Average Slope vs. Cmax: Which Truly Reflects the Drug-Absorption Rate?
Abstract
1. Introduction
2. Materials and Methods
2.1. Strategy of the Analysis
2.2. Pharmacokinetic Properties
2.3. Machine-Learning Algorithms
2.3.1. Principal Component Analysis
2.3.2. Random Forest
2.3.3. Hierarchical Clustering
2.3.4. Artificial Neural Networks
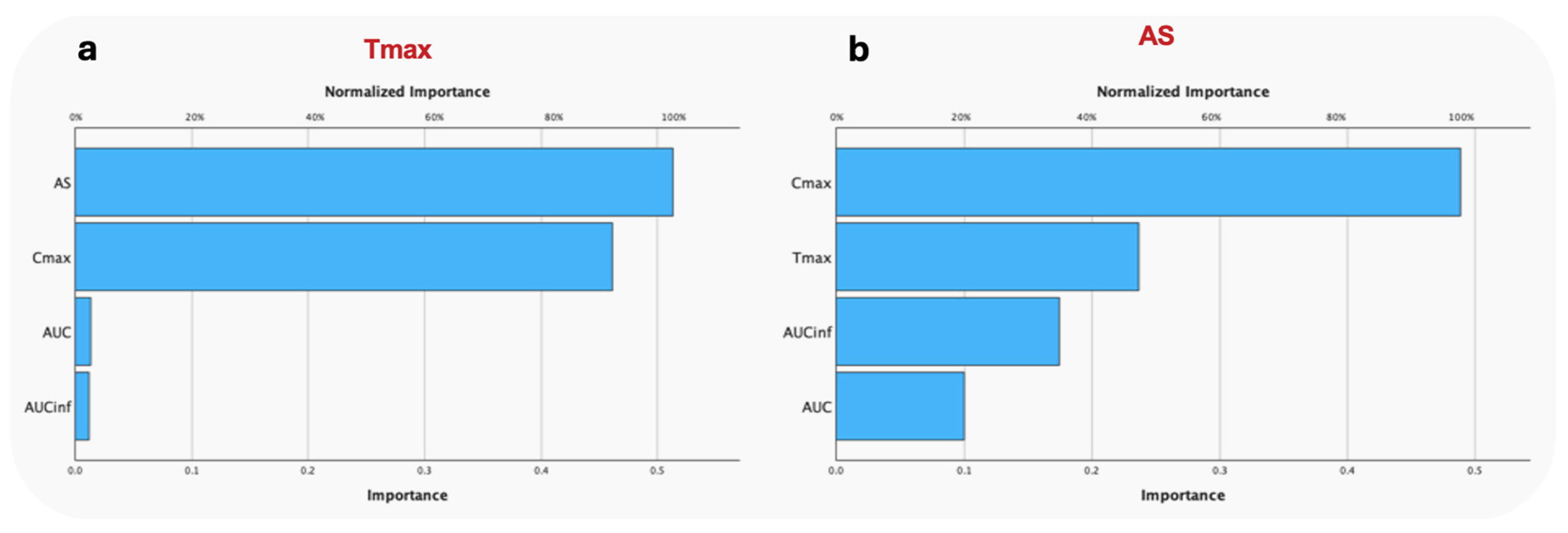
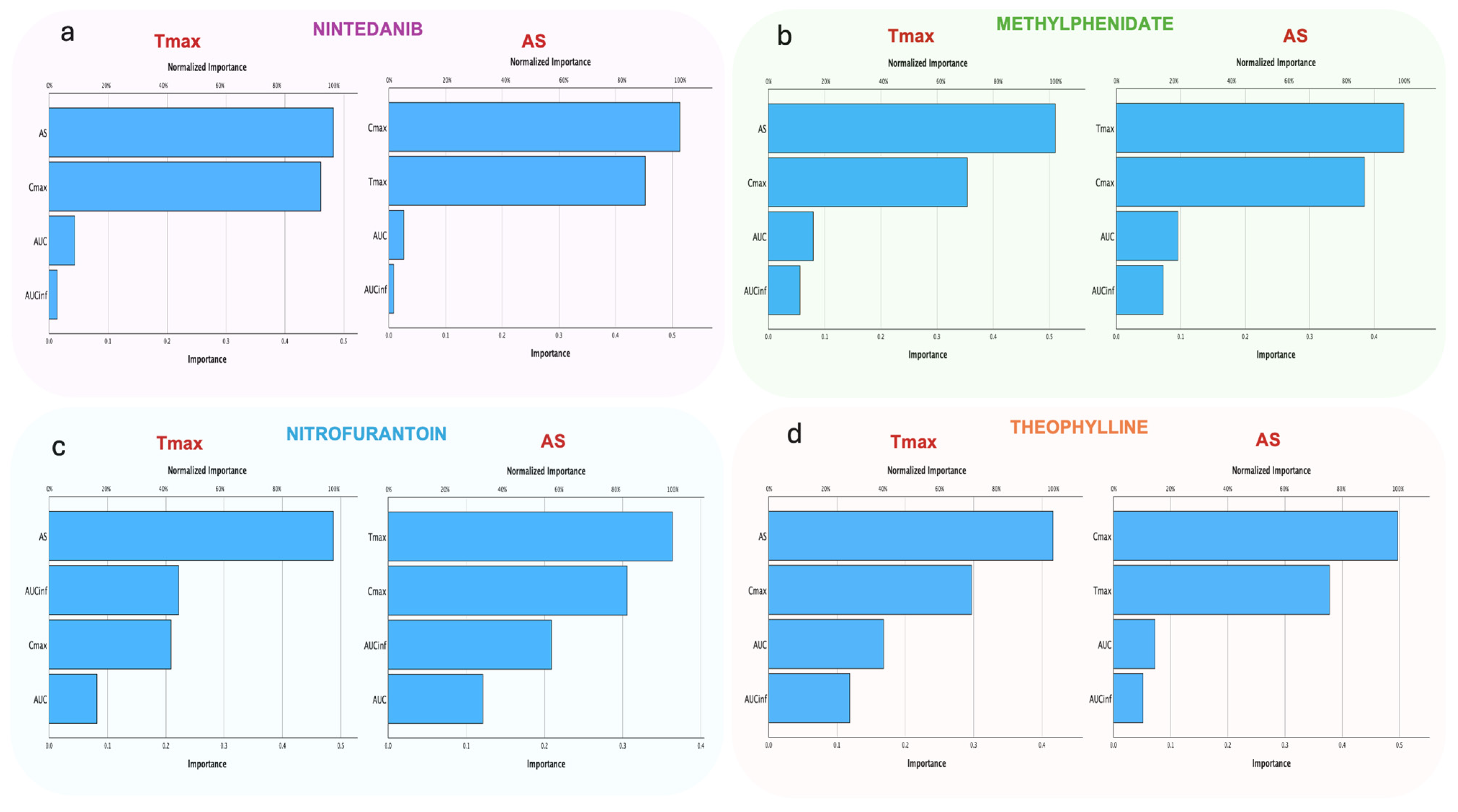
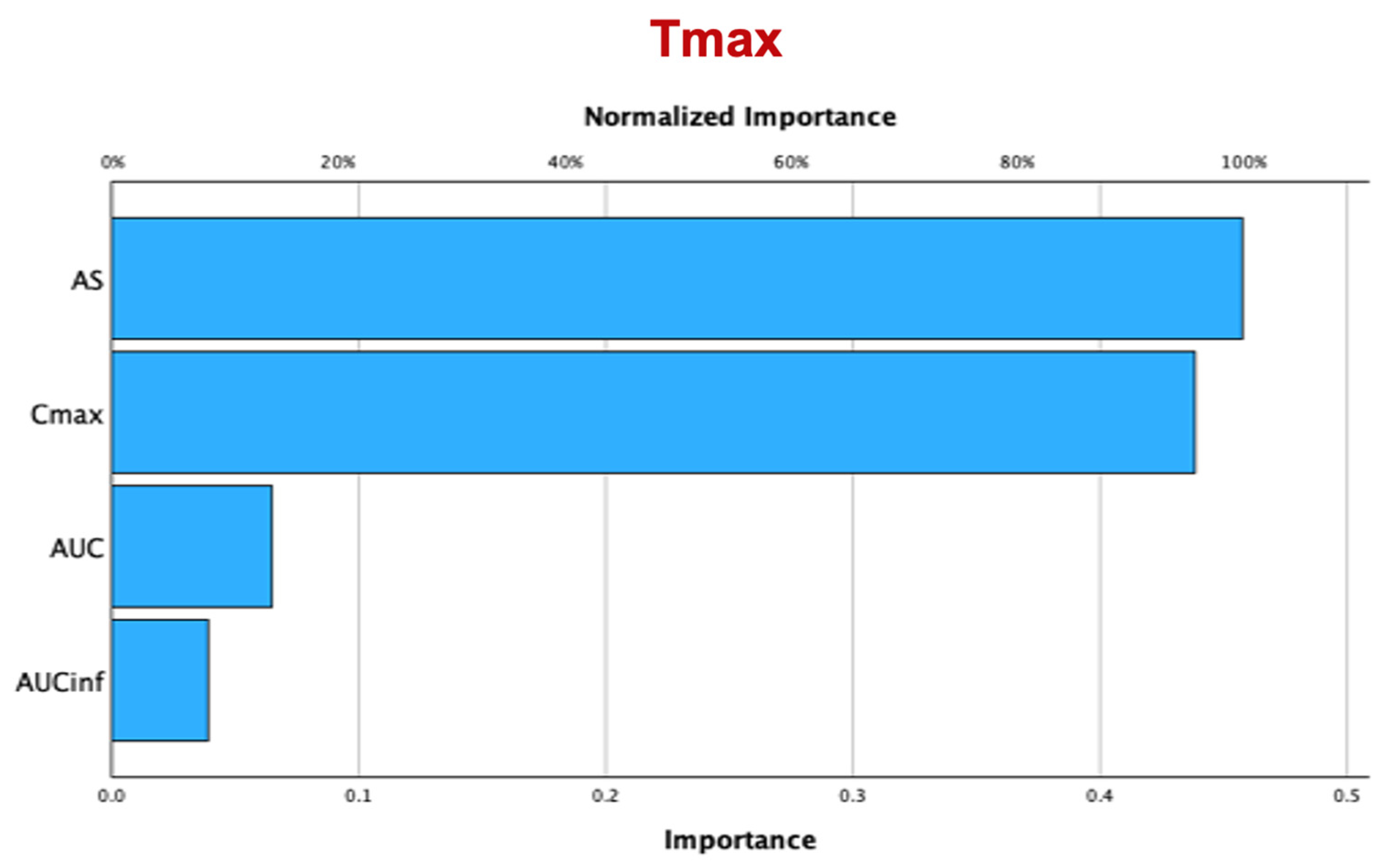
3. Results
3.1. Pharmacokinetic Data
3.2. Comparative Performance between AS and Cmax
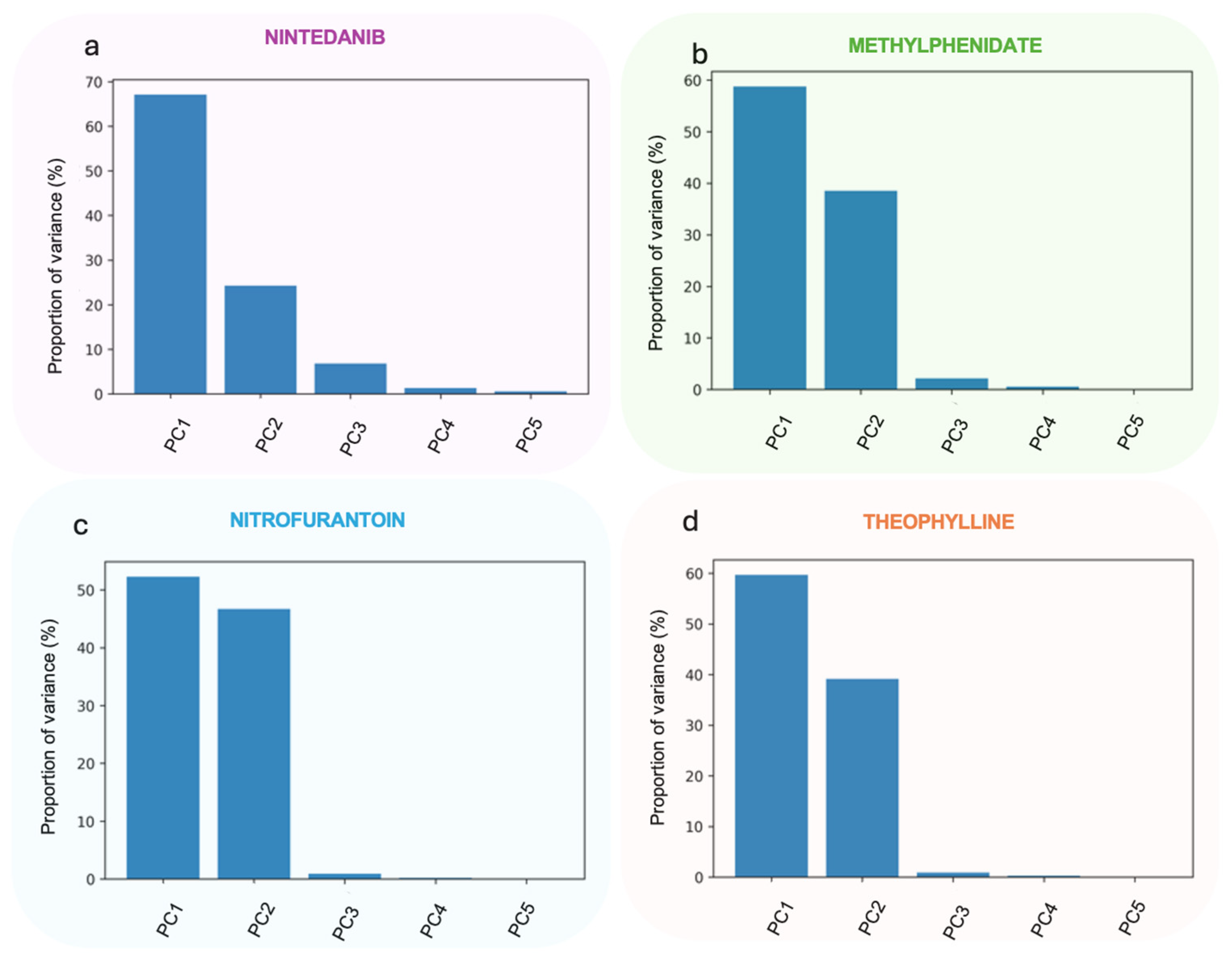
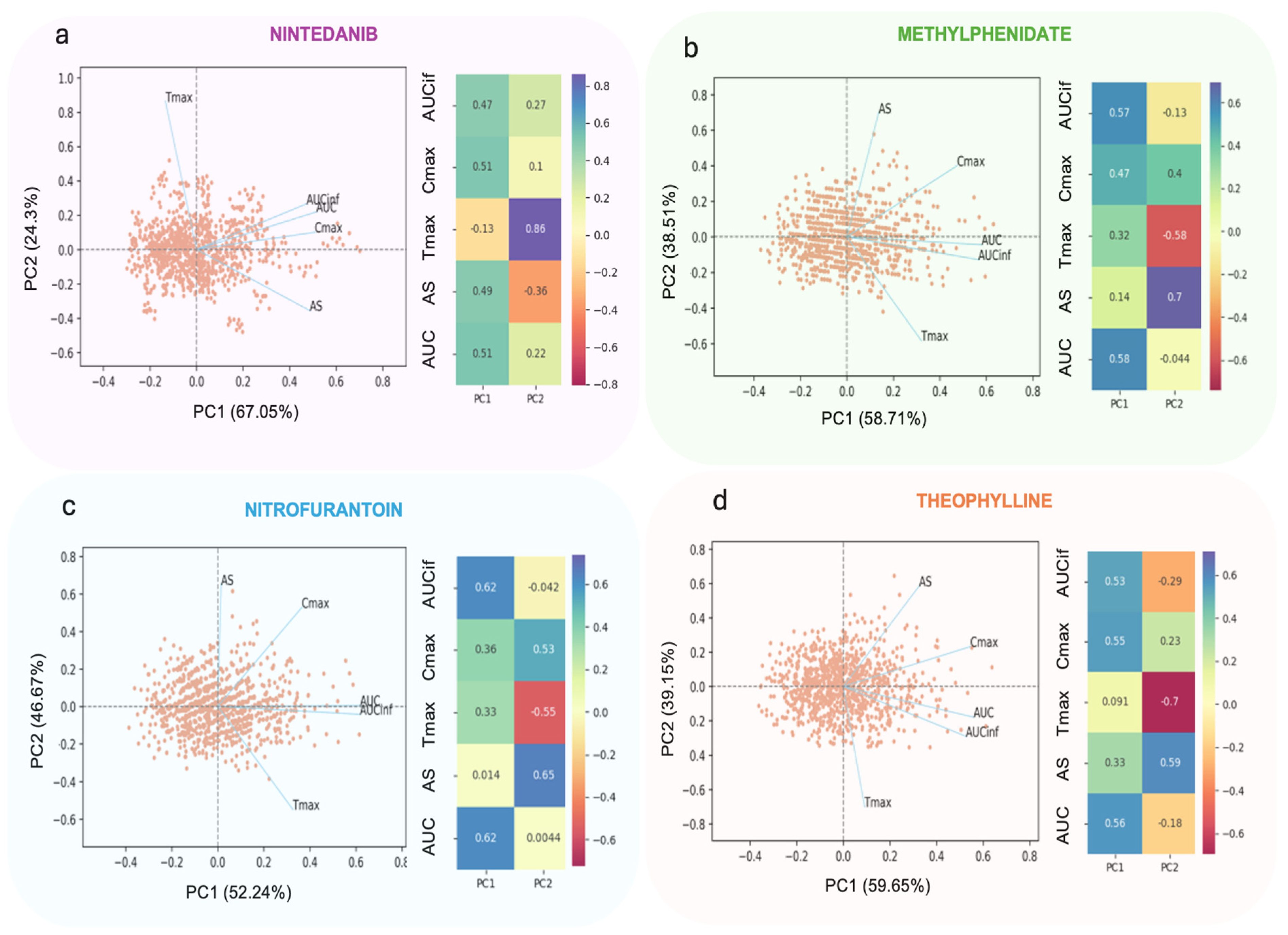
3.2.1. Principal Components Analysis
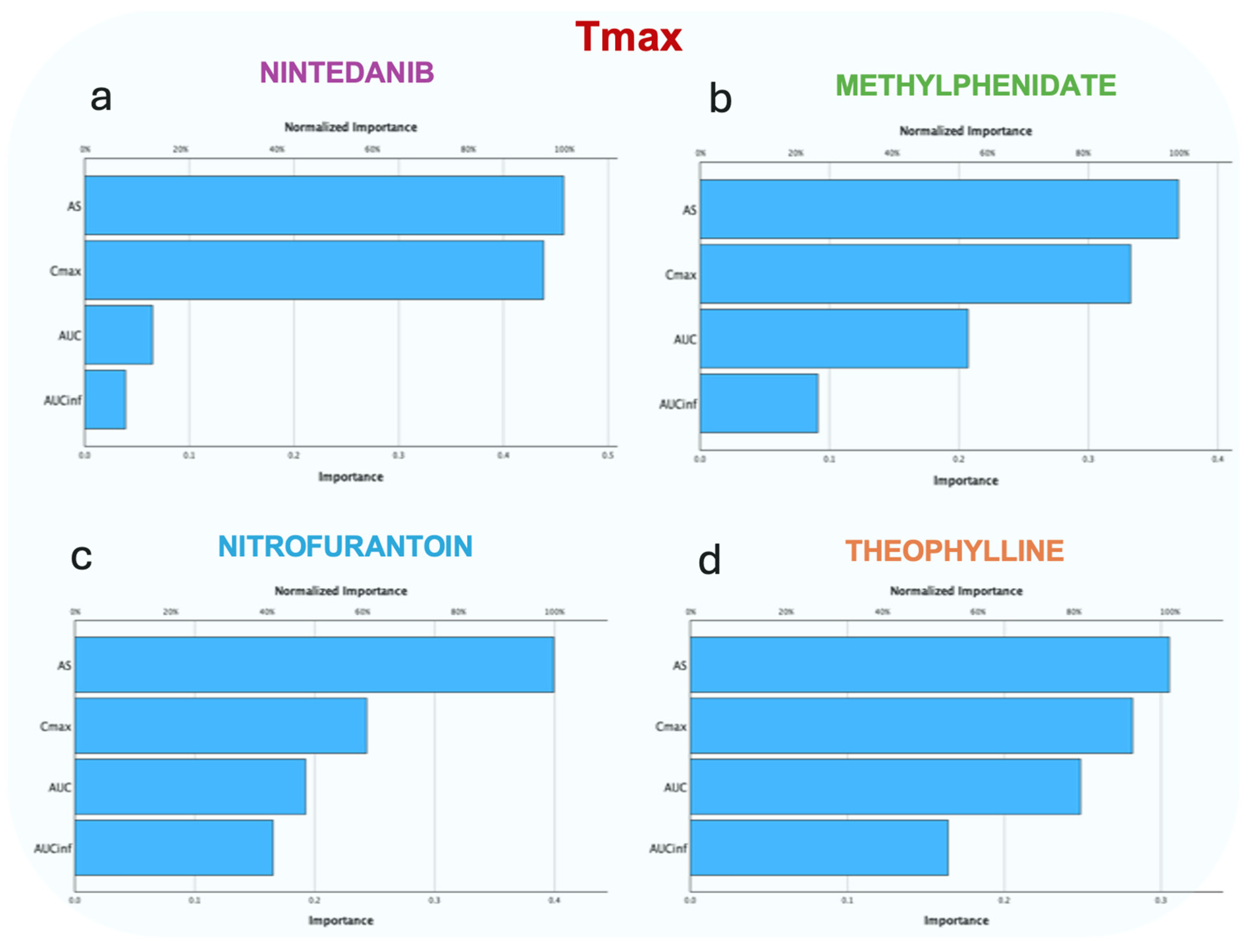
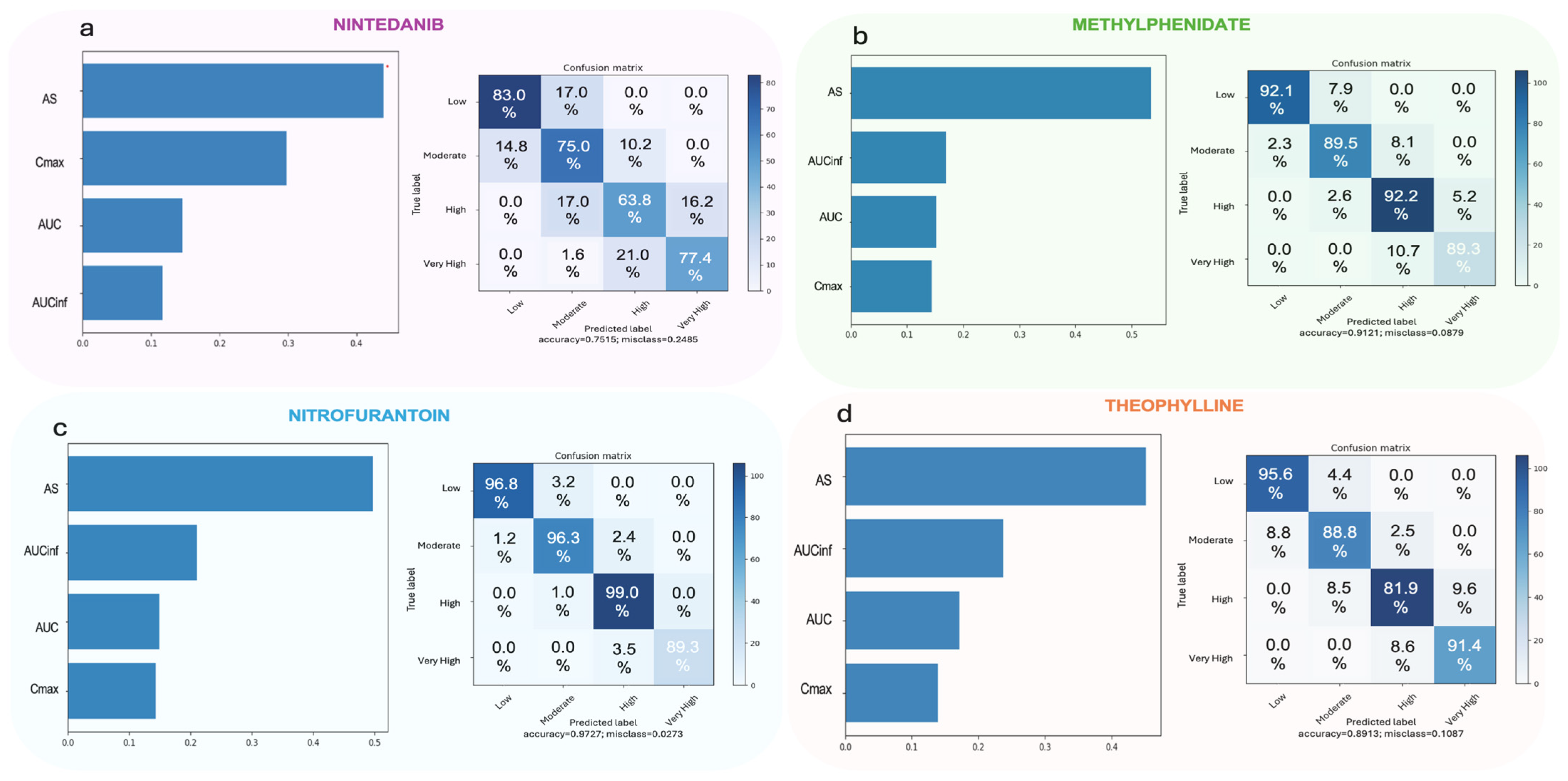
3.2.2. Random Forest
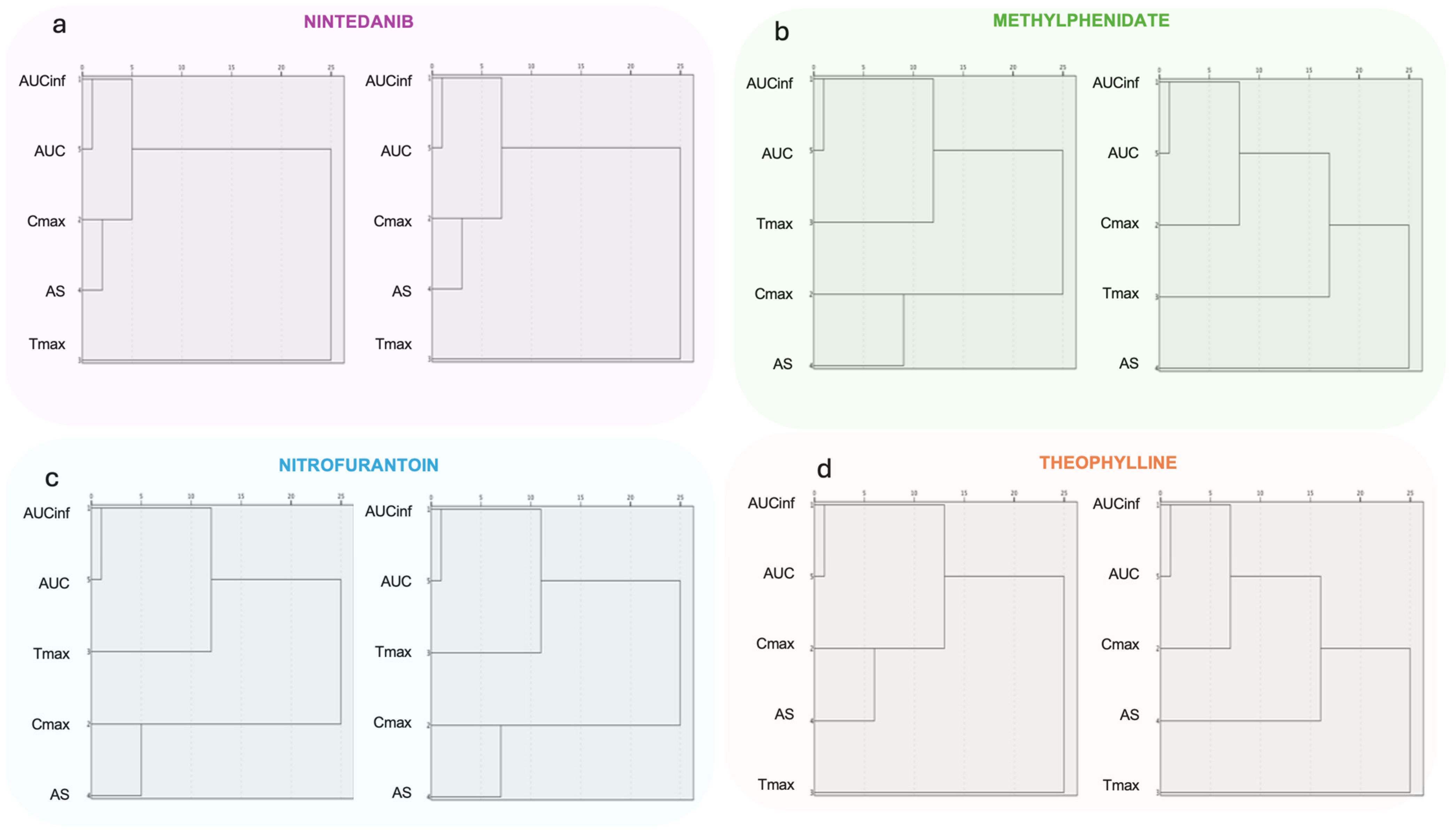
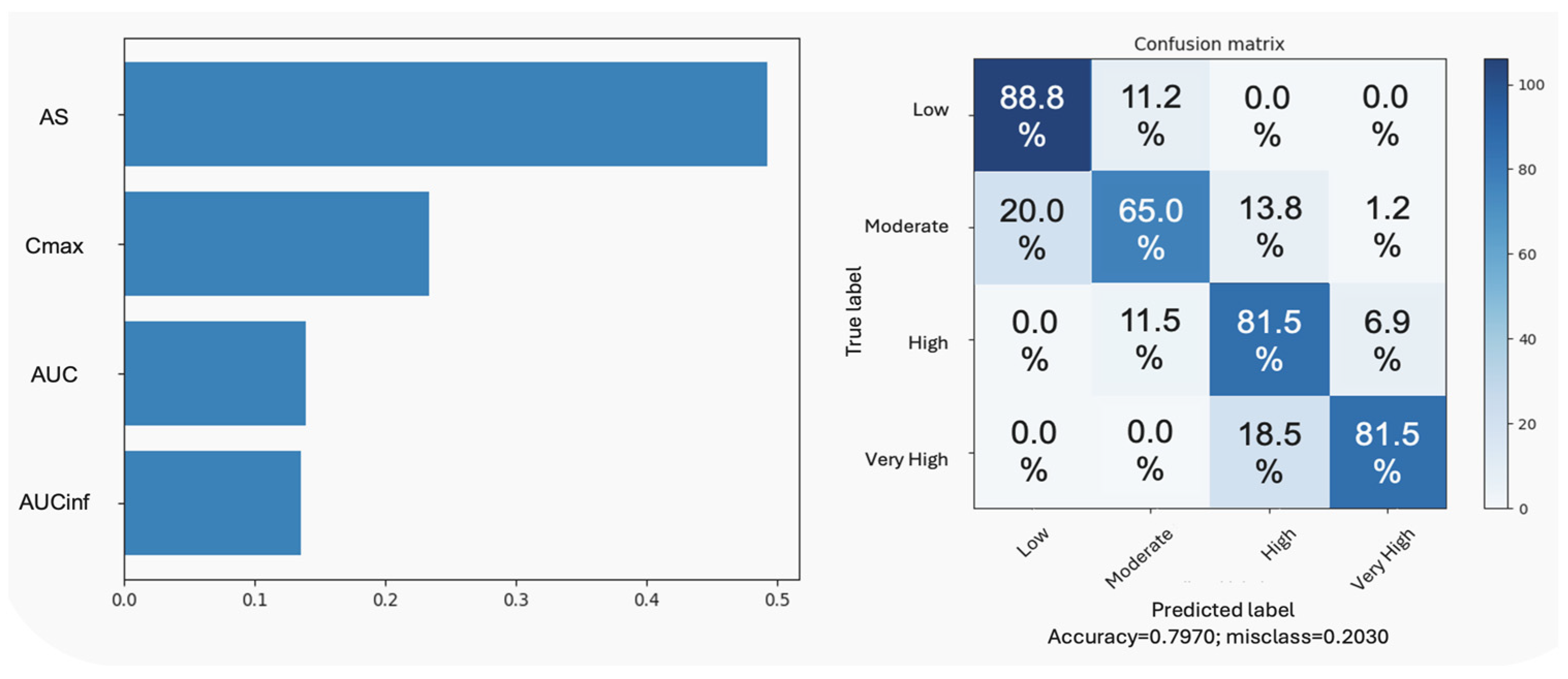
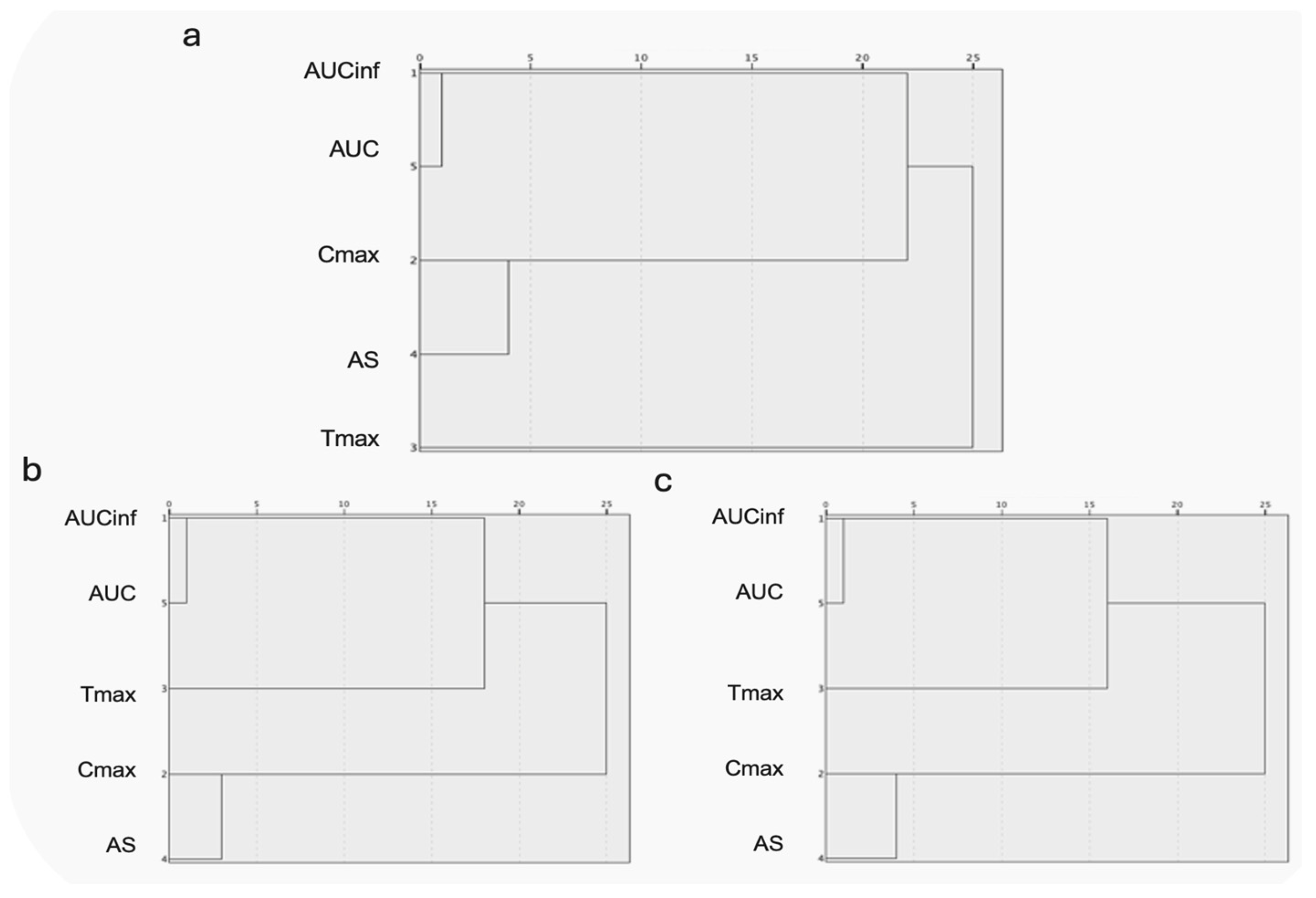
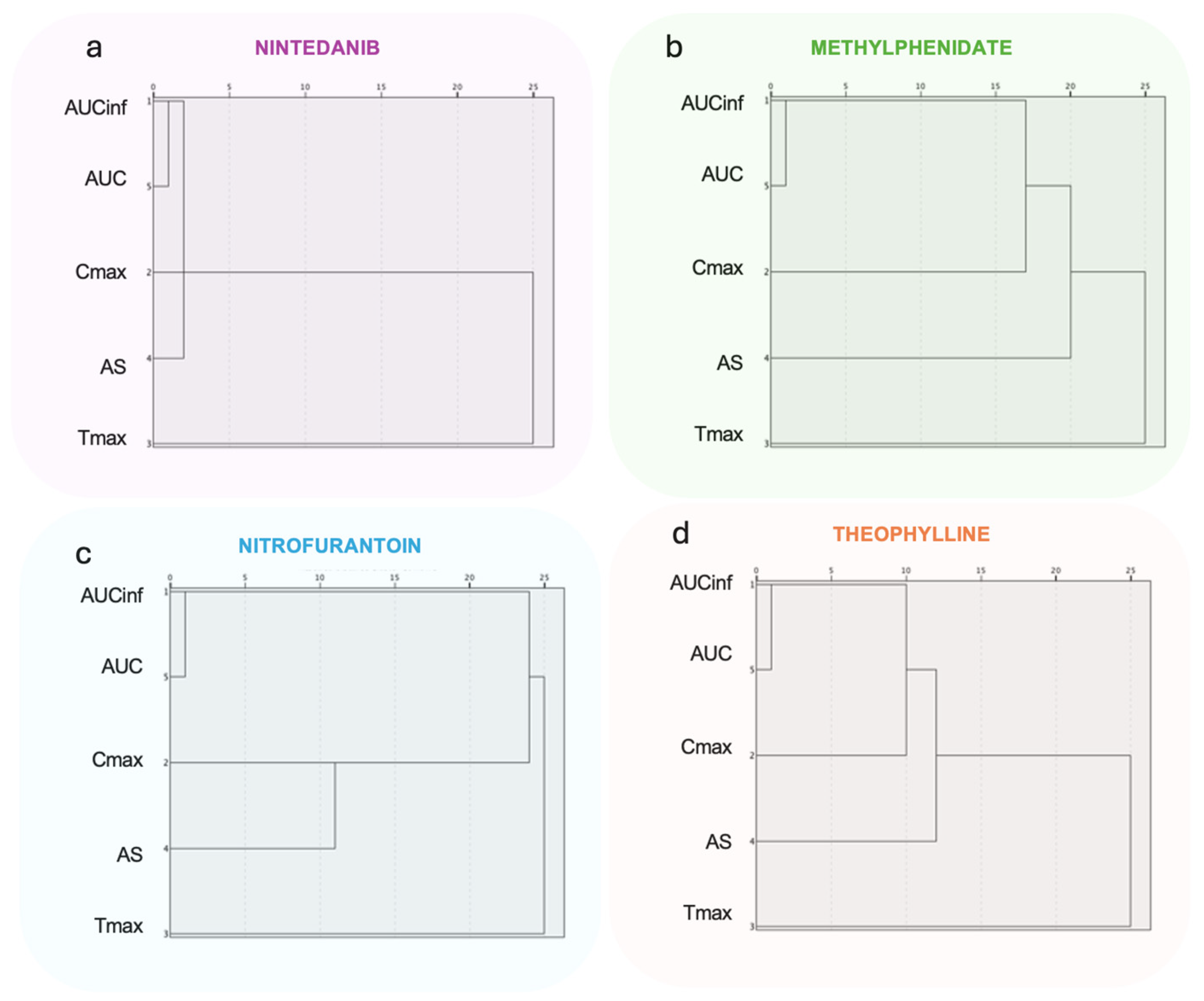
3.2.3. Hierarchical Clustering Analysis
3.2.4. Artificial Neural Networks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- European Medicines Agency; Committee for Medicinal Products for Human Use (CHMP). Guideline on the Investigation of Bioequivalence; London, UK, 20 January 2010. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-bioequivalence-rev1_en.pdf (accessed on 12 June 2024).
- Food and Drug Administration Center for Drug Evaluation and Research (CDER). Guidance for Industry Bioavailability and Bioequivalence Studies Submitted in NDAs or INDs—General Considerations. 2014. Available online: https://www.fda.gov/media/88254/download (accessed on 12 June 2024).
- Niazi, S. Handbook of Bioequivalence Testing (Drugs and the Pharmaceutical Sciences), 2nd ed.; Taylor & Francis Group: Boca Raton, FL, USA, 2014. [Google Scholar]
- Karalis, V. Modeling and Simulation in Bioequivalence. In Modeling in Biopharmaceutics, Pharmacokinetics and Pharmacodynamics. Homogeneous and Heterogeneous Approaches; Springer International Publishing: Cham, Switzerland, 2016; pp. 227–255. [Google Scholar]
- Bois, F.Y.; Tozer, T.N.; Hauck, W.W.; Chen, M.-L.; Patnaik, R.; Williams, R.L. Bioequivalence: Performance of Several Measures of Extent of Absorption. Pharm. Res. 1994, 11, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Reppas, C.; Lacey, L.F.; Keene, O.N.; Macheras, P.; Bye, A. Evaluation of Different Metrics as Indirect Measures of Rate of Drug Absorption from Extended Release Dosage Forms at Steady-State. Pharm. Res. 1995, 12, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-L.; Lesko, L.; Williams, R.L. Measures of Exposure versus Measures of Rate and Extent of Absorption. Clin. Pharmacokinet. 2001, 40, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.J. Determination of in Vivo Bioequivalence. Pharm. Res. 2002, 19, 227–228. [Google Scholar] [CrossRef] [PubMed]
- Endrenyi, L.; Tothfalusi, L. Metrics for the Evaluation of Bioequivalence of Modified-Release Formulations. AAPS J. 2012, 14, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Stier, E.M.; Davit, B.M.; Chandaroy, P.; Chen, M.-L.; Fourie-Zirkelbach, J.; Jackson, A.; Kim, S.; Lionberger, R.; Mehta, M.; Uppoor, R.S.; et al. Use of Partial Area under the Curve Metrics to Assess Bioequivalence of Methylphenidate Multiphasic Modified Release Formulations. AAPS J. 2012, 14, 925–926. [Google Scholar] [CrossRef] [PubMed]
- Rostami-Hodjegan, A.; Jackson, P.R.; Tucker, G.T. Sensitivity of Indirect Metrics for Assessing “Rate” in Bioequivalence Studies—Moving the “Goalposts” or Changing the “Game”. J. Pharm. Sci. 1994, 83, 1554–1557. [Google Scholar] [CrossRef] [PubMed]
- Basson, R.P.; Cerimele, B.J.; DeSante, K.A.; Howey, D.C. Tmax: An Unconfounded Metric for Rate of Absorption in Single Dose Bioequivalence Studies. Pharm. Res. 1996, 13, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-L. An Alternative Approach for Assessment of Rate of Absorption in Bioequivalence Studies. Pharm. Res. 1992, 9, 1380–1385. [Google Scholar] [CrossRef]
- Lacey, L.F.; Keene, O.N.; Duquesnoy, C.; Bye, A. Evaluation of Different Indirect Measures of Rate of Drug Absorption in Comparative Pharmacokinetic Studies. J. Pharm. Sci. 1994, 83, 212–215. [Google Scholar] [CrossRef]
- Endrenyi, L.; Al-Shaikh, P. Sensitive and Specific Determination of the Equivalence of Absorption Rates. Pharm. Res. 1995, 12, 1856–1864. [Google Scholar] [CrossRef] [PubMed]
- Schall, R.; Luus, H.G.; Steinijans, V.W.; Hauschke, D. Choice of Characteristics and Their Bioequivalence Ranges for the Comparison of Absorption Rates of Immediate-Release Drug Formulations. Int. J. Clin. Pharmacol. Ther. 1994, 32, 323–328. [Google Scholar] [PubMed]
- Tothfalusi, L.; Endrenyi, L. Without Extrapolation, Cmax/AUC Is an Effective Metric in Investigations of Bioequivalence. Pharm. Res. 1995, 12, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Vincze, I.; Endrényi, L.; Tóthfalusi, L. Bioequivalence Metrics for Absorption Rates: Linearity, Specificity, Sensitivity. Acta Pharm. Hung. 2019, 89, 17–21. [Google Scholar] [CrossRef]
- Karalis, V.D. On the Interplay between Machine Learning, Population Pharmacokinetics, and Bioequivalence to Introduce Average Slope as a New Measure for Absorption Rate. Appl. Sci. 2023, 13, 2257. [Google Scholar] [CrossRef]
- Karalis, V.D. Machine Learning in Bioequivalence: Towards Identifying an Appropriate Measure of Absorption Rate. Appl. Sci. 2022, 13, 418. [Google Scholar] [CrossRef]
- Karalis, V.D. An in Silico Approach toward the Appropriate Absorption Rate Metric in Bioequivalence. Pharmaceuticals 2023, 16, 725. [Google Scholar] [CrossRef]
- Shamout, F.G.; Zhu, T.; Clifton, D.A. Machine Learning for Clinical Outcome Prediction. IEEE Rev. Biomed. Eng. 2021, 14, 116–126. [Google Scholar] [CrossRef]
- James, G.; Hastie, T.; Tibshirani, R.; Witten, D. An Introduction to Statistical Learning with Applications in R; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Schlender, J.-F.; Meyer, M.; Thelen, K.; Krauss, M.; Willmann, S.; Eissing, T.; Jaehde, U. Development of a Whole-Body Physiologically Based Pharmacokinetic Approach to Assess the Pharmacokinetics of Drugs in Elderly Individuals. Clin. Pharmacokinet. 2016, 55, 1573–1589. [Google Scholar] [CrossRef] [PubMed]
- Watanabe-Uchida, M.; Narukawa, M. Utilization of Population Pharmacokinetics in Drug Development and Provision of the Results to Healthcare Professionals. Int. J. Clin. Pharmacol. Ther. 2017, 55, 25–31. [Google Scholar] [CrossRef]
- Wang, W.-J.; Bansal, A.; Bennette, C.S.; Basu, A. Mimicking Clinical Trials Using Real-World Data: A Novel Method and Applications. Med. Decis. Making 2023, 43, 275–287. [Google Scholar] [CrossRef]
- Zhang, F.; Jia, R.; Gao, H.; Wu, X.; Liu, B.; Wang, H. In Silico Modeling and Simulation to Guide Bioequivalence Testing for Oral Drugs in a Virtual Population. Clin. Pharmacokinet. 2021, 60, 1373–1385. [Google Scholar] [CrossRef]
- European Medicines Agency, Committee for Medicinal Products for Human Use (CHMP). Assessment Report; Of eu International Non-Proprietary Name: Nintedanib. 2014. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/ofev (accessed on 12 June 2024).
- Marzin, K.; Kretschmar, G.; Luedtke, D.; Kraemer, S.; Kuelzer, R.; Schlenker-Herceg, R.; Schmid, U.; Schnell, D.; Dallinger, C. Pharmacokinetics of Nintedanib in Subjects with Hepatic Impairment. J. Clin. Pharmacol. 2018, 58, 357–363. [Google Scholar] [CrossRef]
- Schmid, U.; Doege, C.; Dallinger, C.; Freiwald, M. Population Pharmacokinetics of Nintedanib in Patients with Idiopathic Pulmonary Fibrosis. Pulm. Pharmacol. Ther. 2018, 48, 136–143. [Google Scholar] [CrossRef]
- Wind, S.; Schmid, U.; Freiwald, M.; Marzin, K.; Lotz, R.; Ebner, T.; Stopfer, P.; Dallinger, C. Clinical Pharmacokinetics and Pharmacodynamics of Nintedanib. Clin. Pharmacokinet. 2019, 58, 1131–1147. [Google Scholar] [CrossRef]
- Adjei, A.; Teuscher, N.S.; Kupper, R.J.; Chang, W.-W.; Greenhill, L.; Newcorn, J.H.; Connor, D.F.; Wigal, S. Single-Dose Pharmacokinetics of Methylphenidate Extended-Release Multiple Layer Beads Administered as Intact Capsule or Sprinkles versus Methylphenidate Immediate-Release Tablets (Ritalin®) in Healthy Adult Volunteers. J. Child Adolesc. Psychopharmacol. 2014, 24, 570–578. [Google Scholar] [CrossRef]
- Geneesmiddeleninformatiebank. Public Assessment Report; Medikinet 5/10/20 Mg; Tabletten Medikinet Retard 10/20/30/40 Mg Methylphenidate Hydrochloride. 2006. Available online: https://www.geneesmiddeleninformatiebank.nl/pars/h34030.pdf (accessed on 12 June 2024).
- Gutiérrez-Casares, J.R.; Quintero, J.; Segú-Vergés, C.; Rodríguez Monterde, P.; Pozo-Rubio, T.; Coma, M.; Montoto, C. In Silico Clinical Trial Evaluating Lisdexamfetamine’s and Methylphenidate’s Mechanism of Action Computational Models in an Attention-Deficit/Hyperactivity Disorder Virtual Patients’ Population. Front. Psychiatry 2023, 14, 939650. [Google Scholar] [CrossRef]
- Teuscher, N.; Adjei, A.; Findling, R.; Greenhill, L.; Kupper, R.; Wigal, S. Population Pharmacokinetics of Methylphenidate Hydrochloride Extended-Release Multiple-Layer Beads in Pediatric Subjects with Attention Deficit Hyperactivity Disorder. Drug Des. Devel. Ther. 2015, 9, 2767. [Google Scholar] [CrossRef]
- Lyauk, Y.K.; Stage, C.; Bergmann, T.K.; Ferrero-Milliani, L.; Bjerre, D.; Thomsen, R.; Dalhoff, K.P.; Rasmussen, H.B.; Jürgens, G. Population Pharmacokinetics of Methylphenidate in Healthy Adults Emphasizing Novel and Known Effects of Several Carboxylesterase 1 (CES1) Variants. Clin. Transl. Sci. 2016, 9, 337–345. [Google Scholar] [CrossRef]
- Wijma, R.A.; Huttner, A.; Koch, B.C.P.; Mouton, J.W.; Muller, A.E. Review of the Pharmacokinetic Properties of Nitrofurantoin and Nitroxoline. J. Antimicrob. Chemother. 2018, 73, 2916–2926. [Google Scholar] [CrossRef]
- College ter beoordeling van geneesmiddelen. Public Assessment Report; Scientific Discussion; Nitrofurantoïne 50 Mg and 100 Mg Focus Care, Hard Capsules (Nitrofurantoin as Macrocrystals). 2022. Available online: www.geneesmiddeleninformatiebank.nl/pars/h127575.pdf. (accessed on 12 June 2024).
- Sharma, R.P.; Burgers, E.J.; Beltman, J.B. Development of a Physiologically Based Pharmacokinetic Model for Nitrofurantoin in Rabbits, Rats, and Humans. Pharmaceutics 2023, 15, 2199. [Google Scholar] [CrossRef]
- Food and Drug Administration. CLINICAL REVIEW 2208510; Lisdexamfetamine Dimesylate (Vyvanase) Chewable Tablets. 2009. Available online: https://www.fda.gov/files/drugs/published/208510-Vyvanse-clinical-prea.pdf (accessed on 12 June 2024).
- Läkemedelsverket. Public Assessment Report; Scientific Discussion Elvanse (Lisdexamfetamine Dimesylate); SE/H/1839/01-03. 2019. Available online: https://docetp.mpa.se/LMF/Elvansecapsule,hardENGPAR_09001bee80cde265.pdf (accessed on 12 June 2024).
- AUSPAR. Australian Public Assessment Report for Lisdexamfetamine Dimesilate. 2013. Available online: https://www.tga.gov.au/sites/default/files/auspar-lisdexamfetamine-dimesilate-131023.pdf (accessed on 12 June 2024).
- Comiran, E.; Carlos, G.; Barreto, F.; Pechanksy, F.; Fröehlich, P.E.; Limberger, R.P. Lisdexamfetamine and Amphetamine Pharmacokinetics in Oral Fluid, Plasma, and Urine after Controlled Oral Administration of Lisdexamfetamine. Biopharm. Drug Dispos. 2021, 42, 3–11. [Google Scholar] [CrossRef]
- Ermer, J.; Corcoran, M.; Lasseter, K.; Martin, P.T. Relative Bioavailabilities of Lisdexamfetamine Dimesylate and D-Amphetamine in Healthy Adults in an Open-Label, Randomized, Crossover Study after Mixing Lisdexamfetamine Dimesylate with Food or Drink. Ther. Drug Monit. 2016, 38, 769–776. [Google Scholar] [CrossRef]
- Dolder, P.C.; Strajhar, P.; Vizeli, P.; Hammann, F.; Odermatt, A.; Liechti, M.E. Pharmacokinetics and Pharmacodynamics of Lisdexamfetamine Compared with D-Amphetamine in Healthy Subjects. Front. Pharmacol. 2017, 8, 617. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, L.; Chen, X.; Kang, Y.; Wang, Y.; Wang, L. Population Pharmacokinetics of Theophylline in Adult Chinese Patients with Asthma and Chronic Obstructive Pulmonary Disease. Int. J. Clin. Pharm. 2018, 40, 1010–1018. [Google Scholar] [CrossRef]
- Australian Industrial Chemicals Introduction Scheme. 1H-Purine-2,6-Dione, 3,7-Dihydro-1,3dimethyl (Theophylline) Evaluation Statement. 2022. Available online: https://www.industrialchemicals.gov.au/sites/default/files/2022-06/EVA00032%20-%20Evaluation%20statement%20-%2030%20June%202022.pdf (accessed on 12 June 2024).
- OECD. THEOPHILLINE CAS N°: 58-55-9. 2001. Available online: https://hpvchemicals.oecd.org/UI/handler.axd?id=f9abfd36-c689-46e5-92da-ff5e7a672e00 (accessed on 12 June 2024).
- Zhang, Z.; Castelló, A. Principal Components Analysis in Clinical Studies. Ann. Transl. Med. 2017, 5, 351. [Google Scholar] [CrossRef]
- Yu, W.; Gu, Y.; Li, Y. Efficient Randomized Algorithms for the Fixed-Precision Low-Rank Matrix Approximation. SIAM J. Matrix Anal. Appl. 2018, 39, 1339–1359. [Google Scholar] [CrossRef]
- Wallace, M.L.; Mentch, L.; Wheeler, B.J.; Tapia, A.L.; Richards, M.; Zhou, S.; Yi, L.; Redline, S.; Buysse, D.J. Use and Misuse of Random Forest Variable Importance Metrics in Medicine: Demonstrations through Incident Stroke Prediction. BMC Med. Res. Methodol. 2023, 23, 1–12. [Google Scholar] [CrossRef]
- Zhang, Z.; Murtagh, F.; Van Poucke, S.; Lin, S.; Lan, P. Hierarchical Cluster Analysis in Clinical Research with Heterogeneous Study Population: Highlighting Its Visualization with R. Ann. Transl. Med. 2017, 5, 75. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Abut, S.; Okut, H.; Zackula, R.; James Kallail, K. Deep Neural Networks and Applications in Medical Research. In Artificial Intelligence; IntechOpen: London, UK, 2023. [Google Scholar]
- Karalis, V. A Vector Theory of Assessing Clinical Trials: An Application to Bioequivalence. J. Cardiovasc. Dev. Dis. 2024, 11, 185. [Google Scholar] [CrossRef]

| Medicine Name | Tmax (h) | t1/2 (h) | Sampling Points (h) | References |
|---|---|---|---|---|
| Nintedanib | 2–4 | 10–15 | 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 6, 7, 8, 10, 12, 24 | [28,29,30,31] |
| Methylphenidate | 1–2 | 2–3 | 0, 0.5, 1, 1.5, 2, 2.5, 3, 4, 4.5, 5, 5.5, 6, 6.5, 7, 8, 8.5, 9, 9.5, 10, 10.5, 12, 15, 19, 24 | [32,33,34,35,36] |
| Nitrofurantoin | 2–4 | 0.5–1.3 | 0, 0.5, 1, 1.5, 2, 2.33, 2.67, 3, 3.33, 3.67, 4, 4.5, 5, 6, 8, 12, 24 | [37,38,39] |
| Lisdexamfetamine | 1 | <1 | 0, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, 6, 9, 12, 16, 24 | [40,41,42,43,44,45] |
| Theophylline | 1–2 | 3–9 | 0, 0.33, 0.67, 1, 1.25, 1.55, 1.75, 2, 2.25, 2.50, 2.75, 3, 4, 6, 8, 10, 12, 16, 24, 48, 72 | [46,47,48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokkali, M.; Karalis, V.D. Average Slope vs. Cmax: Which Truly Reflects the Drug-Absorption Rate? Appl. Sci. 2024, 14, 6115. https://doi.org/10.3390/app14146115
Kokkali M, Karalis VD. Average Slope vs. Cmax: Which Truly Reflects the Drug-Absorption Rate? Applied Sciences. 2024; 14(14):6115. https://doi.org/10.3390/app14146115
Chicago/Turabian StyleKokkali, Maria, and Vangelis D. Karalis. 2024. "Average Slope vs. Cmax: Which Truly Reflects the Drug-Absorption Rate?" Applied Sciences 14, no. 14: 6115. https://doi.org/10.3390/app14146115
APA StyleKokkali, M., & Karalis, V. D. (2024). Average Slope vs. Cmax: Which Truly Reflects the Drug-Absorption Rate? Applied Sciences, 14(14), 6115. https://doi.org/10.3390/app14146115







