1. Introduction
In today’s rapidly evolving healthcare landscape, telemedicine stands as a cornerstone in the delivery of medical services, enabling remote diagnosis, treatment, and consultation through integrated telecommunications networks. This paradigm shift has revolutionized healthcare delivery, transcending geographical barriers and enhancing access to quality medical care [
1]. In a digitally connected ecosystem, healthcare professionals and patients can interact seamlessly, exchanging vital health information and receiving timely medical attention irrespective of physical distances. Telemedicine indeed provides convenience and accessibility, but concerns about data security and privacy are substantial [
2]. Images are a rich source of information, recently used in tasks like object detection and medical imaging [
3,
4]. They can also hide information through techniques like steganography, embedding data within images for secure communication. The transmission of sensitive medical images and patient data over digital networks necessitates robust measures to safeguard confidentiality, authenticity, and integrity. This is where image watermarking emerges as a critical component in ensuring the secure exchange of medical information within telemedicine frameworks.
The primary aim of image watermarking in the field of telemedicine is twofold: to authenticate the origin and integrity of medical images and to protect patient confidentiality [
1]. By embedding imperceptible digital watermarks within medical images, healthcare providers can verify the authenticity of images, ensuring they have not been tampered with during transmission or storage. Additionally, watermarking serves as a means to preserve patient privacy by embedding encrypted patient information directly into medical images, facilitating secure data exchange while mitigating the risk of unauthorized access. Within the realm of image watermarking, various techniques and methodologies have been developed to address specific security requirements and operational constraints. These techniques can be broadly categorized into spatial and frequency-based approaches, each offering unique advantages and trade-offs. Spatial domain techniques involve directly modifying pixel values within the image to embed the watermark, with methods like least significant bit (LSB) insertion being prominent examples [
5]. On the other hand, frequency-based techniques operate by embedding the watermark within frequency transforms of the image, such as the Discrete Cosine Transform (DCT), the DWT, Singular-Value Decomposition (SVD), and so on [
6,
7,
8]. These frequency domain methods offer enhanced robustness against certain types of attacks while preserving image quality, making them well suited for applications in telemedicine where image fidelity is crucial for accurate diagnosis. In the subsequent sections, we will delve into the intricacies of image watermarking techniques, exploring their strengths, weaknesses, and applicability in securing medical images within telemedicine frameworks. By understanding the nuances of watermarking methodologies, we aim to shed light on effective strategies for enhancing data security and privacy in the digital healthcare domain.
The proposed medical image watermarking technique, utilizing a combination of the Discrete Wavelet Transform (DWT) and gradients, significantly contributes to sustainability in healthcare. This method ensures patient privacy by offering a secure and reliable way to watermark images, preventing unauthorized access to sensitive data. Additionally, the DWT and gradient approach enhances image robustness and integrity, leading to accurate diagnosis and minimizing the risk of data breaches. Furthermore, by embedding watermarks securely, this technique reduces the need for repeated imaging procedures due to lost or corrupted images. This translates to lower resource consumption and a smaller environmental footprint for healthcare practices. The main contributions of the present work are as follows:
By effectively combining the DWT and gradients, the proposed method achieves superior robustness compared to existing techniques.
We employ image gradients to provide a “topological map” of the image, aiding in the identification of areas susceptible to disruption
The proposed method leverages the relationship between the gradient direction and magnitude to construct a robust histogram for feature extraction. This approach ensures reliable feature recovery even after attacks on the watermarked medical image.
The method is meticulously designed to strike a balance between robustness, imperceptibility, and computational efficiency, making it well suited for practical applications.
The remainder of the paper is organized as follows:
Section 2 reviews existing watermarking techniques. The core of our proposed method is detailed in
Section 3.
Section 4 demonstrates the effectiveness of our approach through experimental results. Finally, we conclude the paper and outline potential areas for future research.
2. Related Work
In the pursuit of robust watermarking systems for telemedicine applications, researchers have explored various methodologies, including spatial and frequency-based approaches, to embed watermarks within medical images. Frequency domain watermarking techniques leverage transforms such as the DCT, the DWT, or SVD to decompose the image into frequency components. These methods offer enhanced robustness against certain types of attacks while preserving image fidelity, making them well suited for applications in telemedicine where image quality is paramount for accurate diagnosis [
9]. By embedding the watermark within the frequency domain, researchers aim to achieve a balance between imperceptibility and robustness. Frequency-based methods exploit the properties of these transforms to ensure that the watermark is resilient to common attacks [
10] such as compression, noise addition, and geometric alterations. However, frequency domain watermarking techniques may also face challenges, such as increased computational complexity and potential perceptual distortion [
11].
Despite these challenges, ongoing research efforts continue to explore novel frequency domain watermarking techniques aimed at bolstering the security and integrity of medical images in telemedicine applications. The Wavelet Transform (DWT) is a mathematical technique used to decompose a signal into frequency components while capturing their locations. Unlike the Discrete Fourier Transform (DFT), which only provides frequency information, the DWT offers additional positional information [
12]. This decomposition into frequency bands with spatial information (low-frequency details, high-frequency details at specific locations) makes the DWT particularly valuable for image watermarking. The ability to target specific frequency bands and locations within the image allows for precise watermark embedding, leading to a wide range of possibilities for robust watermarking techniques, as evidenced by the numerous DWT-based watermarking systems [
6].
Anand et al. [
13] developed a medical image watermarking technique that integrates multiple text and image watermarks. They first applied the DWT to the image, then SVD to the HH, HL, and LH sub-bands. The text watermark is embedded into the singular values of the HH sub-band, while the image watermarks are embedded into the singular values of the LH and HL sub-bands. To enhance security, the watermarked image is then encrypted using hyperchaotic encryption. Devi and Singh [
14] used a DWT to transform the blue component of medical images. The LL sub-band is transformed using a fast Walsh–Hadamard transformation. Next, SVD is performed on the acquired blocks. The watermark bits are then incorporated into the singular values of the S matrix. To further protect the watermarked image, it is scrambled using the Arnold transform. In the paper of Devi and Mohapatra [
6], a robust watermarking scheme for Digital Imaging and Communications in Medicine (DICOM) images is proposed. To ensure security, the binary watermark is first encrypted with a logistic chaotic map, then embedded into the cover DICOM image by dividing it into blocks. Subsequently, the blocks undergo a first-level DWT to decompose them in the frequency domain. A random set of coefficients within each block is then selected using a secret key. Finally, the encrypted watermark is embedded based on a rank-based criterion within these chosen coefficients. The approach of Tiwari et al. [
15] performed zigzag scanning, then reorganized the coefficients within the Lifting Wavelet Transform (LWT) LL sub-band for watermark integration. The method of Thakur et al. [
16] prioritized security through a two-step embedding process. The cover image undergoes a DWT decomposition, followed by the selection of specific sub-bands for watermark integration. To enhance robustness, these sub-bands are further transformed using SVD. This facilitates embedding two watermarks: the highly robust “patient report” and the less robust “patient medical image” directly into the singular values of the chosen DWT sub-bands. The study of Khare and Srivastava [
17] proposed a novel image watermarking in which the HL sub-band of DWT is subjected to Homomorphic Transform (HT) separating the illumination and reflectance components. The watermark is then embedded into the singular values of the reflectance component using SVD. Inspired by the latter, Nawaz et al. [
18] employed a pretrained Deep Neural Network (DNN) to extract key features of the image to be watermarked. The DWT and Discrete Cosine Transform (DCT) are utilized to transform these features into a more compact representation for more robustness. To further enhance the security, the watermark undergoes chaotic scrambling before being XORed with the medical image using a secret key. Soundrapandiyan et al. [
19] developed a method that embeds feature information in the Region of Interest (ROI) using the DWT and tag information in the Non-Interest Region (NIR) using the DCT. The process starts by segmenting the original medical image into the ROI, containing crucial details, and an NIR. A two-level DWT is then applied to the ROI, generating specific coefficients (LL1, LH1, HL1, HH1, LL2, LH2, HL2, HH2). The watermark for quality features is embedded by modifying the low-frequency coefficients (LL2) of the DWT decomposition. Next, the NIR is divided into non-overlapping 8 × 8 blocks, and each block undergoes a DCT. The DCT coefficients are then rearranged using a zigzag scan, and eight intermediate-frequency coefficients are chosen for embedding the private label watermark. Another work of Araghi and Megías [
20] explored two watermarking DWT-based schemes for medical image watermarking. The first scheme employed a single-level SVD on each DWT sub-band, while the second used a two-level SVD to compare the impact of SVD levels on the robustness and imperceptibility of the watermarking.
Among the modern methods of watermarking images is the use of gradients. The method proposed by Luo et al. [
21] addresses medical image watermarking challenges by proposing a zero-watermarking scheme that combines feature extraction and encryption. The method extracted a feature vector using the Histogram of Oriented Gradients (HOG), capturing image’s edge information, which is transformed into a compact feature sequence through a perceptual hashing algorithm. The watermark image undergoes chaotic scrambling via a logical mapping process and then is embedded into the medical image. In the approach of Faheem et al. [
22], the information is embedded in the LSB of the image by first dividing the image into non-overlapping 16 × 16 blocks, then for each block, the gradient magnitude and direction are calculated, and the central pixel’s LSB is selected for embedding. After scrambling with a chaotic system for security, the watermark bits are embedded based on the gradient values (i.e., stronger magnitudes or directions allow for more watermark bits to be embedded). The final watermarked image is constructed by combining the modified LSB with the original most significant bits (MSBs). In the method proposed by Mokhnache et al. [
23], the cover image is decomposed with a specific DWT followed by a targeted transformation, using the DCT, on certain parts. Then, the smoothness of different image regions is analyzed to guide the division and embedding of the watermark, with specific thresholds determining the placement. The efficiency of this method hinges on using image features (smoothness) to select embedding locations. This approach enhances security by requiring a secret key based on the chosen embedding locations for successful watermark extraction.
A key challenge in medical image watermarking lies in achieving a balance between the desired properties of imperceptibility, capacity, and robustness. Existing literature revealed that many watermarking techniques prioritize robustness at the expense of the other two aspects due to inherent trade-offs. To address this limitation, we propose a novel DWT-based approach that leverages gradient analysis. This approach aligns well with existing research that demonstrates the effectiveness of the DWT for robust watermarking in medical images, often alongside additional techniques like feature extraction. Furthermore, gradient information has proven valuable in some watermarking methods, further supporting our focus on the DWT with gradient analysis to optimize the crucial balance between imperceptibility, capacity, and robustness.
3. Proposed Method
Most watermarking techniques share a similar framework, with variations arising in the specific methods used for embedding and extracting the watermark [
24]. In this work, we introduce a new blind and robust watermarking approach that extends the capabilities of the Discrete Wavelet Transform (DWT) by scrutinizing both the gradient direction and magnitude across decomposed sub-bands.
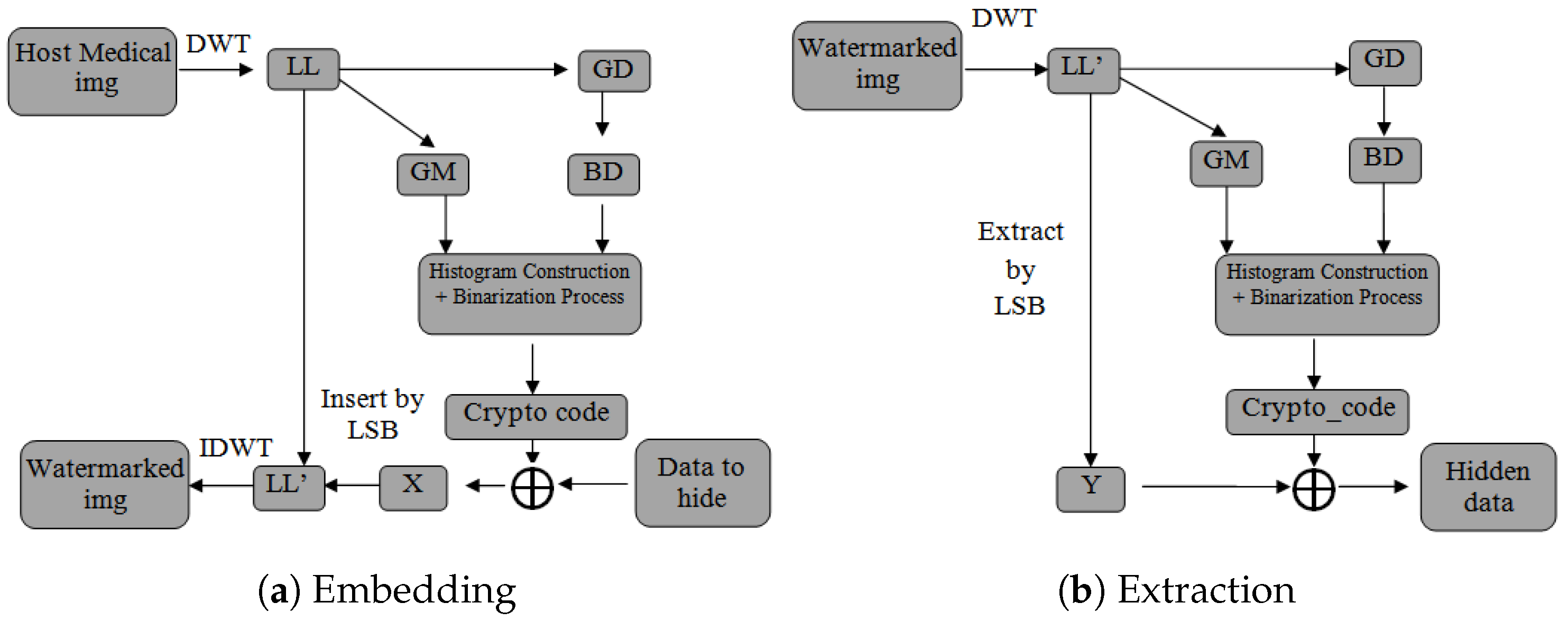
Figure 1 illustrates the schematic representation of the proposed approach.
Our proposed method consists of two main sections, namely (a) Information Embedding in which the information of the patient will be hidden in the cover image and (b) Information Extraction in which the hidden information will be extracted.
Our approach considers extracting three main components, which will be used later on for the embedding, from the cover image. The three components the sub-band for the Haar wavelet, the gradient magnitude, and the gradient direction are extracted as follows:
sub-band: We apply the Haar wavelet transform to the image, resulting in the frequency domain representation with the
sub-band:
This sub-band captures the low-frequency content, representing a smoothed version of the original image. The forthcoming procedures are performed at block-levels of size
.
Gradient direction (D): This provides crucial information about the orientation of edges and structures in the image. The gradient direction at each pixel is calculated using the following function:
To further improve robustness and to alleviate the noise effect, a binning is performed in the gradient direction according to the following formula:
where
c is a constant that determines the size of each interval and
, the ceiling function, returns the smallest integer that is greater than or equal to the input value. In our work,
c was set to
to provide greater flexibility and enhance robustness against noise effects.
Gradient magnitude (M): This represents the rate of change of pixel intensity across the image. It measures the overall intensity variation at each pixel, providing information about the edges. The gradient magnitude at each pixel is calculated using the gradient operators
and
in the horizontal and vertical directions, respectively:
where
and
represent the horizontal and vertical gradients of the image
I at the pixel
, respectively.
After having binned the gradient direction
and extracted the gradient magnitude
, we construct the watermark’s final histogram
H accumulating binarized magnitudes according to the binned directions as follows:
where
D is the set of all possible binarized directions,
is the set of all pixel indices
having gradient directions
d and that fall into the
k-th bin of the histogram, and
is a binarization function defined as follows:
For simplicity,
can be set to the middle of the magnitude value range for each
, calculated as:
It should be mentioned that each block must produce a 1 byte code (8 bits) called the crypto_code. In case the crypto_codes generated are of a length less than or greater than 8 bits, they should be completed or reduced to 8 bits using the same approach uniformly over all blocks. Likewise, the watermark (i.e., information to be hidden) is partitioned into segments comprising 8 bits each and then encrypted with crypto_codes using the XOR operation. Thereafter, the least significant bits (LSBs) from the corresponding encrypted watermark are applied to each coefficient of the watermarked block. Finally, an inverse DWT is then applied to the modified sub-band, then combined with the original ones , , and to reconstruct the watermarked medical image.
For watermark extraction, the reverse operations of the embedding process should be executed. This involves first applying the DWT to the watermarked medical image to decompose it into sub-bands (, , , ). The modified sub-band is then divided into non-overlapping blocks. From each block’s coefficients, the least significant bits (LSBs) are extracted to retrieve the encrypted watermark segments. These segments are decrypted using the corresponding crypto_code with the XOR operation. Finally, the decrypted segments are combined to reconstruct the original watermark. By following these reverse operations, the embedded watermark can be accurately extracted from the watermarked medical image.
4. Experiments and Evaluation
In this section, we present a comprehensive evaluation of our proposed watermarking scheme. Various aspects of the proposed scheme have been meticulously evaluated, including its robustness, imperceptibility, and computational efficiency. Additionally, we perform a comparative analysis against other related works to highlight the advantages and improvements offered by our approach. The results demonstrate the effectiveness and reliability of our watermarking method in practical scenarios.
The images used in our experiments, as represented in
Figure 2, were sourced from COVID-19 dataset (
https://www.kaggle.com/datasets/fusicfenta/chest-xray-for-covid19-detection (accessed on 15 January 2024)). This dataset contains two types of chest X-ray images, one which represents COVID-19 infection and the other being normal. The images are gray-level images (only intensity channel), in the format of
JPG, having variable sizes ranging from 1200 px to 2500 px.
4.1. Metrics
In order to thoroughly evaluate the performance of our watermarking technique, we employed several key metrics. These metrics are crucial for assessing the robustness, imperceptibility, and overall effectiveness of the watermarking scheme. The metrics used in our evaluation include the following:
Peak Signal-to-Noise Ratio (PSNR): The PSNR between the original image
I and the watermarked image
is calculated as:
where MAX is the maximum possible pixel value (typically 255 for 8-bit images) and MSE is the mean-squared error between the original and watermarked images:
Structural Similarity Index (SSIM): The SSIM index between the original image
I and the watermarked image
is given by:
where
and
are the mean intensities of the original and watermarked images,
and
are the standard deviations, and
is the covariance. The constants
and
are small positive constants to avoid division by zero.
Normalized Cross-Correlation (NC): The NC between the original watermark
W and the extracted watermark
is calculated as:
where
N is the number of bits in the watermark and
and
are the
i-th bits of the original and extracted watermarks, respectively.
Bit Error Rate (BER): The BER between the original watermark
W and the extracted watermark
is calculated as:
where
N is the number of bits in the watermark and
is the Kronecker delta function that returns 0 if
and 1 otherwise.
These formulas provide quantitative measures for assessing the quality and fidelity of the watermarking process, aiding in the evaluation and comparison of different watermarking techniques.
4.2. Imperceptibility Assessment
We evaluated the imperceptibility of the watermarked images to the human eye using two key metrics: the Peak Signal-to-Noise Ratio (PSNR) and the Structural Similarity Index (SSIM). Higher PSNR values indicate better quality, with minimal visual distortion caused by the watermark. The PSNR provides a more comprehensive assessment of visual fidelity, as the PSNR can be fooled by noise that appears similar to the image details. The results assessing the effect of different watermark sizes on imperceptibility are summarized in
Table 1.
From
Table 1, it is evident that smaller watermark sizes tend to yield slightly higher PSNR values, indicating better imperceptibility. This aligns with the principle that smaller watermarks are less likely to introduce noticeable visual artifacts in the watermarked images. However, it is noteworthy that, even with larger watermark sizes, the imperceptibility remains high, underscoring the robustness of our watermarking technique. For instance, using a medium watermark size (
bytes) results in an image with almost imperceptible changes. Human eyes generally would not perceive any significant quality difference between the original and watermarked images if the PSNR value is above 50 dB. This demonstrates the effectiveness of our method in preserving the visual quality of the medical images while embedding the watermark. The findings from this analysis reinforce the practical applicability of our watermarking approach in medical imaging scenarios. The ability to embed watermarks with minimal impact on image quality, even with larger sizes, enhances the sensitive medical data without compromising the diagnostic integrity of the images.
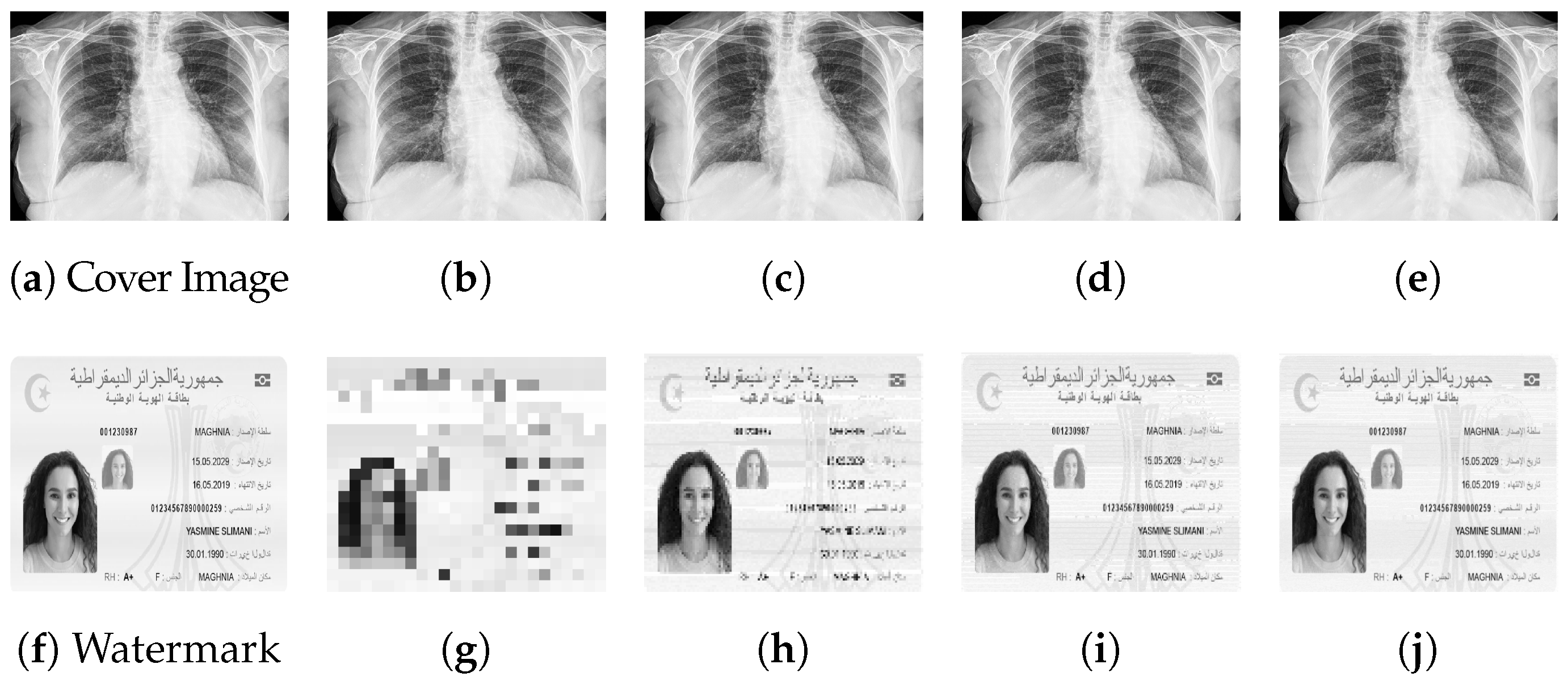
To further elucidate the impact of different sizes of watermarks on the imperceptibility of our watermarking technique, we provide a visual illustrations in
Figure 3. As depicted in
Figure 3, varying sizes of watermark images are embedded within the cover images, ranging from small to large. As it appears, the quality and structural similarity between the watermarked (b)–(c) and the cover image (a) does not degrade as the size of the watermark increases.
To establish the superiority of imperceptibility, we compared various watermarking methods. The primary objective was to evaluate each technique’s effectiveness in concealing watermarks within images while minimizing visual distortion. Notably, our method, as illustrated in
Table 2, surpassed competing approaches by achieving the highest imperceptibility score. This outcome confirms our method’s exceptional ability to seamlessly integrate watermarks into medical images while preserving their original visual quality.
4.3. Robustness Evaluation
In addition to assessing imperceptibility, we also evaluated the robustness of our watermarking technique against common image processing operations and attacks. Robustness testing is crucial to ensure that the embedded watermark remains intact and detectable even after various modifications to the watermarked image. Random noise or intentional tampering could heavily affect the extracted watermark, making it essential to evaluate how well the watermark withstands such conditions. In
Figure 4, we show the watermark extracted before and after applying image attacks using Gaussian and salt and pepper (S & P) noises. These results illustrate the robustness of our watermarking method under different types of noise, demonstrating its ability to maintain watermark integrity even in challenging conditions.
We subjected the watermarked images to a series of attacks, including noise, scaling, filtering, and intensity enhancement, among others. The goal was to determine the resilience of the embedded watermark under different scenarios.
Table 3 summarizes the results of our robustness testing, showing the performance of the watermark under each operation or attack.
From
Table 3, it is evident that the proposed method demonstrates robustness across various attacks, achieving high NC values. Specifically, it exhibits strong resilience against both Gaussian and salt and pepper noises (NC: 0.97), outperforming most of the comparison methods. Despite the challenges posed by histogram equalization, the proposed method maintains a respectable NC value (0.8). Additionally, its robustness against the median filter (NC: 0.86) and scaling (NC: 0.78) underscores its applicability in medical imaging scenarios where image orientation and cropping are common. The Normalized Correlation (NC) metric is not sufficient for evaluating the performance of watermarking techniques, as it only reflects the true positives. A more important metric is the Bit Error Rate (BER), which considers the matching of both positive and negative bits, providing a more comprehensive assessment of the watermark’s performance. Our method outperforms all other methods in this task, demonstrating superior accuracy and reliability in watermark detection and integrity. Overall, while the proposed method exhibits promising robustness against a wide range of attacks, further exploration and refinement in certain areas could enhance its effectiveness in robust medical image watermarking applications.
These results demonstrate the effectiveness of our watermarking method in preserving the integrity and authenticity of medical images, even in the face of potential alterations and attacks. The high detection rates across various tests underscore the reliability and robustness of our approach, making it suitable for real-world applications where image security is paramount. The scaling and sharpening attacks highlight potential limitations of our method, requiring further investigation.
4.4. Computational Efficiency
The computational efficiency of the proposed approach, leveraging the Discrete Wavelet Transform (DWT) alongside image gradient scaling using Sobel edge detection for watermark embedding in the least significant bit (LSB), entails a multi-faceted process. Integrating Sobel edge detection adds a computational layer to identify image gradients efficiently, enhancing the security of watermark embedding. Considering
N as the number of pixels in the image,
M as the number of bits to be embedded, and
L as the number of decomposition levels in the DWT, the overall complexity comprises three main components: DWT processing
, Sobel edge detection
, and LSB embedding
. Thus, the total complexity is
, with the dominant factor contingent on the interplay between
L,
M, and
N. Knowing that
, we can say with certainty that the increase of time consumption over the size of the watermark is Linear. In the subsequent
Figure 5, we delineate the computational watermarking/extraction time over variations in the watermark size.
As anticipated from
Figure 5, the consumed time exhibits a linear correlation with the size of the watermark. This linear relationship suggests that, as the size of the watermark increases, the time required for computation also increases proportionally. This stability implies that the proposed watermarking approach can efficiently handle varying sizes of watermarks, which is crucial for applications such as video processing where watermark sizes may vary.
5. Conclusions
Our research presents a pioneering approach to blind and robust watermarking for medical images, which capitalizes on the Discrete Wavelet Transform (DWT) in conjunction with gradient analysis. By integrating the LL sub-band, gradient magnitude, and gradient direction, our method achieves a balance between imperceptibility and robustness, ensuring the protection of sensitive medical data. Through comprehensive evaluations, including metrics like the PSNR, SSIM, NC, and BER, we have demonstrated the superiority of our technique over existing methods, promising enhanced security without compromising image quality. Moreover, simulations on real healthcare data involving various common attacks, such as median and noise addition, further underscored the resilience of our method, highlighting its efficacy in maintaining watermark integrity under adverse conditions. This innovative solution holds significant potential for practical applications in medical image authentication and secure data transmission within healthcare systems.