The Effectiveness of Semi-Supervised Learning Techniques in Identifying Calcifications in X-ray Mammography and the Impact of Different Classification Probabilities
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
3.1. Subjects
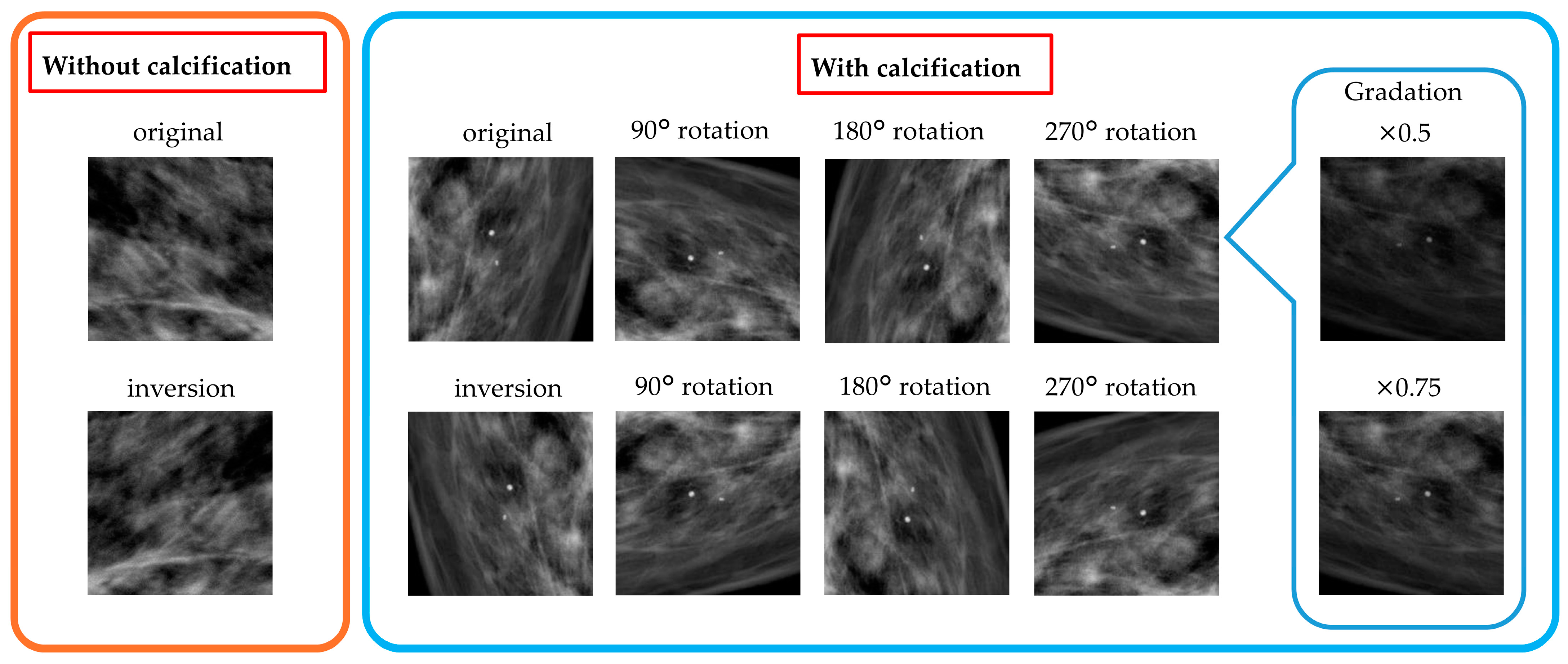
3.2. Data Preprocessing
3.3. Semi-Supervised Learning
3.4. Evaluation of Created Models
3.4.1. Accuracy Evaluation of Each Classifier
- (1)
- Recall
- (2)
- Precision
- (3)
- Overall Accuracy
- (4)
- Area Under the Curve (AUC)
- (5)
- F1 score
3.4.2. Improvement Relative to the Initial Classifier
4. Results and Discussion
4.1. Patches Used for Transfer Learning
4.2. Evaluation of the Accuracy of Each Classifier and Improvement Relative to Initial Classifier
4.3. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Prim. 2019, 5, 66. [Google Scholar] [CrossRef]
- ud din, N.M.; Dar, R.A.; Rasool, M.; Assad, A. Breast Cancer Detection Using Deep Learning: Datasets, Methods, and Challenges Ahead. Comput. Biol. Med. 2022, 149, 106073. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y. A Context-Sensitive Deep Learning Approach for Microcalcification Detection in Mammograms. Pattern Recognit. 2018, 78, 12–22. [Google Scholar] [CrossRef]
- Duffy, S.W.; Yen, A.M.-F.; Tabar, L.; Lin, A.T.-Y.; Chen, S.L.-S.; Hsu, C.-Y.; Dean, P.B.; Smith, R.A.; Chen, T.H.-H. Beneficial Effect of Repeated Participation in Breast Cancer Screening upon Survival. J. Med. Screen. 2023, 31, 3–7. [Google Scholar] [CrossRef]
- Sechopoulos, I.; Teuwen, J.; Mann, R. Artificial Intelligence for Breast Cancer Detection in Mammography and Digital Breast Tomosynthesis: State of the Art. Semin. Cancer Biol. 2021, 72, 214–225. [Google Scholar] [CrossRef]
- Wilkinson, L.; Thomas, V.; Sharma, N. Microcalcification on Mammography: Approaches to Interpretation and Biopsy. Br. J. Radiol. 2017, 90, 20160594. [Google Scholar] [CrossRef]
- Muttarak, M.; Kongmebhol, P.; Sukhamwang, N. Breast Calcifications: Which Are Malignant? Singap. Med. J. 2009, 50, 907–914. [Google Scholar]
- Leong, Y.S.; Hasikin, K.; Lai, K.W.; Mohd Zain, N.; Azizan, M.M. Microcalcification Discrimination in Mammography Using Deep Convolutional Neural Network: Towards Rapid and Early Breast Cancer Diagnosis. Front. Public Health 2022, 10, 875305. [Google Scholar] [CrossRef]
- Demetri-Lewis, A.; Slanetz, P.J.; Eisenberg, R.L. Breast Calcifications: The Focal Group. AJR Am. J. Roentgenol. 2012, 198. [Google Scholar] [CrossRef]
- Szewczuk, M.; Konefał, A. Optimization of Image Quality in Digital Mammography with the Response of a Selenium Detector by Monte Carlo Simulation. Appl. Sci. 2023, 13, 171. [Google Scholar] [CrossRef]
- Kawakami, M.; Hirata, K.; Furuya, S.; Kobayashi, K.; Sugimori, H.; Magota, K.; Katoh, C. Development of Combination Methods for Detecting Malignant Uptakes Based on Physiological Uptake Detection Using Object Detection With PET-CT MIP Images. Front. Med. 2020, 7, 616746. [Google Scholar] [CrossRef]
- Asami, Y.; Yoshimura, T.; Manabe, K.; Yamada, T.; Sugimori, H. Development of Detection and Volumetric Methods for the Triceps of the Lower Leg Using Magnetic Resonance Images with Deep Learning. Appl. Sci. 2021, 11, 12006. [Google Scholar] [CrossRef]
- Manabe, K.; Asami, Y.; Yamada, T.; Sugimori, H. Improvement in the Convolutional Neural Network for Computed Tomography Images. Appl. Sci. 2021, 11, 1505. [Google Scholar] [CrossRef]
- Sugimori, H. Evaluating the Overall Accuracy of Additional Learning and Automatic Classification System for CT Images. Appl. Sci. 2019, 9, 682. [Google Scholar] [CrossRef]
- Chan, H.P.; Samala, R.K.; Hadjiiski, L.M. CAD and AI for Breast Cancer-Recent Development and Challenges. Br. J. Radiol. 2020, 93, 20190580. [Google Scholar] [CrossRef]
- Cai, H.; Huang, Q.; Rong, W.; Song, Y.; Li, J.; Wang, J.; Chen, J.; Li, L. Breast Microcalcification Diagnosis Using Deep Convolutional Neural Network from Digital Mammograms. Comput. Math. Methods Med. 2019, 2019, 2717454. [Google Scholar] [CrossRef]
- Jung, H.; Kim, B.; Lee, I.; Yoo, M.; Lee, J.; Ham, S.; Woo, O.; Kang, J. Detection of Masses in Mammograms Using a One-Stage Object Detector Based on a Deep Convolutional Neural Network. PLoS ONE 2018, 13, e0203355. [Google Scholar] [CrossRef]
- Arefan, D.; Mohamed, A.A.; Berg, W.A.; Zuley, M.L.; Sumkin, J.H.; Wu, S. Deep Learning Modeling Using Normal Mammograms for Predicting Breast Cancer Risk. Med. Phys. 2020, 47, 110–118. [Google Scholar] [CrossRef]
- Baccouche, A.; Garcia-Zapirain, B.; Zheng, Y.; Elmaghraby, A.S. Early Detection and Classification of Abnormality in Prior Mammograms Using Image-to-Image Translation and YOLO Techniques. Comput. Methods Programs Biomed. 2022, 221, 106884. [Google Scholar] [CrossRef]
- Lopez-Almazan, H.; Javier Pérez-Benito, F.; Larroza, A.; Perez-Cortes, J.C.; Pollan, M.; Perez-Gomez, B.; Salas Trejo, D.; Casals, M.; Llobet, R. A Deep Learning Framework to Classify Breast Density with Noisy Labels Regularization. Comput. Methods Programs Biomed. 2022, 221, 106885. [Google Scholar] [CrossRef]
- Sakaida, M.; Yoshimura, T.; Tang, M.; Ichikawa, S. Development of a Mammography Calcification Detection Algorithm Using Deep Learning with Resolution-Preserved Image Patch Division. Algorithms 2023, 16, 483. [Google Scholar] [CrossRef]
- Yan, J.; Wang, X. Unsupervised and Semi-Supervised Learning: The next Frontier in Machine Learning for Plant Systems Biology. Plant J. 2022, 111, 1527–1538. [Google Scholar] [CrossRef]
- Xu, X.; Sanford, T.; Turkbey, B.; Xu, S.; Wood, B.J.; Yan, P. Shadow-Consistent Semi-Supervised Learning for Prostate Ultrasound Segmentation. IEEE Trans. Med. Imaging 2022, 41, 1331–1345. [Google Scholar] [CrossRef]
- Han, C.H.; Kim, M.; Kwak, J.T. Semi-Supervised Learning for an Improved Diagnosis of COVID-19 in CT Images. PLoS ONE 2021, 16, e0249450. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent Advances and Clinical Applications of Deep Learning in Medical Image Analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef]
- Burton, W.; Myers, C.; Rullkoetter, P. Semi-Supervised Learning for Automatic Segmentation of the Knee from MRI with Convolutional Neural Networks. Comput. Methods Programs Biomed. 2020, 189, 105328. [Google Scholar] [CrossRef]
- Calderon-Ramirez, S.; Murillo-Hernandez, D.; Rojas-Salazar, K.; Elizondo, D.; Yang, S.; Moemeni, A.; Molina-Cabello, M. A Real Use Case of Semi-Supervised Learning for Mammogram Classification in a Local Clinic of Costa Rica. Med. Biol. Eng. Comput. 2022, 60, 1159–1175. [Google Scholar] [CrossRef]
- Azary, H.; Abdoos, M. A Semi-Supervised Method for Tumor Segmentation in Mammogram Images. J. Med. Signals Sens. 2020, 10, 12–18. [Google Scholar] [CrossRef]
- Mavroforakis, M.E.; Georgiou, H.V.; Dimitropoulos, N.; Cavouras, D.; Theodoridis, S. Mammographic Masses Characterization Based on Localized Texture and Dataset Fractal Analysis Using Linear, Neural and Support Vector Machine Classifiers. Artif. Intell. Med. 2006, 37, 145–162. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks Mark. arXiv 2019, arXiv:1801.04381. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Al-antari, M.A.; Al-masni, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. A Fully Integrated Computer-Aided Diagnosis System for Digital X-ray Mammograms via Deep Learning Detection, Segmentation, and Classification. Int. J. Med. Inform. 2018, 117, 44–54. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Zhang, Z.; Zhang, X.; Wang, S.H. MIDCAN: A Multiple Input Deep Convolutional Attention Network for Covid-19 Diagnosis Based on Chest CT and Chest X-Ray. Pattern Recognit. Lett. 2021, 150, 8–16. [Google Scholar] [CrossRef]
- Oliver, A.; Odena, A.; Raffel, C.; Cubuk, E.D.; Goodfellow, I.J. Realistic Evaluation of Deep Semi-Supervised Learning Algorithms. In Proceedings of the 32nd International Conference on Neural Information Processing Syste, Montreal, QC, Canada, 2–8 December 2018; pp. 3235–3246. [Google Scholar]
- Cheplygina, V.; de Bruijne, M.; Pluim, J.P.W. Not-so-Supervised: A Survey of Semi-Supervised, Multi-Instance, and Transfer Learning in Medical Image Analysis. Med. Image Anal. 2019, 54, 280–296. [Google Scholar] [CrossRef]
- Watanabe, A.T.; Retson, T.; Wang, J.; Mantey, R.; Chim, C.; Karimabadi, H. Mammographic Breast Density Model Using Semi-Supervised Learning Reduces Inter-/Intra-Reader Variability. Diagnostics 2023, 13, 2694. [Google Scholar] [CrossRef]
- Khaled, R.; Helal, M.; Alfarghaly, O.; Mokhtar, O.; Elkorany, A.; El Kassas, H.; Fahmy, A. Categorized Contrast Enhanced Mammography Dataset for Diagnostic and Artificial Intelligence Research. Sci. Data 2022, 9, 122. [Google Scholar] [CrossRef]
- Altameem, A.; Mahanty, C.; Poonia, R.C.; Saudagar, A.K.J.; Kumar, R. Breast Cancer Detection in Mammography Images Using Deep Convolutional Neural Networks and Fuzzy Ensemble Modeling Techniques. Diagnostics 2022, 12, 1812. [Google Scholar] [CrossRef]
- Marmot, M.G.; Altman, D.G.; Cameron, D.A.; Dewar, J.A.; Thompson, S.G.; Wilcox, M. The Benefits and Harms of Breast Cancer Screening: An Independent Review. Br. J. Cancer 2013, 108, 2205–2240. [Google Scholar] [CrossRef]
- Wanders, J.O.P.; Holland, K.; Veldhuis, W.B.; Mann, R.M.; Pijnappel, R.M.; Peeters, P.H.M.; van Gils, C.H.; Karssemeijer, N. Volumetric Breast Density Affects Performance of Digital Screening Mammography. Breast Cancer Res. Treat. 2017, 162, 95–103. [Google Scholar] [CrossRef]
- Mann, R.M.; Athanasiou, A.; Baltzer, P.A.T.; Camps-Herrero, J.; Clauser, P.; Fallenberg, E.M.; Forrai, G.; Fuchsjäger, M.H.; Helbich, T.H.; Killburn-Toppin, F.; et al. Breast Cancer Screening in Women with Extremely Dense Breasts Recommendations of the European Society of Breast Imaging (EUSOBI). Eur. Radiol. 2022, 32, 4036–4045. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, F.T.H.; van Asselt, A.A.; Dorrius, M.D.; Greuter, M.J.W.; de Bock, G.H. Mammographic Breast Density and the Risk of Breast Cancer: A Systematic Review and Meta-Analysis. Breast 2022, 66, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.M.; Kim, E.H. Breast Density and Risk of Breast Cancer in Asian Women: A Meta-Analysis of Observational Studies. J. Prev. Med. Public Health 2016, 49, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Stanescu, A.; Caragea, D. An Empirical Study of Ensemble-Based Semi-Supervised Learning Approaches for Imbalanced Splice Site Datasets. BMC Syst. Biol. 2015, 9, S1. [Google Scholar] [CrossRef][Green Version]
- Habel, L.A.; Capra, A.M.; Oestreicher, N.; Greendale, G.A.; Cauley, J.A.; Bromberger, J.; Crandall, C.J.; Gold, E.B.; Modugno, F.; Salane, M.; et al. Mammographic Density in a Multiethnic Cohort. Menopause 2007, 14, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Sharma, G. OmniVec: Learning Robust Representations with Cross Modal Sharing. In Proceedings of the 2024 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2024; pp. 1225–1237. [Google Scholar] [CrossRef]
- Sannasi Chakravarthy, S.R.; Bharanidharan, N.; Vinoth Kumar, V.; Mahesh, T.R.; Alqahtani, M.S.; Guluwadi, S. Deep Transfer Learning with Fuzzy Ensemble Approach for the Early Detection of Breast Cancer. BMC Med. Imaging 2024, 24, 82. [Google Scholar] [CrossRef]

| Previous Methods for Mammography | Novelty for This Study |
|---|---|
| -Detection of soft tissue findings | Utilization of semi-supervised learning for mammography |
| -Detection rate of 80–90% [4] | -Reduction of labeling effort |
| -Detection of calcification clusters [8,10] | -Improvement in classification accuracy |
| -Difficulty in detecting microcalcifications [4,9] | -Investigation of classification probability thresholds |
| -Information loss due to compression | -New approach to classification accuracy |
| -High labeling effort | -Combination of patch division and semi-supervised learning |
| Right | Left | |||
|---|---|---|---|---|
| CC a Image | MLO b Image | CC Image | MLO Image | |
| Number of images | 173 | 174 | 183 | 182 |
| Environment | Contents |
|---|---|
| Software | MATLAB 2023a (MathWorks) |
| OS | Windows 11 |
| CPU | Intel Core i9-10920X 3.50 GHz |
| GPU | NVIDIA Quadro P5000 16 GB × 4 |
| Memory | DIMM 2666 MHz 64.0 GB |
| Calcification | Number of Images |
|---|---|
| Yes | 1029 |
| No | 14,020 |
| Training Dataset | Test Dataset | |||
|---|---|---|---|---|
| without Calcification | with Calcification | without Calcification | with Calcification | |
| Number of images | 10,668 | 835 | 3352 | 194 |
| Training (without Calcification) | Training (with Calcification) | |||
|---|---|---|---|---|
| Original Data | Augmented Data | Original Data | Augmented Data | |
| Number of images | 10,668 | 21,336 | 835 | 20,040 |
| Parameters | |
|---|---|
| CNN a | ResNet50 |
| Mini batch size | 128 |
| Max epochs | 10 |
| optimizer | SGDM b |
| Initial learning rate | 0.001 |
| Classification Probability | without Calcification | with Calcification |
|---|---|---|
| 0.80 | 27,526 | 2563 |
| 0.85 | 27,245 | 2444 |
| 0.90 | 26,804 | 2298 |
| 0.95 | 25,960 | 2112 |
| 1.00 | 23,476 | 1754 |
| Classification Probability | without Calcification | with Calcification | Total Number of Images for Transfer Learning |
|---|---|---|---|
| 0.80 | 25,630 | 25,630 | 51,260 |
| 0.85 | 24,440 | 24,440 | 48,880 |
| 0.90 | 22,980 | 22,980 | 45,960 |
| 0.95 | 21,120 | 21,120 | 42,240 |
| 1.00 | 17,540 | 17,540 | 35,080 |
| Classification Probability | Recall (Improvement Ratio) | Precision (Improvement Ratio) | Overall Accuracy (Improvement Ratio) | AUC a (Improvement Ratio) | F1 Score (Improvement Ratio) |
|---|---|---|---|---|---|
| (original ResNet50) | 0.778 | 0.751 | 0.974 | 0.969 | 0.765 |
| 0.80 | 0.866 (111.26%) | 0.636 (84.71%) | 0.966 (99.16%) | 0.974 (100.53%) | 0.734 (95.95%) |
| 0.85 | 0.845 (108.61%) | 0.643 (85.61%) | 0.966 (99.19%) | 0.970 (100.18%) | 0.731 (95.55%) |
| 0.90 | 0.856 (109.93%) | 0.678 (90.19%) | 0.970 (99.59%) | 0.971 (100.28%) | 0.756 (98.92%) |
| 0.95 | 0.856 (109.93%) | 0.675 (89.82%) | 0.970 (99.57%) | 0.976 (100.77%) | 0.755 (98.69%) |
| 1.00 | 0.830 (106.62%) | 0.682 (90.81%) | 0.970 (99.57%) | 0.971 (100.28%) | 0.749 (97.94%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakaida, M.; Yoshimura, T.; Tang, M.; Ichikawa, S.; Sugimori, H.; Hirata, K.; Kudo, K. The Effectiveness of Semi-Supervised Learning Techniques in Identifying Calcifications in X-ray Mammography and the Impact of Different Classification Probabilities. Appl. Sci. 2024, 14, 5968. https://doi.org/10.3390/app14145968
Sakaida M, Yoshimura T, Tang M, Ichikawa S, Sugimori H, Hirata K, Kudo K. The Effectiveness of Semi-Supervised Learning Techniques in Identifying Calcifications in X-ray Mammography and the Impact of Different Classification Probabilities. Applied Sciences. 2024; 14(14):5968. https://doi.org/10.3390/app14145968
Chicago/Turabian StyleSakaida, Miu, Takaaki Yoshimura, Minghui Tang, Shota Ichikawa, Hiroyuki Sugimori, Kenji Hirata, and Kohsuke Kudo. 2024. "The Effectiveness of Semi-Supervised Learning Techniques in Identifying Calcifications in X-ray Mammography and the Impact of Different Classification Probabilities" Applied Sciences 14, no. 14: 5968. https://doi.org/10.3390/app14145968
APA StyleSakaida, M., Yoshimura, T., Tang, M., Ichikawa, S., Sugimori, H., Hirata, K., & Kudo, K. (2024). The Effectiveness of Semi-Supervised Learning Techniques in Identifying Calcifications in X-ray Mammography and the Impact of Different Classification Probabilities. Applied Sciences, 14(14), 5968. https://doi.org/10.3390/app14145968






