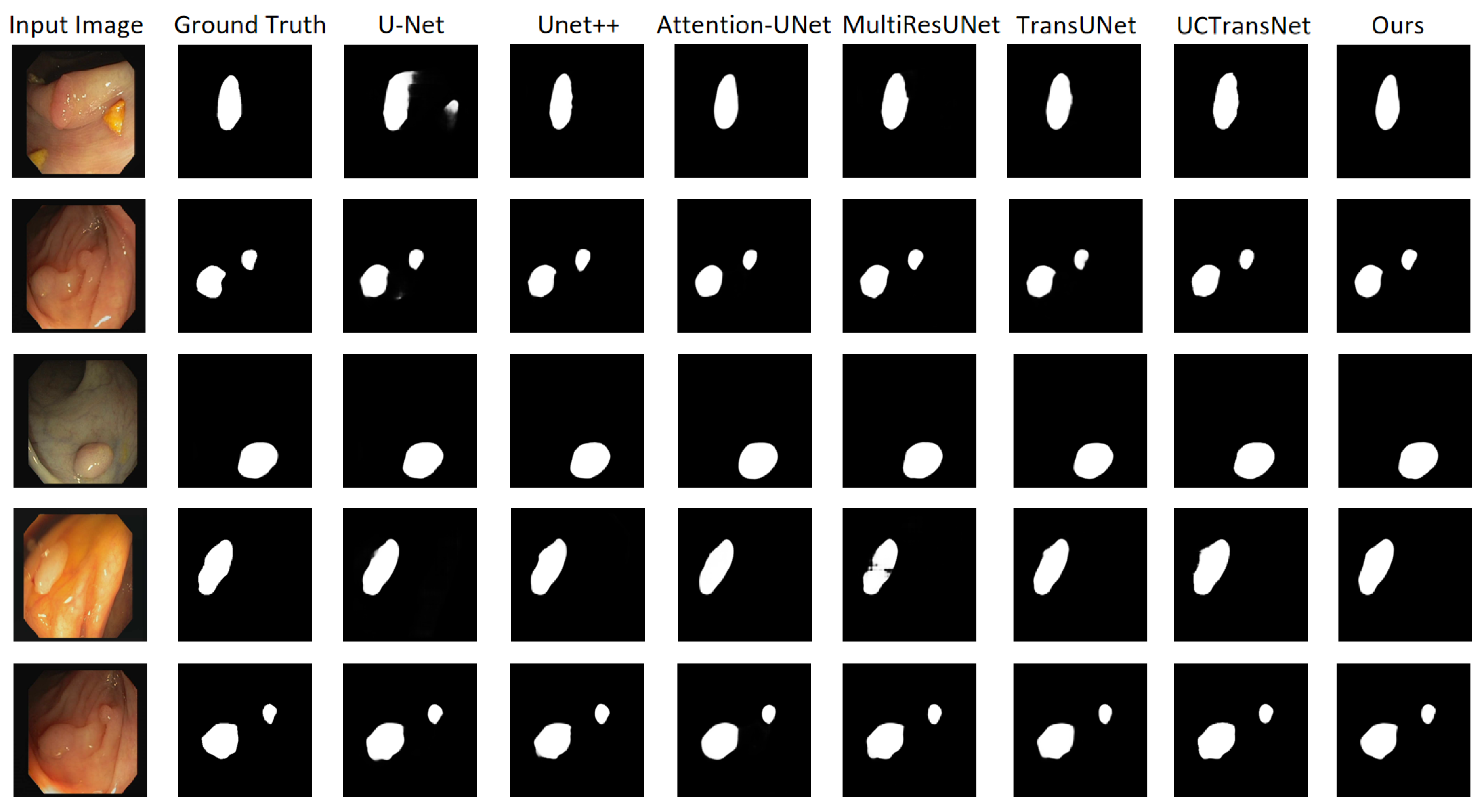
4.3. Results
4.3.1. 2018 Data Science Bowl Dataset Comparison Results
4.3.2. CVC-ClinicDB Dataset Comparison Results
4.3.3. ISIC 2018 Dataset Comparison Results
4.3.4. Colon Cancer Slice Dataset Comparison Results
4.4. Comparison of Parameters and FLOPs
4.5. Ablation Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, Q.; Ma, Z.; He, N.; Duan, W. DCSAU-Net: A deeper and more compact split-attention U-Net for medical image segmentation. Comput. Biol. Med. 2023, 154, 106626. [Google Scholar] [CrossRef] [PubMed]
- Canny, J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, PAMI–8, 679–698. [Google Scholar] [CrossRef]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active contour models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Vincent, L.M.; Soille, P. Watersheds in Digital Spaces: An Efficient Algorithm Based on Immersion Simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 583–598. [Google Scholar] [CrossRef]
- Sandler, M.; Zhmoginov, A.; Luo, L.; Mordvintsev, A.; Randazzo, E. Image segmentation via Cellular Automata. arXiv 2020, arXiv:2008.04965. [Google Scholar]
- Antony, M.; Sathiaseelan, J.G.R. Optimal Cellular Automata Technique for Image Segmentation. Int. J. Innov. Technol. Explor. Eng. 2020, 9, 1474–1478. [Google Scholar]
- Dorigo, M.; Birattari, M.; Stutzle, T. Ant colony optimization. IEEE Comput. Intell. Mag. 2006, 1, 28–39. [Google Scholar] [CrossRef]
- Wang, X.N.; Feng, Y.J.; Feng, Z.R. Ant colony optimization for image segmentation. In Proceedings of the 2005 International Conference on Machine Learning and Cybernetics, Guangzhou, China, 18–21 August 2005; Volume 9, pp. 5355–5360. [Google Scholar]
- Feng, S.; Zhao, H.; Shi, F.; Cheng, X.; Wang, M.; Ma, Y.; Xiang, D.; Zhu, W.; Chen, X. CPFNet: Context Pyramid Fusion Network for Medical Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 3008–3018. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T.J.A. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.J.; Heinrich, M.P.; Misawa, K.; Mori, K.; McDonagh, S.G.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted Res-UNet for High-Quality Retina Vessel Segmentation. In Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; pp. 327–331. [Google Scholar]
- Vaswani, A.; Shazeer, N.M.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All you Need. In Proceedings of the he Thirty-First Annual Conference on Neural Information Processing Systems (NIPS), San Diego, CA, USA, 4–9 December 2017. [Google Scholar]
- Caicedo, J.C.; Goodman, A.; Karhohs, K.W.; Cimini, B.A.; Ackerman, J.; Haghighi, M.; Heng, C.; Becker, T.; Doan, M.; McQuin, C.; et al. Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef]
- Gutman, D.; Codella, N.C.F.; Celebi, E.M.; Helba, B.; Marchetti, M.; Mishra, N.K.; Halpern, A.J.A. Skin Lesion Analysis toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc. 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Mu, C.C.; Li, G. Research progress in medical imaging based on deep learning of neural network. Zhonghua Kou Qiang Yi Xue Za Zhi = Zhonghua Kouqiang Yixue Zazhi = Chin. J. Stomatol. 2019, 54, 492–497. [Google Scholar]
- Philbrick, K.A.; Weston, A.D.; Akkus, Z.; Kline, T.L.; Korfiatis, P.; Sakinis, T.; Kostandy, P.M.; Boonrod, A.; Zeinoddini, A.; Takahashi, N.; et al. RIL-Contour: A Medical Imaging Dataset Annotation Tool for and with Deep Learning. J. Digit. Imaging 2019, 32, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, J.; Hua, Z. MPSH: Multiple Progressive Sampling Hybrid Model Multi-Organ Segmentation. IEEE J. Transl. Eng. Health Med. 2022, 10, 1800909. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C.J.A. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. arXiv 2019, arXiv:1904.00592. [Google Scholar] [CrossRef]
- Liu, J.; Kim, J.H. A Variable Attention Nested UNet++ Network-Based NDT X-ray Image Defect Segmentation Method. Coatings 2022, 12, 634. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J.J.D.L. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. arXiv 2018, arXiv:1807.10165. [Google Scholar]
- Yang, L.; Zhai, C.; Liu, Y.; Yu, H. CFHA-Net: A polyp segmentation method with cross-scale fusion strategy and hybrid attention. Comput. Biol. Med. 2023, 164, 107301. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, S.; Ji, C.; Fan, J.; Li, Y. Boundary-aware Context Neural Network for Medical Image Segmentation. Med. Image Anal. 2020, 78, 102395. [Google Scholar] [CrossRef]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K.J.A. Recurrent Residual Convolutional Neural Network based on U-Net (R2U-Net) for Medical Image Segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Yu, Z.; Yu, L.; Zheng, W.; Wang, S. EIU-Net: Enhanced feature extraction and improved skip connections in U-Net for skin lesion segmentation. Comput. Biol. Med. 2023, 162, 107081. [Google Scholar] [CrossRef] [PubMed]
- Zhan, B.; Song, E.; Liu, H. FSA-Net: Rethinking the attention mechanisms in medical image segmentation from releasing global suppressed information. Comput. Biol. Med. 2023, 161, 106932. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; She, C.; Wang, W.; Huang, Q. LM-Net: A light-weight and multi-scale network for medical image segmentation. Comput. Biol. Med. 2024, 168, 107717. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Dai, D.; Zhang, Q.; Wang, Y.; Xu, S.; Lian, C. MSCA-Net: Multi-scale contextual attention network for skin lesion segmentation. Pattern Recognit. 2023, 139, 109524. [Google Scholar] [CrossRef]
- Yuan, Y.; Chen, X.; Wang, J.J.A. Object-Contextual Representations for Semantic Segmentation. arXiv 2019, arXiv:1909.11065. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. arXiv 2021, arXiv:2103.14030. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation; Springer: Tel Aviv, Israel, 2021. [Google Scholar]
- Huang, X.; Chen, J.; Chen, M.; Chen, L.; Wan, Y. TDD-UNet: Transformer with double decoder UNet for COVID-19 lesions segmentation. Comput. Biol. Med. 2022, 151, 12. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, X.; Huang, L.; Huang, C.; Wei, Y.; Shi, H.; Liu, W. CCNet: Criss-Cross Attention for Semantic Segmentation. arXiv 2018, arXiv:1811.11721. [Google Scholar]
- Fu, J.; Liu, J.; Tian, H.; Fang, Z.; Lu, H. Dual Attention Network for Scene Segmentation. arXiv 2018, arXiv:1809.02983. [Google Scholar]
- Jin, Q.; Meng, Z.-P.; Sun, C.; Wei, L.; Su, R.J. RA-UNet: A Hybrid Deep Attention-Aware Network to Extract Liver and Tumor in CT Scans. arXiv 2018, arXiv:1811.01328. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. arXiv 2017, arXiv:1709.01507. [Google Scholar]
- Qiu, S.; Li, C.; Feng, Y.; Zuo, S.; Liang, H.; Xu, A. GFANet: Gated Fusion Attention Network for skin lesion segmentation. Comput. Biol. Med. 2023, 155, 106462. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liu, Z.; Maaten, L.V.D.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.-S. CBAM: Convolutional Block Attention Module. arXiv 2018, arXiv:1807.06521. [Google Scholar]
- Wei, J.; Wang, S.; Huang, Q. F³Net: Fusion, feedback and focus for salient object detection. Proc. AAAI Conf. Artif. Intell. 2020, 34, 12321–12328. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y.J.A. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Wang, H.; Cao, P.; Wang, J.; Zaiane, O.R. UCTransNet: Rethinking the Skip Connections in U-Net from a Channel-Wise Perspective with Transformer. Proc. AAAI Conf. Artif. Intell. 2022, 36, 2441–2449. [Google Scholar] [CrossRef]






| Model | Parameter | FLOPs | Acc | Dice | Precision | IoU | Recall | Infer-T |
|---|---|---|---|---|---|---|---|---|
| U-Net [11] | 14.75 M | 25.22 G | 0.970 | 0.911 | 0.905 | 0.842 | 0.926 | 1.79 s |
| U-Net++ [25] | 36.63 M | 105.85 G | 0.971 | 0.907 | 0.930 | 0.894 | 0.904 | 2.86 s |
| MultiResUNet [45] | 7.25 M | 14.30 G | 0.966 | 0.896 | 0.926 | 0.822 | 0.885 | 2.36 s |
| AttentionU-Net [12] | 34.88 M | 16.70 G | 0.969 | 0.901 | 0.893 | 0.834 | 0.925 | 3.33 s |
| TransU-Net [46] | 105.28 M | 25.35 G | 0.972 | 0.911 | 0.913 | 0.824 | 0.916 | 4.73 s |
| UCTransNet [47] | 63.36 M | 36.12 G | 0.968 | 0.901 | 0.910 | 0.827 | 0.904 | 4.36 s |
| Ours | 99.47 M | 19.86 G | 0.979 | 0.918 | 0.925 | 0.871 | 0.938 | 3.41 s |
| Model | Acc | Dice | Precision | IoU | Recall | Infer-T |
|---|---|---|---|---|---|---|
| U-Net [11] | 0.982 | 0.903 | 0.904 | 0.839 | 0.916 | 2.61 s |
| U-Net++ [25] | 0.985 | 0.922 | 0.925 | 0.866 | 0.924 | 6.20 s |
| MultiResUNet [45] | 0.982 | 0.900 | 0.886 | 0.835 | 0.923 | 2.10 s |
| AttentionU-Net [12] | 0.984 | 0.915 | 0.902 | 0.858 | 0.932 | 3.12 s |
| TransU-Net [46] | 0.984 | 0.923 | 0.933 | 0.864 | 0.922 | 5.12 s |
| UCTransNet [47] | 0.985 | 0.916 | 0.903 | 0.860 | 0.931 | 4.47 s |
| Ours | 0.992 | 0.917 | 0.911 | 0.870 | 0.934 | 3.45 s |
| Model | Acc | Dice | Precision | IoU | Recall | Infer-T |
|---|---|---|---|---|---|---|
| U-Net [11] | 0.950 | 0.862 | 0.870 | 0.782 | 0.901 | 81.02 s |
| U-Net++ [25] | 0.954 | 0.880 | 0.917 | 0.808 | 0.881 | 116.82 s |
| MultiResUNet [45] | 0.958 | 0.888 | 0.911 | 0.817 | 0.897 | 102.60 s |
| AttentionU-Net [12] | 0.958 | 0.883 | 0.899 | 0.812 | 0.934 | 90.57 s |
| TransU-Net [46] | 0.955 | 0.887 | 0.896 | 0.814 | 0.901 | 91.25 s |
| UCTransNet [47] | 0.953 | 0.869 | 0.875 | 0.794 | 0.904 | 101.48 s |
| Ours | 0.962 | 0.913 | 0.937 | 0.857 | 0.917 | 100.41 s |
| Model | Acc | Dice | Precision | IoU | Recall | Infer-T |
|---|---|---|---|---|---|---|
| U-Net [11] | 0.868 | 0.805 | 0.823 | 0.718 | 0.811 | 2.93 s |
| U-Net++ [25] | 0.849 | 0.790 | 0.795 | 0.656 | 0.811 | 5.96 s |
| MultiResUNet [45] | 0.851 | 0.764 | 0.828 | 0.672 | 0.746 | 2.45 s |
| AttentionU-Net [12] | 0.858 | 0.776 | 0.806 | 0.692 | 0.779 | 3.84 s |
| TransU-Net [46] | 0.849 | 0.819 | 0.795 | 0.736 | 0.889 | 5.36 s |
| UCTransNet [47] | 0.850 | 0.773 | 0.799 | 0.683 | 0.783 | 4.58 s |
| Ours | 0.869 | 0.811 | 0.823 | 0.721 | 0.827 | 3.79 s |
| Dataset | Method | Acc | Dice | Precision | IoU | Recall |
|---|---|---|---|---|---|---|
| CVC-ClinicDB | Baseline | 0.982 | 0.899 | 0.924 | 0.835 | 0.919 |
| Baseline + MRD | 0.982 | 0.908 | 0.895 | 0.852 | 0.928 | |
| Baseline + PDA | 0.984 | 0.913 | 0.908 | 0.848 | 0.935 | |
| MSDA-Net | 0.992 | 0.917 | 0.911 | 0.870 | 0.934 | |
| 2018 Data Science Bowl | Baseline | 0.970 | 0.900 | 0.923 | 0.863 | 0.914 |
| Baseline + MRD | 0.971 | 0.912 | 0.901 | 0.864 | 0.916 | |
| Baseline + PDA | 0.971 | 0.908 | 0.917 | 0.866 | 0.921 | |
| MSDA-Net | 0.979 | 0.918 | 0.925 | 0.871 | 0.938 | |
| ISIC 2018 | Baseline | 0.958 | 0.883 | 0.903 | 0.816 | 0.898 |
| Baseline + MRD | 0.960 | 0.899 | 0.914 | 0.824 | 0.901 | |
| Baseline + PDA | 0.960 | 0.901 | 0.923 | 0.827 | 0.903 | |
| MSDA-Net | 0.962 | 0.913 | 0.937 | 0.857 | 0.917 | |
| Colon Cancer Slice dataset | Baseline | 0.857 | 0.801 | 0.816 | 0.704 | 0.810 |
| Baseline + MRD | 0.861 | 0.806 | 0.815 | 0.711 | 0.811 | |
| Baseline + PDA | 0.864 | 0.804 | 0.818 | 0.714 | 0.816 | |
| MSDA-Net | 0.869 | 0.811 | 0.823 | 0.721 | 0.827 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Cheng, K.; Hua, X. A Medical Image Segmentation Network with Multi-Scale and Dual-Branch Attention. Appl. Sci. 2024, 14, 6299. https://doi.org/10.3390/app14146299
Zhu C, Cheng K, Hua X. A Medical Image Segmentation Network with Multi-Scale and Dual-Branch Attention. Applied Sciences. 2024; 14(14):6299. https://doi.org/10.3390/app14146299
Chicago/Turabian StyleZhu, Cancan, Ke Cheng, and Xuecheng Hua. 2024. "A Medical Image Segmentation Network with Multi-Scale and Dual-Branch Attention" Applied Sciences 14, no. 14: 6299. https://doi.org/10.3390/app14146299
APA StyleZhu, C., Cheng, K., & Hua, X. (2024). A Medical Image Segmentation Network with Multi-Scale and Dual-Branch Attention. Applied Sciences, 14(14), 6299. https://doi.org/10.3390/app14146299






