The DSL’s design incorporates several key features to enhance its functionality and user experience. It features a hierarchical structure for clear navigation, modularity for adaptability to various research scenarios, and unique identifiers for precise element reference. The system supports task-specific models, allowing researchers to tailor their approach to particular aspects of MS research. Flexible data loading capabilities enable users to easily import data from various sources, while comprehensive evaluation metrics provide thorough model assessment. We have also included mechanisms for feature selection, visualization, and handling of logical expressions, all facilitated by an auxiliary table for attribute values. MS experts can leverage this tool without extensive knowledge of ML models or programming. Their primary focus should be on identifying crucial features and defining clear objectives, such as data classification or feature relationship discovery. By utilizing AutoML methods, these experts can generate initial results efficiently. Subsequently, ML researchers can build upon these preliminary findings, refining and enhancing the models to achieve a superior performance. Our tool facilitates a streamlined ML data pipeline improvement process, accommodating users with varying levels of ML expertise. It promotes iterative enhancements through collaborative efforts among multiple stakeholders, fostering a synergistic approach to MS research and model development.
3.1. Meta Model Design
As we discussed, the design of our DSL’s meta model is guided by key requirements ensuring its flexibility and applicability in MS research and treatment. It features a hierarchical structure for clear navigation, modularity for adaptability, and unique identifiers for precise element reference. The DSL supports task-specific models, flexible data loading, comprehensive evaluation metrics, and a rule-based training approach. It also includes mechanisms for feature selection, visualization, and handling of logical expressions, all facilitated by an auxiliary table for attribute values. These requirements ensure the DSL is robust, capable of addressing complex challenges, and user-friendly.
The abstract syntax of our DSL in Listing 1 defines how each construct is interpreted and executed within the context of the ML model and feature selection. Here, we provide a precise description of the key language elements:
RuleModel: This is the root container for all the elements in a model definition. When executing a RuleModel, each contained element is interpreted and executed in sequence.
Model: Defines an ML task with a specific name and type (classification or regression). It encapsulates all the elements required for model training and evaluation. As shown in Listing 2, the model is defined with a regression task. It includes an AutoML model, feature selection from the loaded dataset, specified metrics, visualizations, and instructions for the ML model to start with.
Load: Specifies the data source. When executed, it loads the dataset from the specified path into memory, making it available for subsequent operations.
MLModel: Declares a ML model type with optional parameters. When invoked, it instantiates the specified model with the given parameters.
Metric: Defines the evaluation metrics for model performance. These metrics are calculated after model training and used in decision-making processes within rules.
FeatureSelection: Specifies the features to be used in model training. It can include filtering conditions and feature selections. When executed, it preprocesses the dataset to include only the specified features. Listing 2 provides a comprehensive example of feature selection. This example demonstrates the ability to select multiple features from different datasets while applying specific conditions. Furthermore, for each selected feature, operators can be added to filter feature values, offering fine-grained control over the data used in the model.
Rule: A rule is composed of two parts: the condition (expression) that checks the metrics of the ML models, and the specific action (typically selecting a model or feature set) to be taken.
RuleSet: Contains a collection of rules for model selection and feature engineering. Rules are evaluated sequentially, and their actions are executed when conditions are met. In Listing 3, the example ruleSet contains three rules, describing a workflow for model selection and training. Initially, if the SVM model does not meet the performance criteria, the system switches to training a RandomForest model. Should the RandomForest model perform well, it is further trained with a filtered set of features. However, if the RandomForest model also underperforms, the system initiates training of a DecisionTree model with a different feature selection. This rule-based approach allows for adaptive model selection and feature refinement based on performance metrics, enabling the system to dynamically adjust its strategy to achieve optimal results.
Start: Initiates the model training process with the specified model and feature set. It triggers the actual computation and evaluation of the ML pipeline.
Show: Outputs the results of the model training and evaluation process. It can display model performance metrics, feature importance, or other relevant information.
| Listing 1. Abstract Syntax of the DSL. |
![Applsci 14 06193 i001]() |
The execution flow of our DSL typically follows this sequence:
Data are loaded using the Load statement. This step calls our defined DataManager class, which provides several functions based on pandas’ read_csv or read_sql functions depending on the data source. It can load csv, xlsx, and .db files, and it includes default data cleaning functions like handling empty features and normalization. The DataManager also handles splitting of the training and testing data, though this is applied after feature selection.
MLModels are instantiated with their respective parameters. Each model type is mapped to its corresponding scikit-learn class based on the defined task, such as sklearn.svm.SVR or SVC. If multiple parameters are defined, it maps to scikit-learn’s grid search functionality. The special autoML model maps to either TPOTRegressor or TPOTClassifier.
FeatureSelection is applied to select features from the dataset(s). It can select from multiple loaded datasets, combine different features from various datasets, and apply filters to the data. After feature selection, the DataManager functions are used to split the data into training and test sets.
Metrics are calculated to evaluate model performance, mapping to functions in
sklearn.metrics. Our system logs all parameters and training results in MLflow [
26], which can be retrieved when checking defined rules and generating visualization diagrams.
RuleSets and Rules are evaluated, potentially altering the model or feature selection based on the defined rules. This is implemented as conditional statements in Python, evaluating metrics and executing corresponding actions.
The Start command initiates the training process of MLModels, including training and fitting, and it logs the corresponding results in MLflow.
Finally, the Show command displays the results of the process. We use Matplotlib to visualize the logged results.
The proposed DSL provides a structured framework for defining and executing ML workflows in MS research. It offers clear semantics for model definition, feature selection, and rule-based execution. The DSL supports both automated and manual methods, catering to users with varying expertise levels. This design enables progressive refinement of models and features, allowing domain experts to effectively utilize ML techniques without extensive programming knowledge. The rule-based system facilitates adaptive model development, promoting an iterative approach to ML in MS research.
Figure A1 serves as a structural framework for our DSL design.
3.2. Task-Specific Model Instantiation
In this section, we will give example models based on our current data requirement analysis in the three ways we mentioned. Our DigiPhenoMS [
27] project is a collaborative endeavor between the MS Center at the University Hospital Carl Gustav Carus Dresden and the Digital Health research group. This project aims to investigate the heterogeneous progression of MS by leveraging the extensive data collected throughout a patient’s clinical journey. MS is a complex neurological disorder that is characterized by a highly variable disease course, which differs significantly from one individual to another. Prior to the formal diagnosis of MS, patients undergo a comprehensive anamnestic assessment and a series of necessary examinations, generating a substantial amount of data. Upon confirmation of the diagnosis, further data are accumulated during subsequent treatment sessions and routine check-up appointments. Over the span of a patient’s life, a unique data trail is formed, encompassing the various stages of the clinical pathway within the healthcare system. The data collected includes MRI, motility and cognitive assessments, and neuropsychological evaluations of 1701 patients at different phases of the disease. These diverse datasets offer valuable insights that can aid domain experts in determining the relevance of specific factors and developing predictive models. Regression algorithms are employed in this research because they are well-suited for predicting continuous variables such as disease progression scores or severity measures, and they can help uncover the relationships between various features and MS outcomes, enabling a better understanding of the factors influencing the disease. We also support classification tasks; however, in this context, we focus on regression due to the specific requirements of our data. Additionally, the extensibility of DSLs allows for the easy addition of other tasks. The current scope of the DSL is limited to supporting scikit-learn [
28] compatible regression models and pipelines. By focusing on scikit-learn, the DSL can leverage the extensive ecosystem of models and tools built around this library. With the extensibility of the meta model design, we aim to integrate the model with additional ML technologies and neural networks in the future. Additionally, we intend to invite medical experts to evaluate the usability and accuracy of our DSLs, with a focus on the continuous improvement of the models and validation of the results.
The first example, shown in
Listing A1, showcases how to define AutoML in our real use case model. All these models could be defined in the same file and share the data loader from the
load command. Here, we load the dataset from the sqlite database. Also, we support the loading of multiple datasets from csv files and databases, and we also support joint queries between them, as shown in Listing 2. This model defines an AutoMLModel task for regression, where the
mlModel parameter is set to
autoML, indicating the use of an automated ML algorithm. The select
pst statement specifies that the initial set of features should be selected from the entire dataset about MRI and process speed time features. Additionally, the
metric parameter defines the evaluation metrics to be used, such as mean squared error (mse), R-squared score (r2_score), mean absolute error (mae), and root mean squared error (rmse). Finally, the start
autoML with the
pst statement initiates the AutoML process with the specified initial features and shows the models and features. AutoML techniques have gained significant attention in recent years due to their ability to streamline the ML workflow and achieve competitive performance without extensive manual intervention. Here, we integrate TPOT [
29], which uses genetic programming to automate the optimization of ML pipelines with techniques like feature selection, feature construction, model selection, and hyperparameter tuning. TPOT is an AutoML system that uses genetic programming to optimize ML pipelines. TPOT supports various models provided by the scikit-learn library, including linear models, support vector machines, decision trees, random forests, and ensemble methods. We could set generations and population sizes to fine tune this model.
Generation: This parameter determines the number of iterations that the genetic algorithm will run. Each generation involves creating a new population of ML pipelines by applying genetic operations (crossover and mutation) to the best-performing pipelines from the previous generation.
- −
A higher number of generations allows for more thorough exploration of the solution space, potentially finding better pipelines.
- −
However, it also increases the computation time.
- −
In our DSL, users can specify this parameter as generation = 100
Population size: This parameter sets the number of individuals (ML pipelines) in each generation.
- −
A larger population size increases the diversity of solutions explored in each generation.
- −
It can lead to better results but at the cost of increased computation times.
- −
Users can set this in our DSL as population_size=50
The genetic processes in AutoML, specifically in TPOT, involve operations like crossover (combining parts of two high-performing pipelines) and mutation (randomly altering parts of a pipeline) to evolve increasingly effective ML workflows. These processes explore various combinations of preprocessing steps, feature selection methods, and ML algorithms. As a result, AutoML can discover complex, multi-step pipelines that often outperform manually designed models, potentially including unexpected feature transformations or ensemble methods that a human data scientist might not have considered. The output typically includes the best-performing pipeline architecture, optimized hyperparameters, and performance metrics, providing users with a ready-to-use model and insights into effective feature engineering strategies for their specific dataset. This allows researchers to focus on higher-level tasks such as problem formulations and results interpretation.
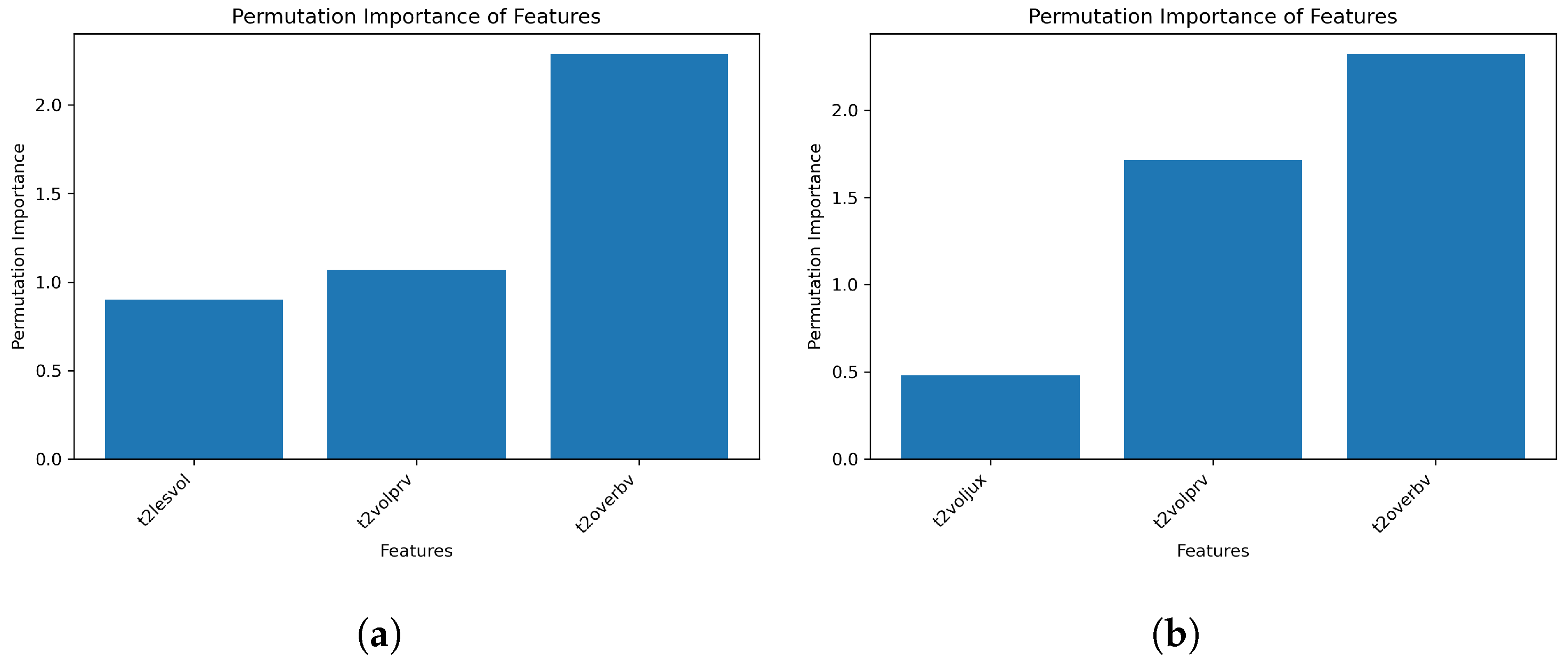
The second model, shown in Listing 3, demonstrates a manual approach to model and feature selection. In this model, three ML algorithms, random forests, decision trees, and SVMs are defined with their respective hyperparameters. The select pst statement specifies a set of initial features based on certain conditions. Additionally, a new set of features, denoted as pst_filter, is defined by utilizing a distinct dataset and specific conditions (e.g., t2lesvol > 3.0) for enhanced screening values. The ruleSet selectionRules defines a set of rules for model selection based on the evaluation metrics. If the r2_score of the SVM model is less than 0.85 and the mean squared error is greater than 0.2, the random forest model is selected. If the r2_score of the random forest model is greater than 0.8, which is a good model, then we further train it with the pst_filter features to see if it could be better. If it does not meet our expectations, we instead use a decision tree. It is worth noting that the ruleSet could also be applied to the other models; however, for the sake of brevity, only one illustrative example is presented here. The output displays the models, which presents a diagram comparing the results of two model runs, and the features, which provide a comparison of feature importance. This model showcases the manual approach to feature and model selection, where domain knowledge and expert intuition play a crucial role. This approach is suitable for further comparison screening and the comparison of customized models and features by obtaining initial results from AutoML and incorporating additional rules and conditions to fine-tune the models and features. By defining specific conditions and rules, researchers and practitioners can meticulously refine the feature selection process and systematically explore various model combinations to achieve optimal performance.
The third model, shown in Listing 4, demonstrates a customizable approach to model selection for a classification task. In this model, a decision tree algorithm is defined with specific hyperparameters, such as max_depth, min_samples_split, min_samples_leaf, and max_features. The selected pst statement specifies the most related features after verification of the last two models. Initiating the decision tree with the pst statement initiates the model training process using the specified features. This model is suitable for re-running the chosen models from coarse screening and attempting to refine them with finer granularity in terms of specific model architectures and features.
| Listing 2. Model example for multiple datasets in AutoML. |
![Applsci 14 06193 i002]() |
Our DSL significantly enhances explainability and facilitates collaboration in MS research through its intuitive, rule-based syntax. For example, a rule like mrt_pst_17_20.t2lesvol > 2.0 is easily interpretable by clinicians, as it uses familiar clinical features and clear thresholds. In terms of collaboration, the DSL serves as a bridge between technical and clinical experts. It allows neurologists to express their domain knowledge in a format that data scientists can directly incorporate into ML pipelines. This shared language reduces miscommunication and accelerates the development of clinically relevant ML models for MS research.
The provided models showcase different approaches to model selection and feature engineering, each with its own strengths and applications. The AutoML model offers a streamlined and automated approach, while the manual selection models provide more control and flexibility for researchers and practitioners to leverage their domain knowledge and expertise. Ultimately, the choice of approach depends on the specific requirements of the problem, the available resources, and the trade-offs between automation and manual interventions. By understanding the capabilities and limitations of each approach, researchers and practitioners can make informed decisions and develop effective ML solutions tailored to their needs. The three approaches presented constitute a layered framework, allowing users to progressively refine and optimize their models according to their expertise and the level of control they desire. This framework also facilitates a better understanding of data flow through the use of more abstract and less technical language. In the following section, we will demonstrate the practical implementation and application of these models on our dataset, showcasing their performance and discussing the insights gained from each approach.
| Listing 3. Model for manual selection. |
![Applsci 14 06193 i003]() |
| Listing 4. Model for manual selection. |
![Applsci 14 06193 i004]() |