Deep-Learning Algorithms for Prescribing Insoles to Patients with Foot Pain
Abstract
1. Introduction
2. Methods
2.1. Study Design and Population
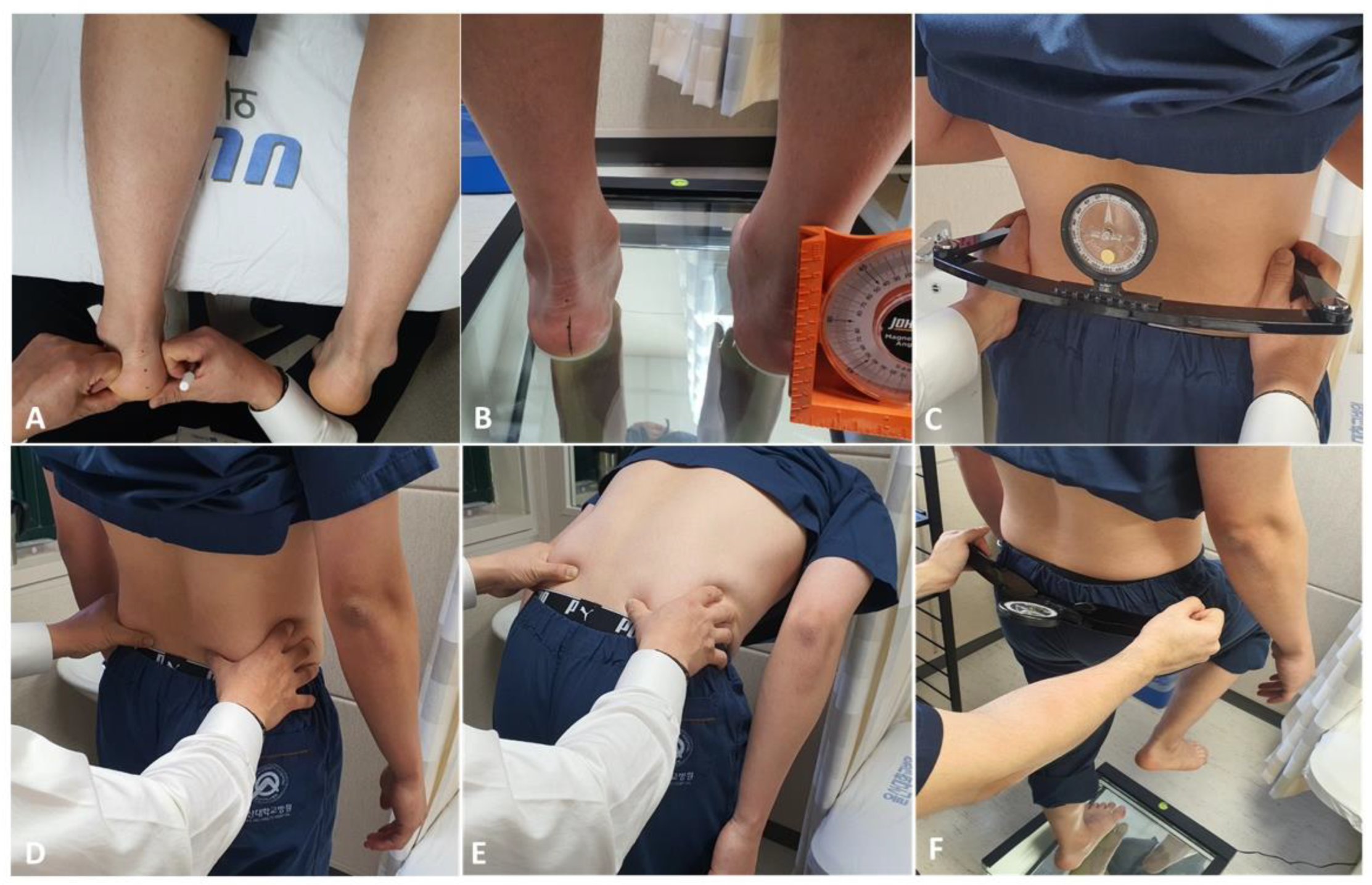
2.2. Input Variables
Resting Calcaneal Stance Position
2.3. Pelvic Elevation, Pelvic Tilt, and Pelvic Rotation
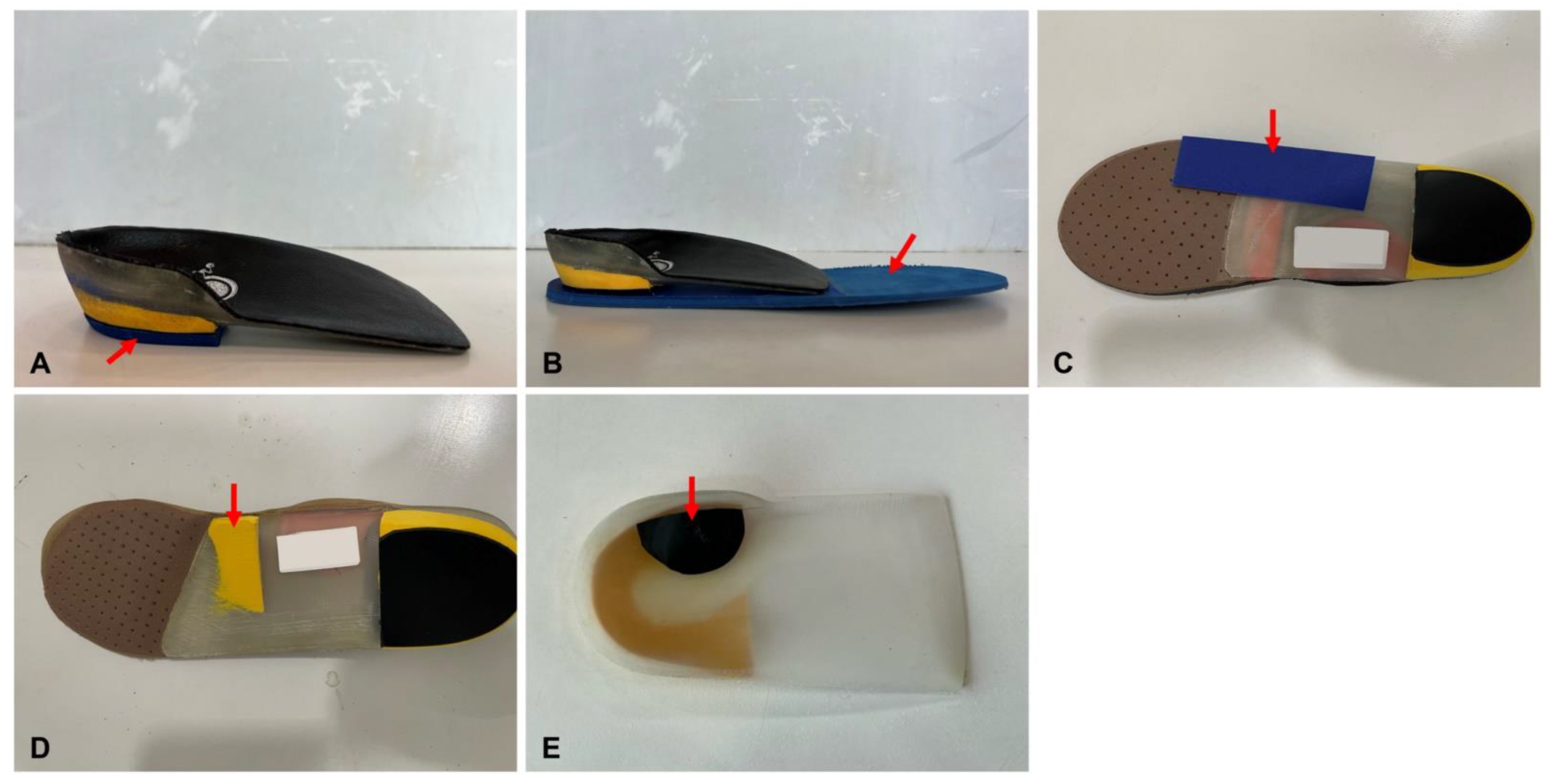
2.4. Target Variables
2.5. Deep-Learning Algorithms
3. Statistical Analysis
4. Results
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Choo, Y.J.; Park, C.H.; Chang, M.C. Rearfoot disorders and conservative treatment: A narrative review. Ann. Palliat. Med. 2020, 9, 3546–3552. [Google Scholar]
- Park, C.H.; Chang, M.C. Forefoot disorders and conservative treatment. Yeungnam Univ. J. Med. 2019, 36, 92–98. [Google Scholar] [CrossRef]
- Alqahtani, T.A. The prevalence of foot pain and its associated factors among Saudi school teachers in Abha sector, Saudi Arabia. Family Med Prim Care. 2020, 9, 4641–4647. [Google Scholar]
- Chang, M.C. The blind spot and challenges in pain management. J. Yeungnam Med. Sci. 2022, 39, 179–180. [Google Scholar]
- Park, C.H.; Boudier-Revéret, M.; Chang, M.C. Tarsal tunnel syndrome due to talocalcaneal coalition. Yeungnam Univ. J. Med. 2023, 40, 106–108. [Google Scholar] [CrossRef]
- Chow, T.H.; Chen, Y.S.; Hsu, C.C. Relationships between Plantar Pressure Distribution and Rearfoot Alignment in the Taiwanese College Athletes with Plantar Fasciopathy during Static Standing and Walking. Int. J. Environ. Res. Public Health 2021, 18, 12942. [Google Scholar]
- Menz, H.B.; Dufour, A.B.; Riskowski, J.L.; Hillstrom, H.J.; Hannan, M.T. Association of planus foot posture and pronated foot function are associated with foot pain: The Framingham foot study. Arthritis Care Res. 2013, 65, 1991–1999. [Google Scholar]
- Elattar, O.; Smith, T.; Ferguson, A.; Farber, D.; Wapner, K. Uses of Braces and Orthotics for Conservative Management of Foot and Ankle Disorders. Foot Ankle Orthop. 2018, 3, 2473011418780700. [Google Scholar] [CrossRef]
- Razeghi, M.; Batt, M.E. Biomechanical analysis of the effect of orthotic shoe inserts: A review of the literature. Sports Med. 2000, 29, 425–438. [Google Scholar] [CrossRef]
- Amer, A.O.; Jarl, G.M.; Hermansson, L.N. The effect of insoles on foot pain and daily activities. Prosthet. Orthot. Int. 2014, 38, 474–480. [Google Scholar]
- Gómez Carrión, Á.; Atín Arratibe, M.L.Á.; Morales Lozano, M.R.; Martínez Rincón, C.; Martínez Sebastián, C.; Saura Sempere, Á.; Nuñez-Fernandez, A.; Sánchez-Gómez, R. Changes in the Kinematics of Midfoot and Rearfoot Joints with the Use of Lateral Wedge Insoles. J. Clin. Med. 2022, 11, 4536. [Google Scholar]
- Paterson, K.L.; Hinman, R.S.; Metcalf, B.R.; McManus, F.; Jones, S.E.; Menz, H.B.; Munteanu, S.E.; Bennell, K.L. Effect of foot orthoses vs sham insoles on first metatarsophalangeal joint osteoarthritis symptoms: A randomized controlled trial. Osteoarthr. Cartil. 2022, 30, 956–964. [Google Scholar]
- Khamis, S.; Dar, G.; Peretz, C.; Yizhar, Z. The Relationship Between Foot and Pelvic Alignment While Standing. J. Hum. Kinet. 2015, 46, 85–97. [Google Scholar] [CrossRef]
- Fukuchi, C.A.; Lewinson, R.T.; Worobets, J.T.; Stefanyshyn, D.J. Effects of Lateral and Medial Wedged Insoles on Knee and Ankle Internal Joint Moments During Walking in Healthy Men. J. Am. Podiatr. Med. Assoc. 2016, 106, 411–418. [Google Scholar] [CrossRef]
- Kerrigan, D.C.; Lelas, J.L.; Goggins, J.; Merriman, G.J.; Kaplan, R.J.; Felson, D.T. Effectiveness of a lateral-wedge insole on knee varus torque in patients with knee osteoarthritis. Arch. Phys. Med. Rehabil. 2002, 83, 889–893. [Google Scholar]
- Mahmoud, A.; Abundo, P.; Basile, L.; Albensi, C.; Marasco, M.; Bellizzi, L.; Galasso, F.; Foti, C. Functional leg length discrepancy between theories and reliable instrumental assessment: A study about newly invented NPoS system. Muscles Ligaments Tendons J. 2017, 7, 293–305. [Google Scholar]
- Choo, Y.J.; Kim, J.K.; Kim, J.H.; Chang, M.C.; Park, D. Machine learning analysis to predict the need for ankle foot orthosis in patients with stroke. Sci. Rep. 2021, 11, 8499. [Google Scholar] [CrossRef]
- Kim, J.K.; Choo, Y.J.; Chang, M.C. Prediction of Motor Function in Stroke Patients Using Machine Learning Algorithm: Development of Practical Models. J. Stroke Cerebrovasc. Dis. 2021, 30, 105856. [Google Scholar]
- Kim, J.K.; Lv, Z.; Park, D.; Chang, M.C. Practical Machine Learning Model to Predict the Recovery of Motor Function in Patients with Stroke. Eur. Neurol. 2022, 85, 273–279. [Google Scholar]
- Kim, J.K.; Chang, M.C.; Park, D. Deep Learning Algorithm Trained on Brain Magnetic Resonance Images and Clinical Data to Predict Motor Outcomes of Patients With Corona Radiata Infarct. Front. Neurosci. 2022, 15, 795553. [Google Scholar]
- Shin, H.; Choi, G.S.; Shon, O.J.; Kim, G.B.; Chang, M.C. Development of convolutional neural network model for diagnosing meniscus tear using magnetic resonance image. BMC Musculoskelet. Disord. 2022, 23, 510. [Google Scholar] [CrossRef]
- Cho, Y.; Park, J.W.; Nam, K. The relationship between foot posture index and resting calcaneal stance position in elementary school students. Gait Posture 2019, 74, 142–147. [Google Scholar] [CrossRef]
- Keenan, A.M.; Bach, T.M. Clinicians’ assessment of the hindfoot: A study of reliability. Foot Ankle Int. 2006, 27, 451–460. [Google Scholar] [CrossRef]
- Kouhkan, S.; Rahimi, A.; Ghasemi, M.; Naimi, S.S.; Baghban, A.A. Postural Changes during First Pregnancy. Br. J. Med. Med. Res. 2015, 7, 744–753. [Google Scholar] [CrossRef]
- Wang, S.; Minku, L.L. AUC estimation and concept drift detection for imbalanced data streams with multiple classes. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–8. [Google Scholar]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef]
- Buchanan, K.R.; Davis, I. The relationship between forefoot, midfoot, and rearfoot static alignment in pain-free individuals. J. Orthop. Sports Phys. Ther. 2005, 35, 559–566. [Google Scholar]
- Kim, P.J.; Peace, R.; Mieras, J.; Thoms, T.; Freeman, D.; Page, J. Interrater and intrarater reliability in the measurement of ankle joint dorsiflexion is independent of examiner experience and technique used. J. Am. Podiatr. Med. Assoc. 2011, 101, 407–414. [Google Scholar]
- Menz, H.B. Foot orthoses: How much customisation is necessary? J Foot Ankle Res. 2009, 2, 23. [Google Scholar] [CrossRef]
- Severin, A.C.; Gean, R.P.; Barnes, S.G.; Queen, R.; Butler, R.J.; Martin, R.; Barnes, C.L.; Mannen, E.M. Effects of a corrective heel lift with an orthopaedic walking boot on joint mechanics and symmetry during gait. Gait Posture 2019, 73, 233–238. [Google Scholar]
- Yen, S.T.; Andrew, P.D.; Cummings, G.S. Short-term effect of correcting leg length discrepancy on performance of a forceful body extension task in young adults. Hiroshima J. Med. Sci. 1998, 47, 139–143. [Google Scholar]
- Kriegeskorte, N.; Golan, T. Neural network models and deep learning. Curr. Biol. 2019, 29, R231–R236. [Google Scholar]
- Sarker, I.H. Deep learning: A comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar]
- Rajula, H.S.; Verlato, G.; Manchia, M.; Antonucci, N.; Fanos, V. Comparison of conventional statistical methods with machine learning in medicine: Diagnosis, drug development, and treatment. Medicina 2020, 56, 455. [Google Scholar] [CrossRef]
- Choo, Y.J.; Chang, M.C. Use of Machine Learning in Stroke Rehabilitation: A Narrative Review. Brain Neurorehabil. 2022, 15, e26. [Google Scholar]
- Lee, H.; Song, J. Introduction to convolutional neural network using Keras; an understanding from a statistician. Commun. Stat. Appl. Methods 2019, 26, 591–610. [Google Scholar] [CrossRef]
- Giulia, A.; Anna, S.; Antonia, B.; Dario, P.; Maurizio, C. Extending association rule mining to microbiome pattern analysis: Tools and guidelines to support real applications. Front. Bioinform. 2022, 1, 794547. [Google Scholar] [CrossRef]
- Mendez, K.M.; Pritchard, L.; Reinke, S.N.; Broadhurst, D.I. Toward collaborative open data science in metabolomics using Jupyter Notebooks and cloud computing. Metabolomics 2019, 15, 125. [Google Scholar] [CrossRef]
| Prescription Left DNN Regression Model | Prescription Right DNN Regression Model | |
|---|---|---|
| DNN model | - Four hidden layers with 256-128-128-64 neurons - RMSProp optimizer, ReLU activation - Learning rate 1 × 10−5, batch size 512 - Batch normalization for regularization | - Five hidden layers with 512-512-1024-1024-512 neurons - RMSProp optimizer, ReLU activation - Learning rate 2 × 10−3, batch size 512 - Batch normalization for regularization |
| Model performance | - MAE 1.460, RMSE 3.539 for training - MAE 1.408, RMSE 3.365 for validation | - MAE 1.560, RMSE 3.860 for training - MAE 1.601, RMSE 3.549 for validation |
| Sample size and ratio Sample class size and ratio | - 70% for training: 586; 30% for validation: 252; total: 838 - Class 0: 392 (66.9%), class 1: 64 (10.9%), class 2: 130 (22.2%) for training - Class 0: 169 (67.1%), class 1: 28 (11.1%), class 2: 55 (21.8%) for validation | |||||
| DNN model | - Five hidden layers with 512-512-1024-1024-512 neurons - Adam optimizer, ReLU activation - Learning rate 1 × 10−2 batch size 32 - Dropout layer for regularization - Training accuracy: 89.1%, validation accuracy: 89.7% | |||||
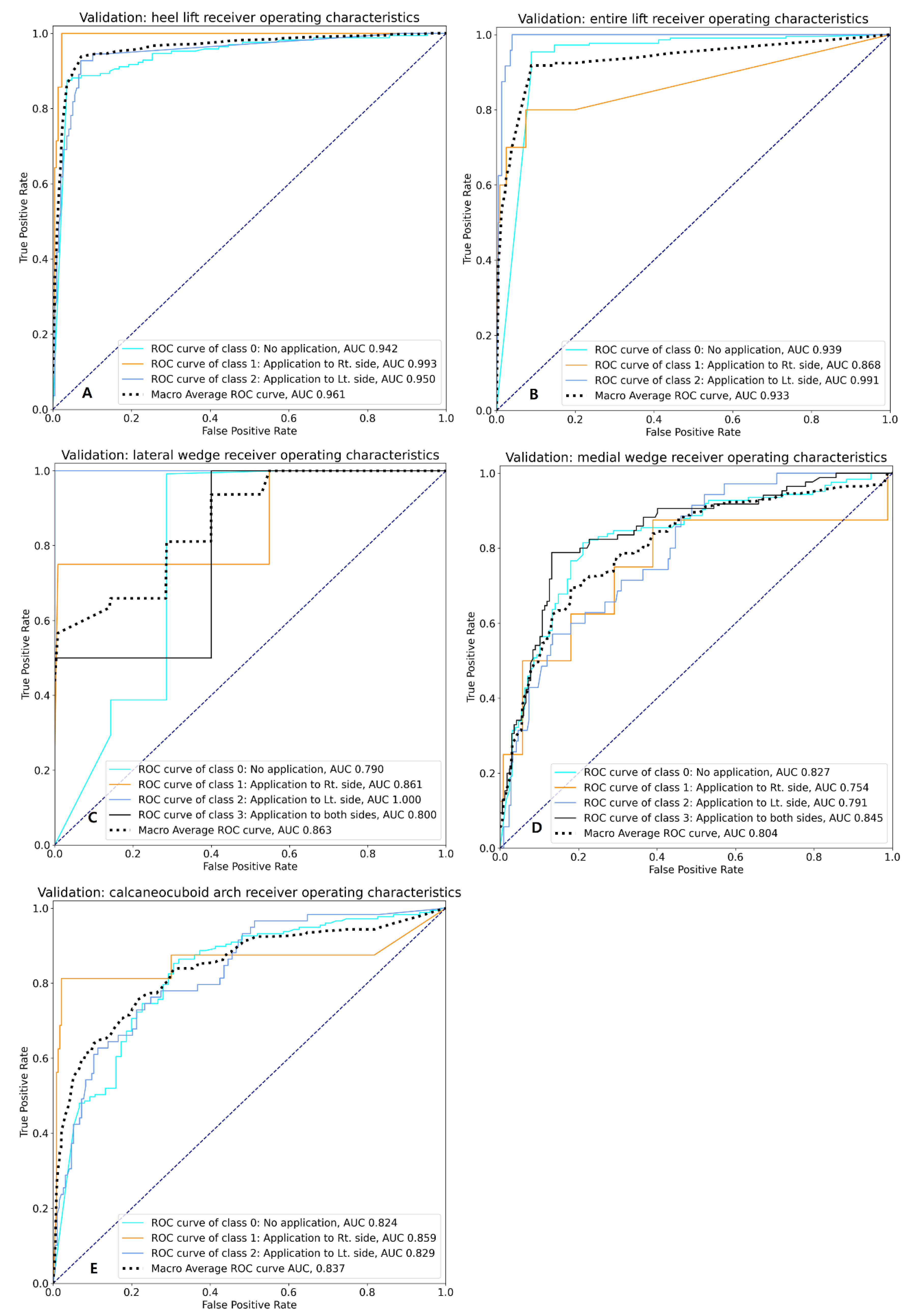
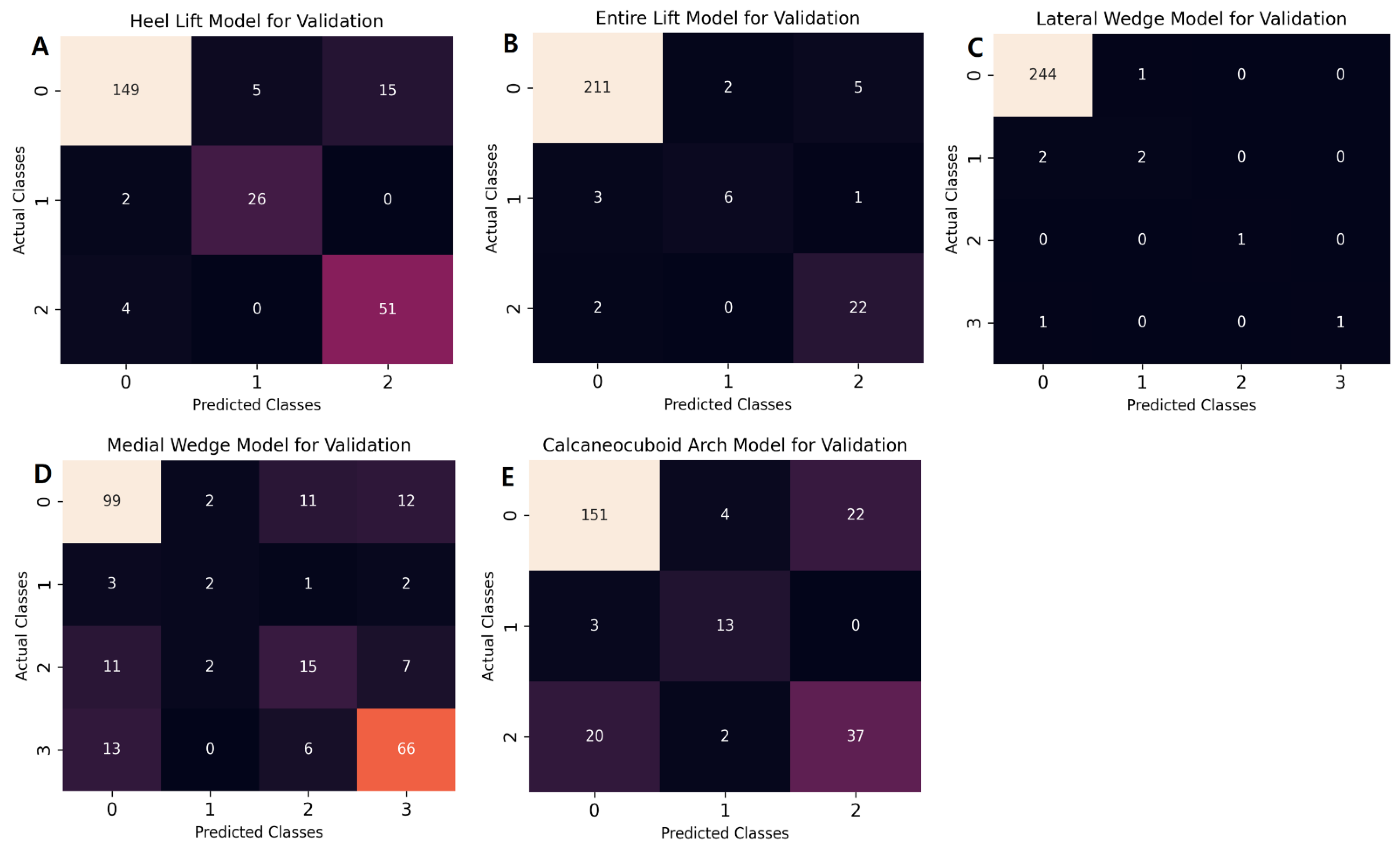
| Model performance (validation data) | Class | Precision | Recall | F1-score | Support | ROC AUC |
| 0 | 0.961 | 0.882 | 0.920 | 169 | 0.942 | |
| 1 | 0.839 | 0.939 | 0.881 | 28 | 0.993 | |
| 2 | 0.773 | 0.927 | 0.843 | 55 | 0.950 | |
| Macro average | 0.858 | 0.913 | 0.881 | 252 | 0.961 | |
| Micro average | 0.907 | 0.897 | 0.899 | 252 | 0.949 | |
| Sample size and ratio Sample class size and ratio | - 70% for training: 586; 30% for validation: 252; total: 838 - Class 0: 508 (86.7%), class 1: 23 (3.9%), class 2: 55 (9.4%) for training - Class 0: 218 (86.5%), class 1: 10 (4%), class 2: 24 (9.5%) for validation | |||||
| DNN model | - Three hidden layers with 256-256-512 neurons - RMSProp optimizer, ReLU activation - Learning rate 5 × 10−3, batch size 2 - Dropout layer for regularization - Training accuracy: 94.7%, validation accuracy: 94.8% | |||||
| Model performance (validation data) | Class | Precision | Recall | F1-score | Support | ROC AUC |
| 0 | 0.977 | 0.968 | 0.972 | 218 | 0.939 | |
| 1 | 0.750 | 0.600 | 0.667 | 10 | 0.868 | |
| 2 | 0.786 | 0.917 | 0.846 | 24 | 0.991 | |
| Macro average | 0.838 | 0.828 | 0.828 | 252 | 0.933 | |
| Micro average | 0.950 | 0.948 | 0.948 | 252 | 0.941 | |
| Sample size and ratio Sample class size and ratio | - 70% for training: 586; 30% for validation: 252; total: 838 - Class 0: 571 (97.4%), class 1: 9 (0.015%), class 2: 3 (0.005%), class 3: 3 (0.005%) for training - Class 0: 245 (97.2%), class 1: 4 (0.016%), class 2: 1 (0.004%), class 3: 2 (0.008%) for validation | |||||
| DNN model | - Two hidden layers with 256-1024 neurons - RMSProp optimizer, ReLU activation - Learning rate 5 × 10−4, batch size 128 - Dropout layer for regularization - Training accuracy: 98.8%, validation accuracy: 98.4% | |||||
| Model performance (validation data) | Class | Precision | Recall | F1-score | Support | ROC AUC |
| 0 | 0.988 | 0.996 | 0.992 | 245 | 0.790 | |
| 1 | 0.667 | 0.500 | 0.571 | 4 | 0.861 | |
| 2 | 1.000 | 1.000 | 1.000 | 1 | 1.000 | |
| 3 | 1.000 | 0.500 | 0.667 | 2 | 0.800 | |
| Macro average | 0.914 | 0.749 | 0.807 | 252 | 0.863 | |
| Micro average | 0.983 | 0.984 | 0.983 | 252 | 0.792 | |
| Sample size and ratio Sample class size and ratio | - 70% for training: 586; 30% for validation: 252; total: 838 - Class 0: 289 (49.3%), class 1: 19 (3.2%), class 2: 80 (13.7%), class 3: 198 (33.8%) for training - Class 0: 124 (49.2%), class 1: 8 (3.2%), class 2: 35 (13.9%), class 3: 85 (33.7%) for validation | |||||
| DNN model | - Two hidden layers with 256-1024 neurons - Nadam optimizer, ReLU activation - Learning rate 5 × 10−5, batch size 64 - Dropout layer for regularization - Training accuracy: 93.0%, validation accuracy: 72.2% | |||||
| Model performance (validation data) | Class | Precision | Recall | F1-score | Support | ROC AUC |
| 0 | 0.786 | 0.798 | 0.792 | 124 | 0.827 | |
| 1 | 0.333 | 0.250 | 0.285 | 8 | 0.754 | |
| 2 | 0.455 | 0.429 | 0.441 | 35 | 0.791 | |
| 3 | 0.759 | 0.776 | 0.767 | 85 | 0.845 | |
| Macro average | 0.583 | 0.583 | 0.572 | 252 | 0.804 | |
| Micro average | 0.716 | 0.722 | 0.719 | 252 | 0.826 | |
| Sample size and ratio Sample class size and ratio | - 70% for training: 586; 30% for validation: 252; total: 838 - Class 0: 412 (70.3%), class 1: 36 (6.1%), class 2: 138 (23.6%) for training - Class 0: 177 (70.2%), class 1: 16 (6.4%), class 2: 59 (23.4%) for validation | |||||
| DNN model | - Four hidden layers with 1024-512-256-128 neurons - RMSProp optimizer, ReLU activation - Learning rate 2 × 10−3, batch size 128 - Dropout layer for regularization - Training accuracy: 88.7%, validation accuracy: 79.8% | |||||
| Model performance (validation data) | Class | Precision | Recall | F1-score | Support | ROC AUC |
| 0 | 0.868 | 0.853 | 0.860 | 177 | 0.824 | |
| 1 | 0.684 | 0.812 | 0.743 | 16 | 0.859 | |
| 2 | 0.627 | 0.627 | 0.627 | 59 | 0.828 | |
| Macro average | 0.726 | 0.764 | 0.743 | 252 | 0.837 | |
| Micro average | 0.800 | 0.798 | 0.798 | 252 | 0.827 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.K.; Choo, Y.J.; Park, I.S.; Choi, J.-W.; Park, D.; Chang, M.C. Deep-Learning Algorithms for Prescribing Insoles to Patients with Foot Pain. Appl. Sci. 2023, 13, 2208. https://doi.org/10.3390/app13042208
Kim JK, Choo YJ, Park IS, Choi J-W, Park D, Chang MC. Deep-Learning Algorithms for Prescribing Insoles to Patients with Foot Pain. Applied Sciences. 2023; 13(4):2208. https://doi.org/10.3390/app13042208
Chicago/Turabian StyleKim, Jeoung Kun, Yoo Jin Choo, In Sik Park, Jin-Woo Choi, Donghwi Park, and Min Cheol Chang. 2023. "Deep-Learning Algorithms for Prescribing Insoles to Patients with Foot Pain" Applied Sciences 13, no. 4: 2208. https://doi.org/10.3390/app13042208
APA StyleKim, J. K., Choo, Y. J., Park, I. S., Choi, J.-W., Park, D., & Chang, M. C. (2023). Deep-Learning Algorithms for Prescribing Insoles to Patients with Foot Pain. Applied Sciences, 13(4), 2208. https://doi.org/10.3390/app13042208








