Abstract
Tolerance to harsh environmental conditions, high growth rates and an amino acid profile adequate for human consumption are beneficial features observed in Dunaliella viridis TAV01, a novel strain isolated from a salt pond in the Algarve, Portugal. TAV01 was identified down to the species level by maximum likelihood and Bayesian phylogenetic analyses of the ribosomal internal transcribed spacers one and two regions (ITS1 and ITS-2) and was supported by ITS2 secondary structure analysis. The biochemical profile revealed high protein (35.7 g 100 g−1 DW; 65% higher than the minimum recommended by the World Health Organization) and lipid contents (21.3 g 100 g−1 DW), a relatively higher proportion of the polyunsaturated fatty acids (PUFAs), α-linolenic (26.3% of total fatty acids (TFA)) and linoleic acids (22.8% of TFA), compared to those of other Dunaliella strains, and a balanced essential amino acids profile containing significant levels of leucine, phenylalanine, valine, and threonine. The major carotenoid was lutein, making up over 85% of total carotenoids. The presence of high-quality natural products in D. viridis TAV01 offers the possibility of using this new strain as a valuable biological resource for novel feed or food products as ingredients or supplements.
1. Introduction
Microalgae are attractive bioresources for several sectors, namely, food, cosmetics, pharmaceutical, and biofuels [1,2]. Nevertheless, microalgae production faces diverse challenges that influence the economic feasibility of the process, including the need for sustainable production, isolation of new strains, and nutrient sourcing [3]. Current commercial species of microalgae are but the “tip of the iceberg” of microalgae’s immense biodiversity, some displaying biochemical profiles suitable for developing innovative health, cosmetic, feed and food products [4,5,6].
The world population is set to reach 10 billion around 2050 (UN, 2019). This perspective suggests that the current capacity to generate food might not be enough to feed the whole human population. In addition, consumption preference trends suggest that sustainable food products might be increasingly demanded by consumers [7]. Meat analogues, particularly those made of microalgal biomass, present themselves as a solution for protein scarcity. In the confection of such analogues, microalgal biomass can be used as a more sustainable and healthier protein source as compared to meat because microalgae are at the bottom of the food webs, decreasing the amount of energy losses, as it avoids the penalties of metabolite conversion upon consumption by an animal feeding on photosynthetic organisms [8]. Moreover, algae, unlike soy and other vegetables rich in protein, can be produced all year round and do not depend on arable land [9]. As they are able to grow in wastewater, microalgae not only can bioremediate these effluents but also they can produce biomass without further addition of fertilizers [10]. Besides a high protein content, microalgae contain other health-linked compounds such as polyunsaturated fatty acids, lipids and pigments [11], which, together with its environmental sustainability, might provide better acceptance by consumers aiding its introduction in the food markets [8]. The market value of microalgae in Europe up to 2020 was around EUR 31,641 k and is expected to increase further in the coming years [12].
Bioprospecting novel microalgal strains combined with their application to a biorefinery concept can be a powerful tool in biotechnology, as the development of multiple co-products from a single microalgal strain allows the improvement of the overall process [3,13].
The genus Dunaliella (Chlorophyta, Chlorophyceae, Chlamydomonadales, Dunaliellaceae), established by Teodoresco (1905), is known for its high adaptability to harsh environments and halotolerance. Indeed, Dunaliella spp. can grow at high salinity levels, withstanding challenging thermal and nutritional conditions combined with high light stress, often lethal to other microalgal species. Interestingly, the pleiotropic effect of salt stress on the microalgal cell has been used as an efficient approach to induce proteins and lipids accumulation in various target species. Because of this robustness, the industrial cultivation of Dunaliella microalgae is relatively easy compared to other species that cannot grow under extreme environments [14]. These features are important since cultivation conditions significantly impact the economic aspect of the whole industrial cultivation process. Firstly, they promote important changes in the biochemistry and physiology of microalgae, leading to changes in the quantity and quality of the produced biocompounds [15]. This might be due to the fact that salinity is associated with three types of stress: ionic, osmotic, and oxidative stress [16,17]. Secondly, high salinity can also be used to limit culture contamination, which is essential for microalgae cultivation at an industrial scale [14,18,19,20,21,22,23]. Therefore, finding microalgal species resistant to these environmental conditions is essential for large-scale production, with salt stress being an auxiliary tool for culture management in open-air systems.
In the present study, the isolation and identification of Dunaliella sp. TAV01 collected from the Algarve coast (Portugal) is reported. Because the identification of Dunaliella down to the species level is notoriously difficult and fraught with errors [24], this strain was identified by means of phylogenetic analyses of 18S ribosomal DNA (rDNA) and internal transcribed spacers ITS1 and ITS2. Furthermore, ITS2 secondary structure analysis was also performed. This combined analysis improves the taxonomical assignment of novel Dunaliella strains and confirms the identification obtained from primary sequences [25,26]. Finally, the biochemical profile of a high-saline culture was determined, revealing high-value compounds, such as essential fatty acids and lutein, and high protein content with a well-balanced amino acids profile.
2. Materials and Methods
2.1. Isolation and Cultivation Conditions
Water samples were collected from the Algarve Coast, Portugal (37°07′15″ N, 7°38′42″ W). Isolated cells of Dunaliella sp. (named TAV01) were obtained by serial dilution in liquid medium and later transferred to 50 mL test tubes containing sterile seawater and concentrated modified algal medium (MAM; 1000:1 respectively) [27], which was the same medium used in the strain isolation step. The isolate was kept in a climate chamber (Panasonic, MLR-352-PE, Osaka, Japan) at 22 °C, with constant aeration and light (100 μmol photons m−2 s−1). Culture salinity was adjusted to 3 M, a concentration 5 times higher than that of the Mediterranean sea [28]. Cultivation was performed in 5 L flasks until the stationary growth phase was reached. Biomass was harvested by centrifugation at 2000× g for 10 min (Beckman Coulter Avanti J-25 High-Performance), freeze-dried, and stored at −20 °C for subsequent analysis.
2.2. Identification
The initial molecular characterization was performed by 18S rDNA sequencing. The DNA extraction of TAV01 was carried out using an EZNA DNA plant extraction kit (Omega Bio-Tek, Norcross, GA, USA) and amplified by PCR using universal 18S primers (Table 1) [29]. Moreover, primers for ITS1 and ITS2, whose amplicon also contained the 5.8S gene (ITS1-5.8S-ITS2), were designed, and the corresponding regions amplified and sequenced to identify the isolate down to the species level. The purified PCR product was sequenced in-house by an Applied Biosystems 3130XL DNA sequencer (Life Technologies BV, Porto, Portugal). The sequence was deposited into GenBank with the accession number (OQ184858).

Table 1.
Primers pairs and sequences. F: Forward, R: Reverse, Tm: Melting temperature, GC: Guanine/Cytosine content.
The sequences were visualized and curated using CLC Sequence Viewer (v.8.0) and compared with the GenBank database using BLAST (https://blast.ncbi.nlm.nih.gov/ accessed on 10 March 2020). Phylogenetic inference was developed by matching the similarity with Dunaliella species using methods of PhyML v. 3.0 [30] with approximate likelihood-ratio test (SH-Like) [31] and bootstrapping as well as Bayesian inference posterior probabilities using MrBayes 3.2.6 [32] by means of a likelihood model using 6(GTR) for the number of substitution types and invariable + gamma to estimate the rates of variation across sites. For the Markov chain Monte Carlo parameters, 100,000 generations were selected with a tree sample every 10 generations and a burn-in value to discard the first 250 sampled trees.
For further understanding of the isolated strain and its taxonomical classification, the secondary structure of ITS2 was modelled using 4Sale software package [33], whose output was confirmed on the RNAfold web server. With this known structure, a specific BLAST search for the ITS2 secondary structure was developed using the ITS2 ribosomal RNA database [34]
2.3. Proximate Composition
The freeze-dried biomass of TAV01 was used to determine the proximate composition. Ash content was determined by incineration of the samples at 550 °C for 8 h, in a muffle furnace until constant weight [35]. Total protein content was analysed using an elemental analyser, model Vario III (Vario EL, Elementar Analysensysteme GmbH, Langenselbold, Germany) according to the manufacturer’s procedure. A nitrogen-to-protein conversion factor (N-Prot factor) of 4.78 was used to obtain total protein content [36]. Total dietary fibre was determined according to the AACC method 32-05.01 and AOAC method 985.29 using an enzyme kit provided by Megazyme (Megazyme International Ireland Limited, Bray, Ireland) [37]. Total lipids content was determined by a modified Bligh and Dyer (1959) method [27]. Briefly, a mixture of methanol, chloroform, and distilled water (2:2:1, v/v/v) was added to the biomass and homogenized with an Ultra-Turrax disperser (IKA-Werke GmbH Staufen, Staufen im Breisgau, Germany). The extracts were centrifuged to achieve phase separation, and a known volume of the organic phase (chloroform) was transferred to pre-weighed tubes and evaporated overnight in a dry bath (60 °C). Total lipids content was estimated by gravimetry.
2.4. Fatty Acids Profile
The profile of fatty acid methyl esters (FAME) was determined by a modified Lepage and Roy (1984) method [38]. Briefly, the biomass was homogenized in a methanol/acetyl chloride solution (20:1, v/v) in derivatization vessels with an Ultra-Turrax (IKA-Werke GmbH Staufen, Staufen im Breisgau, Germany). Subsequently, 1 mL of n-hexane was added, and the samples were derivatized at 70 °C for 60 min. FAME were extracted with n-hexane and anhydrous sodium sulphate was added to remove residual water, followed by filtration with a 0.2 µm PTFE filter (Whatman Puradisc, PTFE). The filtered sample was evaporated under a nitrogen flow and resuspended in 500 µL of chromatography-grade n-hexane. FAME were analysed by a Bruker Gas Chromatographer coupled to a mass spectrometry system (Bruker SCION 456-GC, SCION TQ MS) equipped with a ZB-5MS capillary column (30 × 0.25 mm internal diameter with 0.25 μm film thickness; Phenomenex), with helium as carrier gas (1 mL/min). The temperature program was set to 1 min at 60 °C, 30 °C/min to 120 °C, 4 °C/min to 250 °C, and 20 °C/min to 300 °C, held for 4 min, with an injection temperature of 300 °C in splitless mode. Calibration curves were established for each compound using external standards (Supelco® 37 Component FAME Mix, Sigma-Aldrich, Sintra, Portugal). The results are presented in % of total fatty acids content (% TFA).
2.5. Amino Acid Profile
The amino acid (AA) profile was performed by ultra-high-performance liquid chromatography (UPLC) in a Waters reversed-phase amino acid analysis system, using norvaline as an internal standard. The sample was hydrolysed with 6 M HCl at 120 °C, over 48 h, in nitrogen-flushed glass vials. All the samples were then pre-column derivatized with Waters AccQ Fluor Reagent (6-aminoquinolyl-N-hydroxysuccinimidyl carbamate) using the AccQ Tag method (Waters, Milford, MA, USA). Tryptophan was not determined since it is partially destroyed by the acid hydrolysis. The resultant peaks were analysed with EMPOWER software (Waters, Milford, MA, USA). The composition was then compared with currently available natural protein sources, namely soy protein, spirulina and Chlorella sp., and the available data on other Dunaliella sp.
The essential amino acid index (EAAI) was calculated based on the EAA content and FAO/WHO EAA requirements values for humans as reference [39,40] using Equation (1).
where aan is the content of each specific EAA in the sample, and AAn is the AA content of a standard egg protein as determined by FAO/WHO (mg EAA g−1 protein) EAAI; values ≥1, 0.95–1, 0.86–0.95, 0.75–0.86, and ≤0.75 indicate superior quality, high quality, good quality, useful and inadequate, respectively [41].
2.6. Mineral Content
Mineral content was determined by microwave plasma-atomic emission spectrometer (MP-AES; Agilent 4200 MP-AES, Santa Clara, CA, USA). Calibration curves were established for each compound with external standards (ICP-OES, Agilent, Santa Clara, CA, USA). The samples were digested with concentrated nitric acid (HNO3) and hydrogen peroxide 30% (2:1, v:v), followed by dilution in HNO3 5% and analysed in triplicate.
2.7. Carotenoid Content
Carotenoids were extracted in triplicate from approximately 10 mg dry biomass using methanol as solvent and with cell disruption by beat beating in a Retsch MM400 mixer mill at 30 Hz for 2 min. The supernatant was collected by centrifugation at 21,000× g for 3 min. The remaining biomass was re-extracted until both pellet and supernatant were colourless. The combined extracts were dried under a gentle flow of nitrogen, resuspended in 1 mL of methanol, filtered (0.22 µm), and analysed by HPLC to quantify carotenoids as described previously [42].
2.8. Statistical Analysis
The experiments were performed in triplicate and results were expressed as mean ± standard error deviation, with significant levels at p ≤ 0.05 determined using the Tukey HSD (honestly significant differences) test. Statistical analyses were performed using Statistica 7.0 software (TIBCO Software Inc., Palo Alto, CA, USA).
3. Results and Discussion
3.1. Identification
Microscopic observations of Dunaliella sp. TAV01 cells revealed spherical to pear-shaped cells with flagella (Figure 1) and the presence of carotenoid pigments (Figure 1A,B). The identification based on 18S rDNA did not provide sufficient information to differentiate this isolate from other Dunaliella species (results not shown). Therefore, for identification down to the species level, phylogenetic analysis using sequences of the ITS1-5.8S-ITS2 rDNA region were used. Upon Maximum Likelihood (ML) and Bayesian phylogenetic analyses, congruent trees were obtained (Figure 2). TAV01 ITS1-5.8S-ITS2 nucleotide sequence clustered together with those of Dunaliella viridis, D. polymorpha and D. parva with an ML branch support value of 0.96 and maximum Bayesian posterior probability (PP) support (100%). This cluster was named as “Clade Viridis”.
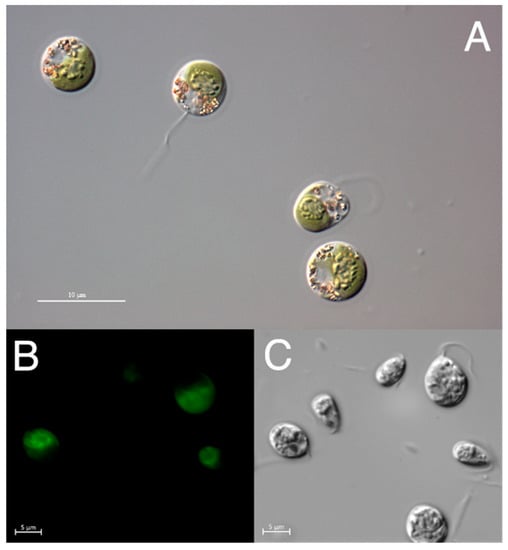
Figure 1.
Microscopic images of Dunaliella sp. TAV01. (A) The differential interference contrast (DIC) on bright field. (B) Cell excitation on natural fluorescence on Ex 470/40; Em 525/50. (C) The same picture as in B but using differential interference contrast (DIC).
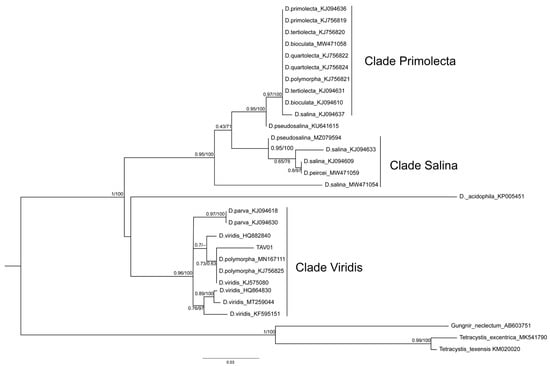
Figure 2.
Maximum likelihood (ML) and Bayesian phylogenetic consensus tree where the TAV01 appears in a cluster together with Dunaliella viridis, D. polymorpha and D. parva ITS1-5.8S-ITS2 sequences. The latter sequence cluster was named as “Clade Viridis”. Other sequences clustered together into two major clades: “Clade Salina” and “Clade Primolecta”, which seem to be sister clades of each other. ML and Bayesian branch support values are given on the left (≥0.43) and on the right (≥63%), respectively. Lower support values were omitted.
The remaining sequences clustered into two additional clades, namely “Clade Primolecta”, containing sequences of D. primolecta, D. bioculata, D. quartolecta, D. polymorpha and D. salina; and “Clade Salina”, containing several D. salina sequences and one sequence of D. pseudosalina (MZ079594). As with clade Viridis, clades Primolecta and Salina displayed high to maximum branch support values (ML > 0.94/PP = 100%). A few sequences did not fall into of these three clades, namely that of D. acidophila and a sequence classified as belonging to D. pseudosalina (KU641615). As previously described by other authors [43], classifying a novel Dunaliella strain down to the species level is notoriously difficult. However, our results do support the idea that the genus Dunaliella has been subdivided into more species than the molecular data suggests [24]. This has caused confusion and misnaming of strains and species, which may explain why sequences classified as D. salina and D. pseudosalina appear in different branches of the consensus tree (Figure 2). The division of the genus Dunaliella into three major clades as suggested here is further supported by recent phylogenetic data [24,43,44]. Thus, it is here proposed that not only TAV01 should be classified as D. viridis, but also the strains previously classified as D. polymorpha (MN167111 and KJ756825) should be reclassified as D. viridis. Such a conclusion seems to be supported by the fact that internal branches within the clade Viridis have lower support except the branch containing D. parva sequences (ML = 0.97/PP = 100%). Moreover, the secondary structure modelling of TAV01 ITS2 showed a 99% similarity with D. viridis ITS2 structure (Figure 3), again strongly suggesting that TAV01 should be classified as D. viridis. Indeed, the inclusion of RNA secondary structures is known to improve the accuracy and robustness of strain assignment to specific taxa using phylogenetics [25].
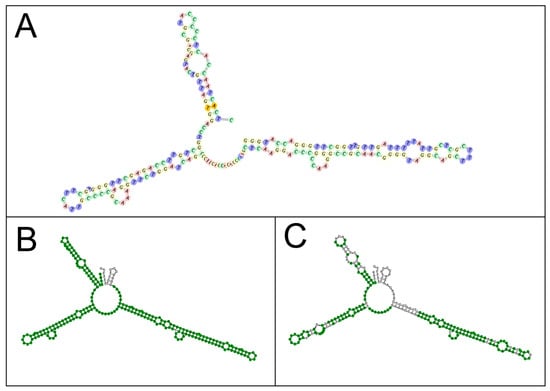
Figure 3.
Comparison of ITS2 secondary structures of Dunaliella viridis TAV01 (A), showing a 99% similarity with that of Dunaliella viridis (ID: AF313419.1; (B)) and Dunaliella salina (ID: EU932917; (C)).
3.2. Proximate Composition
D. viridis TAV01 biochemical characterization is presented in Table 2. The results obtained were compared with other Dunaliella strains. Ash content (23.7 g 100 g−1 DW) was similar to that of D. bardawil and may be related to the high salt content of the medium [45]. Total dietary fibre (TDF) content (9.06 g 100 g−1 DW) was higher than other Dunaliella species but lower than species from other genera, such as Haematococcus pluvialis, Arthrospira platensis, or Chlorella vulgaris: 58.52, 42.82, 35.04 g 100 g−1 DW, respectively. Comparing with other species, D. viridis TAV01 showed the highest protein and lipid contents (35.75 and 21.31 g 100 g−1 DW, respectively) (Table 2). These results may be related to the cultivation conditions, since medium salinity can influence the proximate composition of microalgae [46]. Overall, Dunaliella species have high protein contents in relation to those of lipids and carbohydrates, as shown by other studies in which Dunaliella microalgae displayed protein contents of 36.2% [14], or Microchloropsis gaditana with 33.2 g 100 g−1 DW. Porphyridium cruentum, however, exhibited significantly lower protein levels (19.6 g 100 g−1 DW) [47]. Instead of a rigid cell wall, Dunaliella cells are known to have a glycocalyx-type structure, which is fragile and easily breakable; this may increase the bioavailability of the biomass nutrients as they would be more accessible for human absorption [48]. TAV01 presented a higher lipid content (21.3%) than other Dunaliella spp. (3.49–18.02%). This could be related to the cultivation conditions applied in this study, as several authors reported that high salinity significantly induced lipid production compared to normal seawater salinity (≈0.3 M) [14]. This conclusion is corroborated by results shown by Rismani and Chariati [21], who obtained higher lipid contents for D. salina grown under salt stress (3 M NaCl) [21]. Furthermore, an increase in NaCl concentration from 0.5 to 1 M allowed for increased intracellular lipids and triacylglycerol concentration in Dunaliella tertiolecta [23]. Dunaliella spp. are known to be halotolerant and capable of easily adapting to high salinity, leading to increased contents of some biocompounds such as fatty acids and lutein, that can be used as nutrients in a response to abiotic stress, which is vital for the survival under such detrimental growth conditions [23].

Table 2.
Proximate composition (g 100 g−1 DW) of Dunaliella viridis TAV01. Values are means ± SEM. TDF: total dietary fibre.
The mechanisms involved in saline stress are complex and affect different metabolic and physiological pathways. Furthermore, the lack of a rigid cell wall in these microalgae might facilitate the maintenance of the cell shape and the osmotic balance, allowing an early response and the development of strategies to adapt to high salt stress, which is typical of D. viridis and D. salina cells [52,53,54].
3.3. Fatty Acids Profile
The fatty acids profile of D. viridis TAV01 is presented in Table 3 and was compared with that of other Dunaliella strains. The highest values correspond to palmitic acid (C16:0, 32.1% of total fatty acids (TFA)), followed by α-linolenic acid (ALA, C18:3n-3, 26.3% of TFA), and linoleic acid (C18:2n-6, 22.8% of TFA).

Table 3.
Fatty acids profile of Dunaliella viridis TAV01. Values are means ± SEM shown in percentage of total fatty acids (TFA).
The response to salt stress in terms of the fatty acids profile seems to vary among Dunaliella taxa, which can change the contents of unsaturated fatty acids upon a shift to higher NaCl concentration, in particular in salt-tolerant species [18,22]. Comparing the TAV01 fatty acid profile with that of Dunaliella viridis described in the literature, PUFA concentrations were relatively higher (62.2%) [55] than those obtained by D. viridis TAV01 (49.7%). Although the nutrient composition of the growth media used were different, a possible factor might have been the higher salinity at which TAV01 was grown (3 M NaCl) compared to the D. viridis isolated from the Dead Sea, which was grown at 2 M NaCl [58]. It is thus possible that if TAV01 is grown in the future at lower salinities, higher PUFA contents will be attained. In comparison, Dunaliella salina [59] and Dunaliella sp. [57], which were grown at significantly lower salinities (0.85 M and 1.02 M, respectively) also presented lower PUFA concentrations, suggesting that either the growth conditions or the strains used were not ideal for achieving higher PUFA contents. Indeed, other authors have reported that the fatty acids profile of D. salina grown under stress conditions show increased PUFA contents at increased salinity. The total omega-3 fatty acids, mainly C18:3 and C16:4, represented 92% of the total fatty acids in cells in a higher salinity medium (2.6 M) with relatively low amounts of nitrogen (2.5 mM). Conversely, under standard conditions of growth (1.3 M NaCl, 5 mM N), total omega-3 fatty acids accounted for only about 50% of TFA [56]. The authors attributed these results to an increase in enzyme activity causing elongation and desaturation of fatty acids due to a regulatory response to NaCl levels. This observation agrees with other studies on D. salina, which suggests that high salinity can promote the aforementioned biochemical modifications [21,60]. For D. tertiolecta, the total lipids increased by 26% at high salinities (2 M NaCl) accompanied by a 10% increase in ALA [18]. Hence, it can be concluded that the manipulation of the growth conditions can be used to change the FA profile of microalgal cells; however, the optimal conditions for obtaining target metabolites must be carefully researched [61].
3.4. Amino Acid Profile
The amino acids (AA) profile of D. viridis TAV01 is presented in Table 4 as well as that of other species. The highest AA content corresponds to arginine (13.0% of total AA (TAA)), followed by leucine (9.27% of TAA) and proline (8.34% of TAA). Taurine was found in a minimum amount, whereas this AA was not detected in other strains. TAV01 presented a high amount of essential amino acids (EAA) (leucine, phenylalanine, valine) and good protein quality, as indicated by an EAAI of 1.80. Observing the AA profile, similar results were reported for soy protein, whereas studies with D. tertiolecta, D. salina, spirulina, and Chlorella sp. showed a lower EAA. Spirulina has a similar EAAI value, being well known for its great protein quality and as an excellent source of EAA [62].

Table 4.
Amino acids concentration (g 100 g−1) of Dunaliella sp. TAV01 compared with other Dunaliella spp., other microalgal species, and soy protein. Values are means ± SEM.
Transforming the data into a radial chart (Figure 4), it can be seen that the Chlorella sp. is the largest lysine producer, whereas D. salina showed the highest phenylalanine contents. Nevertheless, the total EAA content of D. viridis TAV01 was 65% higher than the minimum amount recommended by the World Health Organization (WHO): 27.1 g 100 g−1 [67]. Also, spirulina [65], one of the algae displaying consistently high protein contents, shows lower EAA values (42.51 g 100 g−1) than those of D. viridis TAV01 (44.88 g 100 g−1). Furthermore, when compared to those of D. tertiolecta (34.78 g 100 g−1) [63], D. salina (33.05 g 100 g−1) [64], Chlorella sp. (42.60 g 100 g−1) [66], or soy protein (39.00 g 100 g−1), TAV01 shows a better EAA value [66]. In addition, all individual EAA contents in TAV01 were equally higher, with threonine and phenylalanine being 2.47- and 1.95-fold higher than WHO requirements, respectively, and leucine, isoleucine, and valine about 1.6-fold higher than the required level, confirming that this strain has a high-quality protein profile, which might be excellent for human consumption. Moreover, the levels of several EEAs, such as valine, methionine, threonine, and isoleucine match human nutritional needs.
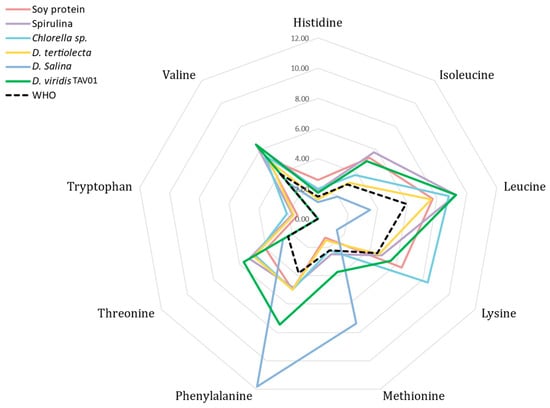
Figure 4.
Radial chart of the essential amino acids.
Although the information regarding D. viridis AA contents and how the cultivation condition affects the AA profile is limited, several studies on other species have shown that stress conditions can significantly increase AA content. Studies varying the growing conditions of D. salina suggest that nitrogen limitation might increase all EAA contents, which might be a consequence of changes in the proteome to cope with the imposed stress [68,69]. In addition, other limiting conditions, such as low concentrations of phosphorus and sulphur, can also increase stress-related compounds such as proline, lysine, and tyrosine in D. salina [69]. Besides nutrient-limited cultivation conditions, accumulation of proline has also been reported with cultivation under NaCl stress, the most extensively accumulated molecule, which might serve as a stress indicator [70]. Interestingly, proline was one of the most produced AA in D. viridis TAV01, which might have been caused by the NaCl stress imposed by the growth conditions used in this study.
3.5. Mineral Profile
As can be seen in Table 5, sodium (41.45 mg g−1 DW) was the major mineral in the D. viridis TAV01 biomass, which was expected due to the high saline culture medium. The WHO recommends a maximum Na daily intake of 2 g. However, even in the case of a high microalgae incorporation in food, 20 g, for example, Na intake will be lower than the maximum recommended levels [71]. However, this is a concern even when discussing other microalgae such as spirulina which was reported to accumulate 151.02 mg g−1 DW of Na [72]. Phosphorus concentration was 9.79 mg g−1, which was close to the value reported by [73] for spirulina and Chlorella (11.9 mg g−1 and 12.4 mg g−1, respectively). Potassium concentration (5.1 mg g−1) was similar to the values described for D. tertiolecta (5.3 and 4.29 mg g−1 DW) and D. salina (5.1 mg g−1 DW) [74,75,76]. Iron content (3.67 mg g−1) was higher compared to that reported for D. tertiolecta (1.37 mg g−1) and similar to that of C. vulgaris (3.6 mg g−1 DW) [75,76]. Considering the intake range recommended for adults, with a minimum daily intake recommendation of 8 mg.d−1 and a maximum nontoxic limit of 40 mg.d−1 DW [77], D. viridis TAV01 shows an interesting profile for possible iron supplementation. The presence of toxic metals may be related to the high concentration of these metals in the environment from which the strain was isolated and its bioaccumulation capacity, where some metals are preferentially sequestered over others [73,78]. Some studies showed that Dunaliella sp. can significantly uptake Pb in a 6 day period [58,79]. Thus, care should be exerted to monitor Dunaliella biomass for the possible accumulation of this and other metals, such as Cd, whose concentration was above the maximum permitted levels in food (FAO/WHO—0.5 mg 100 g−1) [80]. Nonetheless, considering the maximum daily intake recommended for each metal, the daily consumption of 50 mg of this alga should not pose a health risk [77], and countermeasures ensuring the absence of such metals from the growth medium could be taken in the future. The elements Ag, Be, Co, Hg, Mo, Ni, Sb, Se, Sn, Th, Ti, and U were not detected.

Table 5.
Mineral profile of Dunaliella sp. TAV01. Values are means ± SEM.
3.6. Carotenoid Content
The major carotenoid in D. viridis TAV01 was lutein, making over 85% of the total carotenoids. However, neoxanthin, violaxanthin, and β-carotene could also be detected in minor quantities (Table 6). This carotenoid profile agrees with other studies on D. viridis. The lutein content in D. viridis TAV01 was 2.15 ± 0.12 mg g−1 DW, after 15 days of growth. However, improved growth conditions and shorter cultivation times could lead to higher contents, as shown in a different study on another D. viridis strain, in which lutein contents as high as 8.87 mg g−1 DW were obtained after 5 days [81]. Furthermore, sulphate deprivation could be a good inducer for lutein accumulation; in D. salina, sulphate deprivation in combination with bicarbonate supplementation, led to a significant increase in lutein, reaching contents of 0.26 mg g−1 DW [82,83]. As the literature suggests, D. viridis TAV01 does not accumulate β-carotene under high salinity nor under nitrogen depletion as seen for D. salina, the major β-carotene microalgal producer [84]. β-carotene supplements from D. salina are already approved for human consumption by the EU; this might facilitate the introduction of D. viridis as a source of lutein. This might be particularly relevant as the global lutein market value is expect to reach USD 170 M in 2029 [85].

Table 6.
Comparison of carotenoid contents (mg g−1 DW) of Dunaliella viridis TAV01 (average ± SEM, n = 3) with other species of the genus Dunaliella reported in the literature.
4. Conclusions
A novel strain Dunaliella viridis TAV01 was successfully isolated from water samples from the Algarve coast. This strain can be cultivated under high salt conditions, thus preventing the proliferation of competing species and/or predators so that culture collapse is avoided and growth of the target species is promoted. When cultivating at these conditions, Dunaliella viridis TAV01 biomass displayed an interesting biochemical profile, rich in proteins, lipids, and pigments, and a high degree of PUFAs with α-linolenic and linoleic acids as the main fatty acids. Protein analysis revealed a well-balanced AA profile, adequate for human consumption and comparable to that of traditional plant or algae-based protein sources. The characteristics and qualities of this novel strain may provide new paths for developing high-quality algae-based nutritional solutions, making it a promising target to be commercialized as supplements and/or food products.
Author Contributions
Conceptualization, G.B., N.L.C., L.B. and J.V.; Data curation, H.P., L.B. and J.V.; Formal analysis, T.F.S., L.S. and I.B.M.; Methodology, G.B., N.L.C., T.F.S., L.S. and I.B.M.; Validation, H.P., L.B. and J.V.; Writing—original draft, G.B. and N.L.C.; Writing—review and editing, T.F.S., L.S., I.B.M., H.P., L.B. and J.V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with funding from the Portuguese Foundation for Science and Technology (FCT) through UIDB/04326/2020, UIDP/0436/2020, LA/P/0101/2020, SFRH/BD/149395/2019, SFRH/BD/115325/2016, SFRH/BD/140143/2018, and 2021.06332.BD. Additional funding was provided by the European Union’s Horizon 2020 Research and Innovation program under grant agreement No. 862980.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Özçimen, D.; İnan, B.; Koçer, A.T.; Vehapi, M. Bioeconomic Assessment of Microalgal Production. In Microalgal Biotechnology; Jacob-Lopes, E., Zepka, L.Q., Queiroz, M.I., Eds.; IntechOpen: London, UK, 2018; ISBN 978-1-78923-332-2. [Google Scholar]
- Pereira, H.; Gangadhar, K.N.; Schulze, P.S.C.; Santos, T.; de Sousa, C.B.; Schueler, L.M.; Custódio, L.; Malcata, F.X.; Gouveia, L.; Varela, J.C.S.; et al. Isolation of a euryhaline microalgal strain, Tetraselmis sp. CTP4, as a robust feedstock for biodiesel production. Sci. Rep. 2016, 6, 35663. [Google Scholar] [CrossRef] [PubMed]
- Tarhan, S.Z.; Koçer, A.T.; Özçimen, D.; Gökalp, I. Cultivation of green microalgae by recovering aqueous nutrients in hydrothermal carbonization process water of biomass wastes. J. Water Process. Eng. 2020, 40, 101783. [Google Scholar] [CrossRef]
- Mendes, M.C.; Navalho, S.; Ferreira, A.; Paulino, C.; Figueiredo, D.; Silva, D.; Gao, F.; Gama, F.; Bombo, G.; Jacinto, R.; et al. Algae as Food in Europe: An Overview of Species Diversity and Their Application. Foods 2022, 11, 1871. [Google Scholar] [CrossRef] [PubMed]
- Stirk, W.A.; van Staden, J. Bioprospecting for bioactive compounds in microalgae: Antimicrobial compounds. Biotechnol. Adv. 2022, 59, 107977. [Google Scholar] [CrossRef]
- Jain, P.; Minhas, A.K.; Shukla, S.; Puri, M.; Barrow, C.J.; Mandal, S. Bioprospecting Indigenous Marine Microalgae for Polyunsaturated Fatty Acids Under Different Media Conditions. Front. Bioeng. Biotechnol. 2022, 10, 13. [Google Scholar] [CrossRef]
- Fu, Y. The potentials and challenges of using microalgae as an ingredient to produce meat analogues. Trends Food Sci. Technol. 2021, 13, 188–200. [Google Scholar] [CrossRef]
- Michel, F.; Knaapila, A.; Hartmann, C.; Siegrist, M. A multi-national comparison of meat eaters’ attitudes and expectations for burgers containing beef, pea or algae protein. Food Qual. Prefer. 2021, 91, 104195. [Google Scholar] [CrossRef]
- Sharif, M.; Zafar, M.H.; Aqib, A.I.; Saeed, M.; Farag, M.R.; Alagawany, M. Single cell protein: Sources, mechanism of production, nutritional value and its uses in aquaculture nutrition. Aquaculture 2020, 531, 735885. [Google Scholar] [CrossRef]
- Yadav, G.; Shanmugam, S.; Sivaramakrishnan, R.; Kumar, D.; Mathimani, T.; Brindhadevi, K.; Pugazhendhi, A.; Rajendran, K. Mechanism and challenges behind algae as a wastewater treatment choice for bioenergy production and beyond. Fuel 2020, 285, 119093. [Google Scholar] [CrossRef]
- Ullmann, J.; Grimm, D. Algae and their potential for a future bioeconomy, landless food production, and the socio-economic impact of an algae industry. Org. Agric. 2021, 11, 261–267. [Google Scholar] [CrossRef]
- Vazquez Calderon, F.; Sanchez Lopez, J. An Overview of the Algae Industry in Europe: Producers, Production Systems, Species, Biomass Uses, Other Steps in the Value Chain and Socio-Economic Data; Publications Office of the European Union: Luxembourg, 2022; ISBN 978-92-76-54516-3.
- Archer, L.; Mc Gee, D.; Paskuliakova, A.; McCoy, G.R.; Smyth, T.; Gillespie, E.; Touzet, N. Fatty acid profiling of new Irish microalgal isolates producing the high-value metabolites EPA and DHA. Algal Res. 2019, 44, 101671. [Google Scholar] [CrossRef]
- Khatoon, H.; Haris, N.; Banerjee, S.; Rahman, N.A.; Begum, H.; Mian, S.; Abol-Munafi, A.B.; Endut, A. Effects of different salinities on the growth and proximate composition of Dunaliella sp. isolated from South China Sea at different growth phases. Process. Saf. Environ. Prot. 2017, 112, 280–287. [Google Scholar] [CrossRef]
- Molino, A.; Rimauro, J.; Casella, P.; Cerbone, A.; Larocca, V.; Chianese, S.; Karatza, D.; Mehariya, S.; Ferraro, A.; Hristoforou, E.; et al. Extraction of astaxanthin from microalga Haematococcus pluvialis in red phase by using generally recognized as safe solvents and accelerated extraction. J. Biotechnol. 2018, 283, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Anand, V.; Kashyap, M.; Samadhiya, K.; Ghosh, A.; Kiran, B. Salinity driven stress to enhance lipid production in Scenedesmus vacuolatus: A biodiesel trigger? Biomass-Bioenergy 2019, 127, 105252. [Google Scholar] [CrossRef]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Salinity induced oxidative stress alters the physiological responses and improves the biofuel potential of green microalgae Acutodesmus dimorphus. Bioresour. Technol. 2017, 244, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- El Arroussi, H.; Benhima, R.; Bennis, I.; El Mernissi, N.; Wahby, I. Improvement of the potential of Dunaliella tertiolecta as a source of biodiesel by auxin treatment coupled to salt stress. Renew. Energy 2015, 77, 15–19. [Google Scholar] [CrossRef]
- Paliwal, C.; Mitra, M.; Bhayani, K.; Bharadwaj, S.V.V.; Ghosh, T.; Dubey, S.; Mishra, S. Abiotic stresses as tools for metabolites in microalgae. Bioresour. Technol. 2017, 244, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Qiao, T.; Zhao, Y.; Zhong, D.-B.; Yu, X. Hydrogen peroxide and salinity stress act synergistically to enhance lipids production in microalga by regulating reactive oxygen species and calcium. Algal Res. 2021, 53, 102017. [Google Scholar] [CrossRef]
- Rismani, S.; Shariati, M. Changes of the Total Lipid and Omega-3 Fatty Acid Contents in two Microalgae Dunaliella Salina and Chlorella Vulgaris under Salt Stress. Braz. Arch. Biol. Technol. 2017, 60. [Google Scholar] [CrossRef]
- Schwenk, D.; Seppälä, J.; Spilling, K.; Virkki, A.; Tamminen, T.; Oksman-Caldentey, K.-M.; Rischer, H. Lipid content in 19 brackish and marine microalgae: Influence of growth phase, salinity and temperature. Aquat. Ecol. 2013, 47, 415–424. [Google Scholar] [CrossRef]
- Takagi, M.; Karseno; Yoshida, T. Effect of salt concentration on intracellular accumulation of lipids and triacylglyceride in marine microalgae Dunaliella cells. J. Biosci. Bioeng. 2006, 101, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Assunção, P.; Jaén-Molina, R.; Caujapé-Castells, J.; de la Jara, A.; Carmona, L.; Freijanes, K.; Mendoza, H. Molecular taxonomy of Dunaliella (Chlorophyceae), with a special focus on D. salina: ITS2 sequences revisited with an extensive geographical sampling. Aquat. Biosyst. 2012, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Förster, F.; Müller, T.; Dandekar, T.; Schultz, J.; Wolf, M. Including RNA secondary structures improves accuracy and robustness in reconstruction of phylogenetic trees. Biol. Direct 2010, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Beuzenberg, V.; Smith, K.; Packer, M. Isolation and characterisation of halo-tolerant Dunaliella strains from Lake Grassmere/Kapara Te Hau, New Zealand. N. Z. J. Bot. 2014, 52, 136–152. [Google Scholar] [CrossRef]
- Pereira, H.; Barreira, L.; Mozes, A.; Florindo, C.; Polo, C.; Duarte, C.V.; Custódio, L.; Varela, J. Microplate-based high throughput screening procedure for the isolation of lipid-rich marine microalgae. Biotechnol. Biofuels 2011, 4, 61. [Google Scholar] [CrossRef]
- Borghini, M.; Bryden, H.; Schroeder, K.; Sparnocchia, S.; Vetrano, A. The Mediterranean is becoming saltier. Ocean Sci. 2014, 10, 693–700. [Google Scholar] [CrossRef]
- Pereira, H.; Barreira, L.; Custódio, L.; Alrokayan, S.; Mouffouk, F.; Varela, J.; Abu-Salah, K.M.; Ben-Hamadou, R. Isolation and Fatty Acid Profile of Selected Microalgae Strains from the Red Sea for Biofuel Production. Energies 2013, 6, 2773–2783. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Seibel, P.N.; Müller, T.; Dandekar, T.; Schultz, J.; Wolf, M. 4SALE—A tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform. 2006, 7, 498. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Wolf, M. ITS2 sequence–structure analysis in phylogenetics: A how-to manual for molecular systematics. Mol. Phylogenetics Evol. 2009, 52, 520–523. [Google Scholar] [CrossRef]
- Sluiter, A.; Hanes, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D. Determination of Ash in Biomass Laboratory Analytical Procedure (LAP); National Renewable Energy Laboratory: Golden, CO, USA, 2005; p. 8.
- Lourenço, S.O.; Barbarino, E.; Lavin, P.; Marquez, U.M.L.; Aidar, E. Distribution of intracellular nitrogen in marine microalgae: Calculation of new nitrogen-to-protein conversion factors. Eur. J. Phycol. 2004, 39, 17–32. [Google Scholar] [CrossRef]
- Lima, A.R.; Castañeda-Loaiza, V.; Salazar, M.; Nunes, C.; Quintas, C.; Gama, F.; Pestana, M.; Correia, P.J.; Santos, T.; Varela, J.; et al. Influence of cultivation salinity in the nutritional composition, antioxidant capacity and microbial quality of Salicornia ramosissima commercially produced in soilless systems. Food Chem. 2020, 333, 127525. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.; Barreira, L.; Figueiredo, F.; Custódio, L.; Vizetto-Duarte, C.; Polo, C.; Rešek, E.; Engelen, A.; Varela, J. Polyunsaturated Fatty Acids of Marine Macroalgae: Potential for Nutritional and Pharmaceutical Applications. Mar. Drugs 2012, 10, 1920–1935. [Google Scholar] [CrossRef] [PubMed]
- Oser, B.L. An Integrated Essential Amino Acid Index for Predicting the Biological Value of Proteins. In Protein and Amino Acid Nutrition; Elsevier: Amsterdam, The Netherlands, 1959; pp. 281–295. [Google Scholar]
- Protein and Amino Acid Requirements in Human Nutrition: Report of a Joint WHO/FAO/UNU Expert Consultation; [Geneva, 9–16 April 2002]; Weltgesundheitsorganisation; FAO; Vereinte Nationen (Eds.) WHO Technical Report Series; WHO: Geneva, Switzerland, 2007; ISBN 978-92-4-120935-9.
- Kent, M.; Welladsen, H.M.; Mangott, A.; Li, Y. Nutritional Evaluation of Australian Microalgae as Potential Human Health Supplements. PLoS ONE 2015, 10, e0118985. [Google Scholar] [CrossRef]
- Schüler, L.M.; Gangadhar, K.N.; Duarte, P.; Placines, C.; Molina-Márquez, A.M.; Léon-Bañares, R.; Sousa, V.S.; Varela, J.; Barreira, L. Improvement of carotenoid extraction from a recently isolated, robust microalga, Tetraselmis sp. CTP4 (chlorophyta). Bioprocess Biosyst. Eng. 2020, 43, 785–796. [Google Scholar] [CrossRef]
- Tran, D.; Vo, T.; Portilla, S.; Louime, C.; Doan, N.; Mai, T.; Tran, D.; Ho, T. Phylogenetic study of some strains of Dunaliella. Am. J. Environ. Sci. 2013, 9, 317–321. [Google Scholar] [CrossRef]
- Henley, W.J.; Cobbs, M.; Novoveská, L.; Buchheim, M.A. Phylogenetic analysis of Dunaliella (Chlorophyta) emphasizing new benthic and supralittoral isolates from Great Salt Lake. J. Phycol. 2018, 54, 483–493. [Google Scholar] [CrossRef]
- Vanitha, A.; Narayan, M.S.; Murthy, K.N.C.; Ravishankar, G.A. Comparative study of lipid composition of two halotolerant alga, Dunaliellabardawil and Dunaliellasalina. Int. J. Food Sci. Nutr. 2007, 58, 373–382. [Google Scholar] [CrossRef]
- Richmond, A. (Ed.) CRC Handbook of Microalgal Mass Culture (1986), 1st ed.; CRC Press: Boca Raton, FL, USA, 2017; ISBN 978-0-203-71240-5. [Google Scholar]
- Di Lena, G.; Casini, I.; Lucarini, M.; del Pulgar, J.S.; Aguzzi, A.; Caproni, R.; Gabrielli, P.; Lombardi-Boccia, G. Chemical characterization and nutritional evaluation of microalgal biomass from large-scale production: A comparative study of five species. Eur. Food Res. Technol. 2019, 246, 323–332. [Google Scholar] [CrossRef]
- Borowitzka, M.A. Biology of Microalgae. In Microalgae in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2018; pp. 109–131. ISBN 978-0-12-811405-6. [Google Scholar]
- Molino, A.; Iovine, A.; Casella, P.; Mehariya, S.; Chianese, S.; Cerbone, A.; Rimauro, J.; Musmarra, D. Microalgae Characterization for Consolidated and New Application in Human Food, Animal Feed and Nutraceuticals. Int. J. Environ. Res. Public Health 2018, 15, 2436. [Google Scholar] [CrossRef] [PubMed]
- Muhaemin, M.; Kaswadji, R. Biomass Nutrient Profiles of Marine Microalgae Dunaliella salina. J. Penelit. Sains 2010, 13, 64–67. [Google Scholar]
- Murthy, K.N.C. Production of Beta-Carotene from Cultured Dunaliella sp. and Evaluation Biological Activities. Ph.D. Thesis., University of Mysore, Mysuru, India, 2005. [Google Scholar]
- Wang, Y.; Cong, Y.; Wang, Y.; Guo, Z.; Yue, J.; Xing, Z.; Gao, X.; Chai, X. Identification of Early Salinity Stress-Responsive Proteins in Dunaliella salina by isobaric tags for relative and absolute quantitation (iTRAQ)-Based Quantitative Proteomic Analysis. Int. J. Mol. Sci. 2019, 20, 599. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Gao, X.; Wang, M.; Cong, Y.; Chai, X. Identification of salt-responsive genes using transcriptome analysis in Dunaliella viridis. J. Appl. Phycol. 2020, 32, 2875–2887. [Google Scholar] [CrossRef]
- Borowitzka, M.A.; Siva, C.J. The taxonomy of the genus Dunaliella (Chlorophyta, Dunaliellales) with emphasis on the marine and halophilic species. J. Appl. Phycol. 2007, 19, 567–590. [Google Scholar] [CrossRef]
- Evans, R.W.; Kates, M. Lipid composition of halophilic species of Dunaliella from the dead sea. Arch. Microbiol. 1984, 140, 50–56. [Google Scholar] [CrossRef]
- El-Baky, H.H.A.; El Baz, F.K.; El-Baroty, G.S. Production of Lipids Rich in Omega 3 Fatty Acids from the Halotolerant Alga Dunaliella salina. Biotechnology 2003, 3, 102–108. [Google Scholar] [CrossRef]
- Talebi, A.F.; Tohidfar, M.; Derazmahalleh, S.M.M.; Sulaiman, A.; Baharuddin, A.S.; Tabatabaei, M. Biochemical Modulation of Lipid Pathway in Microalgae Dunaliella sp. for Biodiesel Production. BioMed Res. Int. 2015, 2015, 1–12. [Google Scholar] [CrossRef]
- Muhaemin, M. Toxicity and Bioacumulation of lead in Chlorella and Dunaliella. J. Coast. Dev. 2004, 8, 27–33. [Google Scholar]
- Fakhry, E.M.; El Maghraby, D.M. Fatty Acids Composition and Biodiesel Characterization of Dunaliella salina. J. Water Resour. Prot. 2013, 05, 894–899. [Google Scholar] [CrossRef]
- Al-Hasan, R.H.; Ghannoum, M.A.; Sallal, A.-K.; Abu-Elteen, K.H.; Radwan, S.S. Correlative Changes of Growth, Pigmentation and Lipid Composition of Dunaliella salina in Response to Halostress. Microbiology 1987, 133, 2607–2616. [Google Scholar] [CrossRef]
- Zhila, N.O.; Kalacheva, G.S.; Volova, T.G. Effect of salinity on the biochemical composition of the alga Botryococcus braunii Kütz IPPAS H-252. J. Appl. Phycol. 2010, 23, 47–52. [Google Scholar] [CrossRef]
- de Morais, E.G.; Cristofoli, N.L.; Bombo, G.d.C.; Maia, I.B.; Cassuriaga, A.P.A.; Costa, J.A.V.; de Morais, M.G.; Bar, S.Y.; Avni, D.; Varela, J.; et al. Spirulina as Immune System Potentiator. In Spirulina and its Health Benefits; Nova Science Publishers: Hauppauge, NY, USA, 2021; pp. 1–44. ISBN 978-1-68507-018-2. [Google Scholar]
- da Silva Gorgonio, C.M.; Aranda, D.A.G.; Couri, S. Morphological and chemical aspects of Chlorella pyrenoidosa, Dunaliella tertiolecta, Isochrysis galbana and Tetraselmis gracilis microalgae. Nat. Sci. 2013, 5, 783–791. [Google Scholar] [CrossRef]
- Sui, Y.; Harvey, P. Effect of Light Intensity and Wavelength on Biomass Growth and Protein and Amino Acid Composition of Dunaliella salina. Foods 2021, 10, 1018. [Google Scholar] [CrossRef] [PubMed]
- Shioji, Y.; Kobayashi, T.; Yoshida, T.; Otagiri, T.; Onoda, K.; Yoshioka, Y.; Sasada, T.; Miyoshi, N. Nitrogen Balance and Bioavailability of Amino Acids in Spirulina Diet-Fed Wistar Rats. J. Agric. Food Chem. 2021, 69, 13780–13786. [Google Scholar] [CrossRef] [PubMed]
- Becker, E. Micro-algae as a source of protein. Biotechnol. Adv. 2007, 25, 207–210. [Google Scholar] [CrossRef] [PubMed]
- E Holt, L.; E Snyderman, S. Protein and amino acid requirements of infants and children. Nutr. Abstr. Rev. 1965, 35, 1–13. [Google Scholar]
- Sui, Y.; Muys, M.; Van de Waal, D.; D’Adamo, S.; Vermeir, P.; Fernandes, T.V.; Vlaeminck, S.E. Enhancement of co-production of nutritional protein and carotenoids in Dunaliella salina using a two-phase cultivation assisted by nitrogen level and light intensity. Bioresour. Technol. 2019, 287, 121398. [Google Scholar] [CrossRef]
- Sui, Y.; Muys, M.; Vermeir, P.; D’Adamo, S.; Vlaeminck, S.E. Light regime and growth phase affect the microalgal production of protein quantity and quality with Dunaliella salina. Bioresour. Technol. 2018, 275, 145–152. [Google Scholar] [CrossRef]
- Fal, S.; Aasfar, A.; Rabie, R.; Smouni, A.; Arroussi, H.E. Salt induced oxidative stress alters physiological, biochemical and metabolomic responses of green microalga Chlamydomonas reinhardtii. Heliyon 2022, 8, e08811. [Google Scholar] [CrossRef]
- World Health Organization. Regional Office for the Eastern Mediterranean Eastern Mediterranean Health Journal. East. Mediterr. Health J. 2018, 24, 17–62. [Google Scholar]
- Morsy, O.M. Production and evaluation of some extruded food products using spirulina algae. Ann. Agric. Sci. Moshtohor 2014, 52, 495–510. [Google Scholar] [CrossRef]
- Rzymski, P.; Budzulak, J.; Niedzielski, P.; Klimaszyk, P.; Proch, J.; Kozak, L.; Poniedziałek, B. Essential and toxic elements in commercial microalgal food supplements. J. Appl. Phycol. 2018, 31, 3567–3579. [Google Scholar] [CrossRef]
- Diprat, A.B.; Menegol, T.; Boelter, J.F.; Zmozinski, A.; Vale, M.G.R.; Rodrigues, E.; Rech, R. Chemical composition of microalgae Heterochlorella luteoviridis and Dunaliella tertiolecta with emphasis on carotenoids. J. Sci. Food Agric. 2017, 97, 3463–3468. [Google Scholar] [CrossRef]
- Varela-Bojórquez, N.; Vélez-De La Rocha, R.; Ángel, M.; Escalante, A.; Sañudo-Barajas, J.A. Production of Bioethanol from Biomass of Microalgae Dunaliella tertiolecta. Int. J. Environ. Agric. Res. 2016, 2, 110–116. [Google Scholar]
- Gong, X.; Zhang, B.; Zhang, Y.; Huang, Y.; Xu, M. Investigation on Pyrolysis of Low Lipid Microalgae Chlorella vulgaris and Dunaliella salina. Energy Fuels 2014, 28, 95–103. [Google Scholar] [CrossRef]
- Institute of Medicine (U.S.) (Ed.) DRI: Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc: A Report of the Panel on Micronutrients … and the Standing Committee on the Scientific Evaluation of Dietary Reference Intakes, Food and Nutrition Board, Institute of Medicine; National Academy Press: Washington, DC, USA, 2001; ISBN 978-0-309-07279-3. [Google Scholar]
- Rzymski, P.; Niedzielski, P.; Karczewski, J.; Poniedziałek, B. Biosorption of toxic metals using freely suspended Microcystis aeruginosa biomass. Cent. Eur. J. Chem. 2014, 12, 1232–1238. [Google Scholar] [CrossRef]
- Kutlu, B.; Mutlu, E. Growth and Bioaccumulation of Cadmium, Zinc, Lead, Copper in Dunaliella sp. Isolated from Homa Lagoon, Eastern Aegean Sea; NISCAIR-CSIR: New Delhi, India, 2017; Volume 46. [Google Scholar]
- Wu, Y. Genral Standard for Contaminants and Toxins in Food and Feed (DOEX STAN 193-1995) Adopted in 1995. Revised in 1997, 2006, 2008, 2009. Amendment 2010, 2012, 2013, 2014. 2014. Joint FAO/WHO Codex Alimentarius Commission. Codex Aliment. 2014. [Google Scholar] [CrossRef]
- Fu, W.; Paglia, G.; Magnusdottir, M.; A Steinarsdóttir, E.; Gudmundsson, S.; Palsson, B.; Andrésson, S.; Brynjolfsson, S. Effects of abiotic stressors on lutein production in the green microalga Dunaliella salina. Microb. Cell Factories 2014, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Mageswari, A.; Subramanian, P.; Suganthi, C.; Chaitanyakumar, A.; Aswini, V.; Gothandam, K.M. Bicarbonate supplementation enhances growth and biochemical composition of Dunaliella salina V-101 by reducing oxidative stress induced during macronutrient deficit conditions. Sci. Rep. 2018, 8, 6972. [Google Scholar] [CrossRef]
- Lv, H.; Cui, X.; Wahid, F.; Xia, F.; Zhong, C.; Jia, S. Analysis of the Physiological and Molecular Responses of Dunaliella salina to Macronutrient Deprivation. PLoS ONE 2016, 11, e0152226. [Google Scholar] [CrossRef] [PubMed]
- Moulton, T.P.; Burford, M.A. The mass culture of Dunaliella viridis (Volvocales, Chlorophyta) for oxygenated carotenoids: Laboratory and pilot plant studies. Hydrobiologia 1990, 204–205, 401–408. [Google Scholar] [CrossRef]
- MMR Lutein Market—Global Industry Analysis and Forecast (2022–2029); Maximize Market Research: Maharashtra, India, 2023.
- Hu, C.-C.; Lin, J.-T.; Lu, F.-J.; Chou, F.-P.; Yang, D.-J. Determination of carotenoids in Dunaliella salina cultivated in Taiwan and antioxidant capacity of the algal carotenoid extract. Food Chem. 2008, 109, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Colusse, G.A.; Mendes, C.R.B.; Duarte, M.E.R.; de Carvalho, J.C.; Noseda, M.D. Effects of different culture media on physiological features and laboratory scale production cost of Dunaliella salina. Biotechnol. Rep. 2020, 27, e00508. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Cui, Y.; Yan, M.; Wang, Y.; Gao, Z.; Meng, C.; Qin, S. Construction of astaxanthin metabolic pathway in the green microalga Dunaliella viridis. Algal Res. 2019, 44, 101697. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).