Abstract
Aptamers are short, single-stranded oligonucleotides synthesized in vitro from a randomized oligonucleotide library against a specific target. These molecules are capable of binding to a wide range of biological targets with high specificity and affinity. They present great advantages over antibodies with potential applications in research, diagnosis, and therapeutics. Specifically for tumors with late-stage identification and poor prognosis, like pancreatic cancer, the study of novel aptamers holds tremendous potential for cancer diagnosis and treatment. Along with cancer treatment, aptamers have also shown high potential in regulating the immune response and modulating several critical steps of signaling cascades, such as in immune checkpoints. In the context of microbiota and infection, aptamers are being studied to identify microbes and their metabolites. This assessment has the potential to improve the detection and management of infectious diseases while assisting us in better understanding health risks and treatment outcomes by tracking changes in the microbiota. In this review, the potential of aptamers is explored regarding their applications in cancer, immune, and microbiota therapy.
1. Introduction
Aptamers are short synthetic single-stranded ribonucleic acid (ssRNA) or desoxyribonucleic acid (ssDNA) oligonucleotides, selected through an in vitro method called Systematic Evolution of Ligands by Exponential enrichment (SELEX) [1,2]. They can display high specificity and affinity to a broad spectrum of target molecules, from ions or small molecules to proteins, viruses, bacteria/pathogens, cells, and tissues.
The SELEX process was developed in 1990, simultaneously by two independent groups, Tuerk and Gold [3] and Ellington and Szostak [4], with the goal of isolating RNA sequences that would specifically recognize target molecules. Since then, many improvements have been made in this procedure to increase its selectivity and shorten the selection process [5]. Aptamers possess a vast field of applications, including diagnosis, therapy and drug delivery systems, food safety, and environment monitoring [1,2,6].
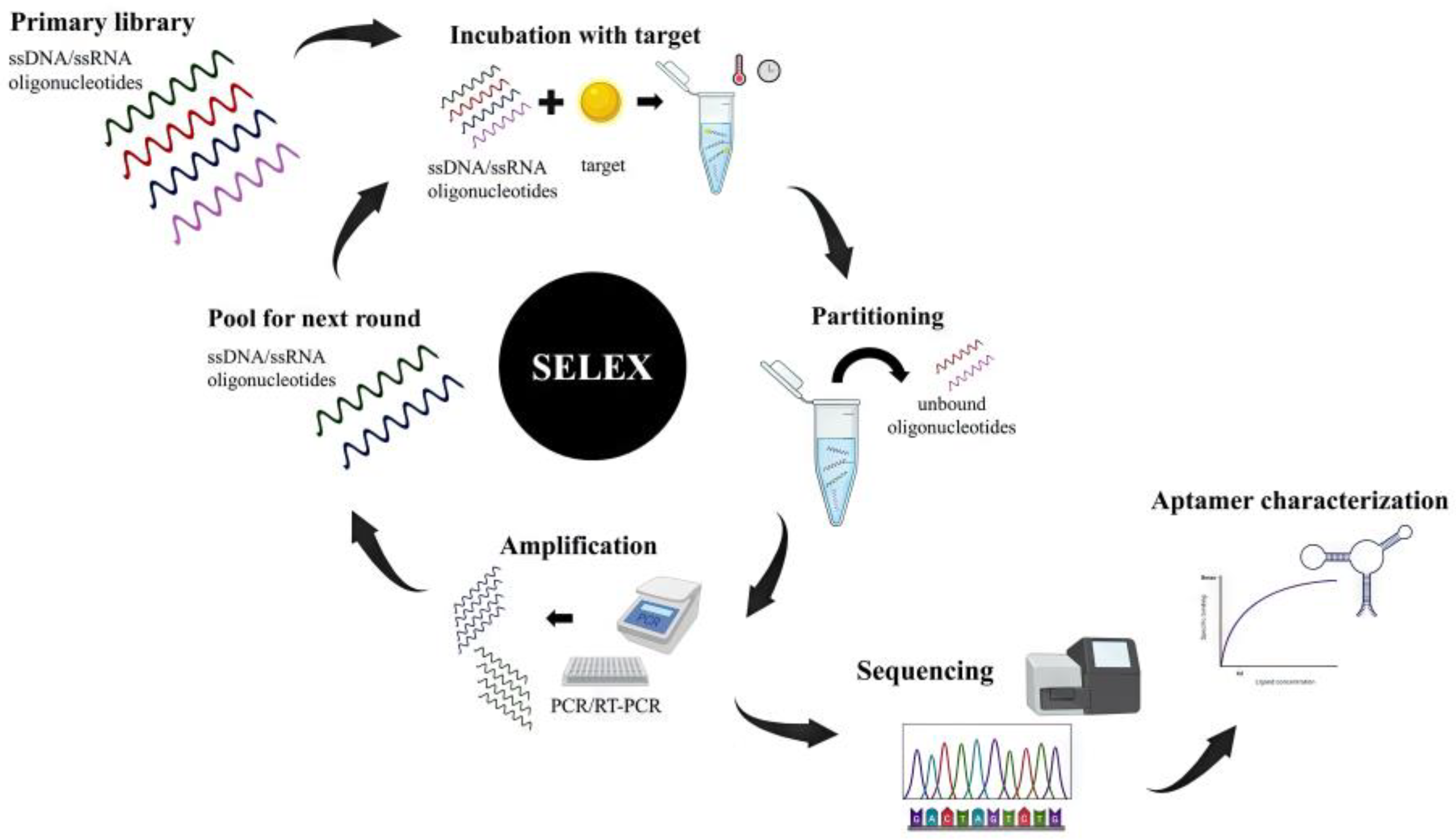
The conventional SELEX involves subsequent cycles of incubation, binding, partitioning, and amplification (Figure 1). It usually starts with a synthetic pool of ssDNA or RNA oligonucleotides called the primary library, which can comprise up to 1015 different sequences. This initial pool is incubated with the target molecule (in a free or immobilized form), under specific conditions of incubation. Unbound or weakly bounded sequences are eluted (partitioning) from the target and the target-bounded sequences are then retrieved and amplified by PCR or RT-PCR, for DNA or RNA libraries, respectively. The process is repeated for several rounds (between four and 20), with increasingly stringent conditions, in order to obtain a final pool with low heterogeneity and high affinity of sequences is achieved. The enriched oligonucleotides obtained are finally sequenced and their binding affinities to the target are assessed to find the best sequence [1,2].

Figure 1.
SELEX usually starts with the incubation of the target molecule and a primary library composed of ssRNA/ssDNA oligonucleotides under specific conditions. Unbound sequences are eluted, and target-bounded sequences are amplified by PCR or RT-PCR. These selected sequences are used in subsequent SELEX rounds until the population achieves the required affinity. The final product is sequenced and the affinity of aptamer-target complex is characterized.
The affinity of aptamer–target complex can be characterized by label-free or label-based strategies, through the calculation of dissociation constant (Kd), conformational changes, kinetics, and thermodynamics [7].
Aptamers can exhibit high affinity and low Kd (usually in pico- to nanomolar range) [2], with a similar or even superior target binding affinity than antibodies. In fact, they are often mentioned as chemical antibodies and offer several inherent advantages, as they can be produced over a wide range of targets, display low toxicity and immunogenicity, low molecular weight, good solubility and tissue permeability, high tolerance to environmental conditions (temperature, pH), conformational flexibility, easy modification and conjugation with other molecules, as well as an economical and reproducible synthesis, avoiding animals’ dependency for production. Once an aptamer is selected and characterized, it can be produced at a large scale, at low cost, and with low batch inter variability [1,2]. Therefore, numerous aptamers have been selected and applied for cancer, immune, and microbiota research.
In this review, the use of aptamers in research was assessed, especially in the area of cancer, immune activation, and microbiota.
2. Cancer Therapeutics
Aptamers can be applied as therapeutic agents acting mostly as antagonists, binding to the target, and blocking its function, but they can also be applied as agonists, stimulating the targets’ function [8,9,10]. As therapeutic agents, the features of aptamers potentiate their utilization on drug delivery systems for human disease treatment [9,11], as they are capable of transporting a variety of therapeutic agents as small molecules, peptides, or toxins [11].
However, in vivo applications of aptamers actually possess some issues. As an example, the conformation of the target during in vitro selection could be different to the conformation under in vivo conditions, affecting its ability in binding. Also, they have short half-life in vivo due to nuclease sensitivity and fast renal filtration [9,10,12,13].
Nowadays, cancer remains one of the deadliest diseases, with a limited number of reliable biomarkers, as well as reduced specificity, affecting healthy tissues. Examples such as chemotherapy or antagonism, like DNA damage repair drugs, can induce modification in the microenvironment, which can lead to an opposite effect, triggering invasion and/or tumor metastases [6].
In view with this, more efficient, specific, and refined therapies are being studied, targeting different hallmarks of the cancer. One of the most promising is the use of oligonucleotide aptamers, which can bind to specific membrane receptors to start the intended action, having already shown solid results against tumor growth and metastatic capacity, as well as modulating the tumor microenvironment to become more susceptible and easier to treat. This methodology also allows combination with other molecules, such as antibodies, which can enhance and potentiate the response. In fact, studies have shown higher biocompatibility and more refined targeting toward the tumor site, also reducing the possibility of developing resistance to treatment [6].
There are several aptamers being tested in different cancer types, as presented in Table 1, as well as the target and their application (diagnostic, therapy, or both).

Table 1.
Main aptamers used in different cancer sites, their targets and applications.
For breast cancer, some aptamers have been studied. HeA2-3 is an aptamer used to target HER2, one of the most relevant receptors against this cancer, which inhibits the growth and survival of breast cancer cells [14]. PNDA-3 aptamer targets periostin and effectively inhibits breast cancer cell invasion and metastasis in vivo, despite limited in vitro growth effects [15] Two aptamers target the MNK1, an essential component for cancer growth, apMNK2F, which has proven to be effective in decreasing both tumor size and the incidence of metastases in murine breast cancer models [16], and apMNK3R [17] The aptamer ex-50.T is used to target the exosomes released by cancer tissue, reducing the molecular pathways that promote cancer proliferation such as AKT/Pi3K pathway [18]. In addition, 2′-Fluoropyrimidines-RNA aptamer is used conjugated with nanoparticles and allows treatment with photothermal therapy in triple-negative breast cancer cells [19]. The AS1411 aptamer targets nucleolin, which leads to the reduction in angiogenesis capacity, i.e., the capacity of producing new vessels to create a better microenvironment for tumoral development [20].
In colorectal cancer, four aptamers with the highest potential were identified. These were aptamer YJ-1, which binds to Carcinoembryonic antigen (CEA) and death receptor 5 and leads to the inhibition of tumor growth and migration potential [21]; apPDGF-BB, which is an aptamer for PDGF-BB and is important in reducing proliferation capacity [22]; another aptamer studied is apPD-1, which targets the PD-1 receptor, which leads to the activation of immune system [23]; and the last aptamer is NOX-A12, which inactivates CXCL12, showing promising results in the following studies [24].
Lung cancer is another cancer with high concern for new therapeutics due to its aggressiveness and lack of early detection strategies. However, several candidates have been identified as promising recognition elements. For example, aptPD-L1 targets the PD-L1 receptor and regulates the tumor microenvironment, stimulating the immune system cells [25] and AP-74 M-545 aptamer is used against galectin-1, also stimulating the immune system. In LL/2 tumors in vivo, it effectively inhibits tumor growth by reducing T cell apoptosis [26]. Two aptamers known as TBA353 and TBA535 target TBA, leading to reduced proliferation and invasive potential in lung cancer cell lines [27] and a PEGylated aptamer called RA16 is used specifically to bind to non-small-cell lung cancer cells, leading to tumor growth suppression [28].
Cancer of the liver also has some potential aptamers. For example, aptamer AS1411 [29] and AS1411-modified [30] target nucleolin and galectin-14, respectively, leading to a reduction in the tumoral features, in both in vitro and in vivo models, and the CL-4RNV616 aptamer targets EGFr, reduces proliferation, and promotes cell apoptosis [31].
Considering renal cancer, some aptamers are described. SW-4 and SW-4b are two aptamers used to modulate the endocytosis process of renal cancer cells, inducing their apoptosis and cell cycle [32]. The aptamer AS1411, currently in phase II clinical trials which have shown high potentiality in other cancers, can also induce a reduction in aggression [33].
Regarding prostate cancer, several studies have focused on the finding of new aptamers, since this cancer affects millions of men. Apt63 aptamer targets APT5B to reduce its metastatic potential and leads to cancer cell death, with no toxicity in non-tumoral cells, both in cell lines and in xenograft models [34]; two aptamer target PSMA, namely A9g [35] and other PSMA-specific aptamers [36], which is considered to have high potential against this cancer type, since PSMA is specific to prostate cancer tissue; combining A9g with AGRO100, the inhibition of cancer features seems to be highly improved [37]; CG3 is an aptamer that inhibits the proliferation of blood cancer cells originated in prostate, showing potential both as a diagnostic tool for prostate cancer and as a drug delivery therapy system [38]; the AS1411 also expresses impact on prostate cancer [39]; finally, the shRNA/PEI-PEG-APT/DOX conjugates can detect and inactivate PSMA-positive cancer cells [40].
Regarding other types of cancer, there are aptamers that exhibit potential in treatment and/or diagnosis. For glioblastoma, 40L and A40s target CD133 and allow the reduction in metastasis process [41]; in oral cancer, the inhibition of Heparinase by the anti-HPSE1 seemed to be effective to reduce invasion capacity and lead to cytotoxicity of cancer cells [42]; regarding cervical cancer, A2 aptamer targets E6 and E7 oncogenes, which lead to tumor regression and growth inhibition [43]; concerning bladder cancer, alncRNA aptamer inhibits TF transcription and oncogene activity, leading to reduced cancer features [44]; and, in view of gastric cancer, the use of trimeric aptamer targets to ErbB-2/HER2, showing high efficiency when combined in the trimeric form, reducing endocytosis and growth rate [45]. Leukemia has some potential aptamers, such as AS1411 [46], apβ-arrestin2 for the reduction in the progression of chronic myeloid leukemia [47], and ssRNA MLL-1 to target MLL-1, leading to cancer growth reduction [48]. For Hodkin’s lymphoma, an aptamer for CD30 is used to diagnose the disease, namely the CD-RNA aptamer with gold nanospheres called HAuNS [49]. In the myeloma disease, the use of NOX-A12, which targets CXCL12, leads to a reduction in cell adhesion and signaling processes, as well as cancer cell growth and proliferation [17], a phase IIa study in 28 relapsed/refractory multiple myeloma patients found NOX-A12, whether given alone or with bortezomib/dexamethasone, to be safe and well-tolerated [24]. Regarding skin cancer, F3B aptamers inhibit MMP-9, which reduces cell proliferation and extracellular matrix remodeling as well as allowing of the detection, being a theragnostic aptamer [50].
Pancreatic cancer is one of the leading causes of cancer death worldwide and is often diagnosed at an advanced stage with local invasion and distant metastases, which contribute to the high mortality rates associated with the disease [75,76]. The lack of effective treatments and diagnostic methods further compounds the problem, underscoring the need to develop targeted therapeutics and diagnostic approaches to improve patient outcomes [63]. Aptamers are, therefore, an attractive option for developing more accurate diagnostic tests and more effective treatments for pancreatic cancer.
The E07 aptamer, which targets the epidermal growth factor receptor (EGFR), can be used to improve the effectiveness of gemcitabine, having the potential to be a more effective treatment for pancreatic cancer than current therapies using gemcitabine alone [51]. Two RNA aptamers (P19 and P1) were selected to specifically recognize pancreatic cancer cells and deliver a duplex C/EBPα-saRNA molecule to target tumor growth suppression. In animal models, P19-C/EBPα-saRNA induced a 40% decrease in tumor growth with no toxicity [52]. A TfR aptamer-C/EBPα-saRNA conjugate used to treat advanced PDAC in a mouse model, also showed a significant reduction in tumor size [53]. Aptamer XQ-2d enables the conjugate to specifically recognize and enter PDAC cells, minimizing its side-effects on normal tissues. XQ-2d-His-SH2 CM-(Arg)9 conjugate impairs the ability of pancreatic cancer cells to undergo epithelial-mesenchymal transition (EMT), inhibiting the proliferation, invasion, and metastasis of PDAC cells [54].
SQ2, is an RNA aptamer with the capacity to be internalized by tumoral cells through ALPPL2-mediated clathrin-independent pathways [55]. This aptamer is capable of carrying a cytotoxic drug, demonstrating potential applications in imaging and therapeutics for all cancers that have aberrant expression of ALPPL2 [55,56]. The aptamers (1 and 146) colocalized with existing Cancer Stem Cell (CSC) markers, such as CD44, CD24, EpCAM, and CD133, which can recognize CSC marker-enriched Circulating Tumor Cells (CTCs) in pancreatic cancer, suggesting potential applications in pancreatic cancer diagnosis and treatment [57]. The X-aptamer targeting the Thy-1 membrane glycoprotein (THY1), overexpressed in pancreatic cancer cells, provides a non-immunogenic and specific option in the context of biomarker development and clinical application [58].
Additionally, some other aptamers are being tested. The aptamer aptacoy binds to the TfR, leading to an enhancement in the therapeutic response of Doxorubicin [59]; NOX-E36 aptamer connects to CCL2 and can be used not only in pancreatic tumors, but also in Diabetes mellitus type 2 (T2DM) [60]; cancer cell secretome can be detected and differentiated from healthy cells using the cyclophilin B aptamer [61]; the P12FR2 aptamer binds to PAUF and reduces the growth and metastatic capacity of cancer cells [62]; the use of NOX-A12, which targets CXCL12, leads to the reduction in cell adhesion and signaling processes, as well as cancer cell growth and proliferation [63].
Aside from the specificity for cancer sites, there are some aptamers that are not specific to target tissues, but cancer cells. The aptamers 4-1BB, which targets TNFSF9 [64], and OX40, which binds to TNFSF9 [65], promote immunotherapy treatment. GL21.T is another aptamer which allows the improvement of cancer cells to endocytose drugs, as well as to promote tumor selectivity [66]; Two different aptamers, Apt-siRNA [67] and CAR-like multivalent aptamer [68], do not have a specific target, although they have the capacity to lead to anticancer effects; bi-functional aptamers target opsonin C3b/iC3b, resulting in cancer cell destruction [69]; the AS1411 aptamer and cyclic peptide RGD bind to Alpha-v beta 3 (αvβ3), inhibiting cell growth and proliferation [70]; the H02 aptamer targets α5β1, reducing invasion capacity [71]; and dox-incorporated multivalent aptamer-siRNA conjugates produce powerful anticancer effects [72].
Conversely, tumor cells express shared tumor-specific antigens, which can induce immune responses when presented with MHC molecules on the cell surface. MAGE-A3 is a tumor-specific antigen that is highly expressed in several types of cancer, including melanoma, esophageal carcinoma, gastric carcinoma, multiple myeloma, breast cancer, and pancreatic cancer, often associated with malignancy and poor prognosis. Wang et al. (2016) described a DNA aptamer against the shared tumor-specific antigen MAGE-A3, which discriminates multiple types of cancer cells from normal cells (Ap52) [73]. In 2021, Wang and his team demonstrated that Ap52 can effectively penetrate cancer cells and selectively deliver the anticancer drug Doxorubicin (Dox), causing cytotoxicity to multiple types of cancer cell, becoming a promising therapeutic delivery system for various tumors [74]. In addition, aptamers may provide combinatorial manipulation of cell immunity and drug targeting for cancer therapy, and a combination of aptamers targeting various shared tumor-specific antigens may be used to combat tumor heterogeneity [74,77].
2.1. Applications in Immune Diseases
The versatility of aptamers created an important opportunity for diverse application in the field of immunology both in diagnosis and therapy. Considering diagnostic techniques aptamers might be used for two main purposes: detection [49,78,79] and separation [80,81,82]. In therapy, aptamers have two other principal functions that include being a therapeutic drug itself [83] or being responsible for taking fewer drugs to treat the affected cell or organ and, in this way, reducing the need for higher doses of drugs that have relevant secondary effects [84,85,86].
The gold standard techniques for immune diagnosis are flow cytometry and Enzyme-linked Immunosorbent Assay (ELISA) [87] and aptamers present features useful for both strategies, offering a broad range of solutions for the detection of biomolecules like proteins, including immunoglobulins or membrane receptors [49,83,85,88], cells [80,81,89], or even Extracellular Vesicles (EVs) that have gained popularity in fighting diseases like cancer. For EVs, CD63 is being consistently used as one of the most promising targets for aptamers on the surfaces of EVs [79,82]. Aptamers might also be used for cell separation using, as an example, sorting equipment accoupled to flow cytometry, which is useful both as a diagnosis technique [80,81] and also for therapy purposes in techniques like immunotherapy or HIV therapy [80,90].
Nozari et al. already reported a broad range of immune cell markers that include the most common markers, like CD4 or CD8, and also some unusual ones. This might be useful for diagnosis, cell sorting, and even therapy in a huge variety of diseases like cancer, HIV, or autoimmune conditions [49] (Table 2).
Considering therapeutical strategies, aptamers might be a drug by themselves and although there is potential for exclusive use of aptamers as a therapy [83], the most common approach is using aptamers together as co-adjuvant therapy. For example, aptamer- T cells together with anti-PD1 immune-checkpoint monoclonal antibodies have a synergetic effect in reducing cancer, in vitro and in rats, and aptamer-linked small-interfering RNA chimeras to selectively knock down genes in mouse breast cancers [91,92]. This approach allows a decrease in dose in immunotherapy applications and, in this way, reduces the secondary effects of therapies that might cause a lot of discomfort and distress in patients [83,93,94]. Aptamers are used not only in order to destroy the disease agent, like a cancer cell [89], but are often used as immune modulators, reducing immune activity [86], targeting or influencing immune checkpoint inhibitors [89,95,96], and/or promoting immune cell activity and infiltration [91,97].

Table 2.
Main aptamers used in different immune diseases.
Table 2.
Main aptamers used in different immune diseases.
| Disease | Aptamer | Target | Technique | Application | References | ||
|---|---|---|---|---|---|---|---|
| Diagnostic | Therapy | Theranostic | |||||
| Auto-immune Diseases | NA | IgE | Electrochemical Sensor | X | [98,99,100] | ||
| NA | IgE | Optical Biosensor | X | [101,102] | |||
| NA | IL-2 | - | X | [103] | |||
| Ap52 | MAGE-A3 | Drug Delivery | X | [73] | |||
| Multiple Cancers | rvCD71apt | CD71 | Cell Separation | X | [81] | ||
| rvCD8apt | CD8 | X | |||||
| NA | CD63 (Exossome) | Exossome Separation | X | [82] | |||
| NA | CD63 (Exossome) | Flow Cytometry | X | [104] | |||
| Acute Promyelocytic Leukemia | NA | NB4 Cell Line | Flow cytometry | X | [78] | ||
| Lung Cancer | NA | CD63 (Exossome) | Flow Cytometry | X | [79] | ||
| HIV/Liquid Tumors | CD4 Aptamer | CD4 | Cell Separation | X | [80] | ||
| HIV | CCR5 Aptamer | CCR5 | Drug | X | [83] | ||
| Melanoma | AS1411 | Nucleoline | Drug Delivery | X | [84] | ||
| Graft versus Host Disease | CD8AP17s | CD8 | Drug Delivery | X | [86] | ||
| Breast Cancer | PDGFRβ | Lymphocytes | Drug | X | [89] | ||
| Glioblastoma | 4-1BB–OPN | Osteopontin | Drug | X | [97] | ||
| Infection and Inflammation | NA | C-Reactive Protein | Optical Biosensor | X | [105] | ||
Abbreviations: NA: not applicable; IgE: Immunoglobulin E; IL-2: Interleukin-2; MAGE-A3: Melanoma antigen family A, 3.
Aptamers are also showing a lot of potential for drug delivery therapies, not only in immune context, but in general. This happens because aptamers are very versatile as drug-targeting tools and can be used with at least three relevant functions. Aptamers might be carriers for other drugs, allowing the encapsulation of drugs [106], might be used for targeting, allowing the drug to reach in higher quantity the compromised cell/organ [84,85,86], and also might be used as a gating system that induces the release of the therapeutical drug when arriving at the target [106].
The scientific community has already gathered a few examples of how useful strategies using aptamers can be. Despite that, the scale of production for aptamers is far away from competing against the scale and optimization that big pharmaceutical companies have already developed for antibodies, making the use of aptamers more of an industrial problem than a scientific issue [90,95,107].
2.2. Microbiota and Infection
The human microbiome is a vast collection of microorganisms that live inside the human body, with bacteria, viruses, fungi, protozoa, and archaea all involved in this environment, playing a crucial role in human health and disease [108]. The microbiota is involved in a variety of physiological processes, including digestion, metabolism, immune system development, and protection against pathogens, and their composition is influenced by several factors such as diet, age, genetics, and environment [108,109]. However, disruption of the microbiota can lead to an increased risk of infection, which is a major cause of morbidity and mortality worldwide, and it has also been associated with several diseases like cardiovascular and gastrointestinal diseases, neurological disorders, cancer, among others [110]. Understanding the microbiota and its interactions with the host is becoming an active area of research, with potential applications in the diagnosis and treatment of diseases. Therefore, the development of new tools for the detection and treatment of infections is of utmost importance.
The microbiota plays a crucial role in protecting against pathogens and preventing infections, competing with pathogenic microorganisms for resources and space and preventing them from colonizing and infecting the host, producing antimicrobial peptides that can kill or inhibit the growth of pathogenic microorganisms and modulate the immune system’s response to pathogens [111,112]. However, when a disruption of this microbiota occurs, it can lead to harmful situations, particularly respiratory, gastrointestinal, and skin infections [111]. One example is antibiotics that can disrupt this balance, promoting pathogenic bacteria overgrowth and increased risk of infection, with Streptococcus and Staphylococcus being among the most common [113]. Also, the overgrowth of Clostridium difficile can lead to a severe infection, which can cause symptoms ranging from diarrhea to life-threatening complications [114].
Aptamers have been increasingly used in microbiome diagnostics, where they can identify and characterize the microorganisms present in a specific environment, such as the human gut, skin, or oral cavity, because of their unique binding properties and their ability to detect specific microbial biomarkers in complex biological samples [1]. Their use in the gut microbiota field has several advantages, including high sensitivity and specificity, easy modulation and synthesis, low cost, and high stability, making them valuable tools for studying the complex interactions between the microbiota and the host and help to develop personalized microbiota-based therapies [1,2].
One application of aptamers in microbiota research is the identification and quantification of specific microbial species or strains [115]. Aptamers can be designed to specifically bind to the surface molecules or antigens of certain bacteria or other microorganisms, facilitating the identification and quantification of those organisms [115]. This method can be particularly useful in identifying and monitoring specific pathogenic or beneficial microorganisms in complex microbial communities, with studies by Hu et al. and Song et al. describing this novel approach in strains like Bifidobacterium bifidum (CCFM641-5) and Lactobacillus casei, at the same time, demonstrating the potential of aptamer-based strategies for the rapid and accurate detection and quantification of bacteria in food products [116,117].
Another application of aptamers in microbiome research is the study of microbial metabolites and signaling molecules, where aptamers can be designed to specifically recognize and bind to small molecules, such as short-chain fatty acids, neurotransmitters, and other signaling molecules produced by the microbiota, where aptamers can profile and characterize at the species, strain, or function level. They can be used in combination with high-throughput sequencing technologies, such as 16S rRNA sequencing to identify and quantify specific microbial populations in the gut microbiota and to identify and quantify specific microbial metabolites, which are important for gut health [118]. This approach allows researchers to better understand the interactions between the microbiome and the host and how they influence overall health and disease, providing a more accurate diagnosis and better understanding of the disease [119,120,121,122,123].
Aptamers can also be used in therapy to target and modulate specific microorganisms, or to target drug delivery in the gut microbiota. For example, Wandtke et al. (2015) designed aptamers to selectively bind to viral proteins or other molecules involved in viral replication and infection with high specificity and sensitivity, preventing their replication or disrupting their functions [124]. Here, the authors discuss several examples of aptamer-based assays developed for the detection of hepatitis C and Human Immunodeficiency Virus (HIV) and also how this novel method could be used as a therapeutic agent for viral infections, such as influenza, with A-20 and V46 aptamers [119,120], dengue fever, with S15G3 aptamer [121], and Zika virus [122]. Additionally, aptamers can be used to deliver therapeutic molecules, such as antibiotics, antivirals, antifungals, or probiotics, specifically to the site of infection or dysbiosis in the human microbiota, reducing side effects and increasing efficacy [123]. They can also be used to deliver therapeutic nucleic acids, such as small interfering RNA (siRNA) or microRNA (miRNA), to downregulate the expression of viral genes or host cell genes that are essential for infection [124]. Several studies have also demonstrated the use of aptamers in detecting specific microbial targets in gastrointestinal dysbiosis, such as detecting Clostridium difficile with the ARH1t6 aptamer [125] in addition to using others to detect gut pathogens such as Salmonella [126] and Escherichia coli [127]. Aptamers have also shown potential in the treatment of gastrointestinal diseases, specifically targeting and inhibiting the growth or virulence of pathogens such as Helicobacter pylori, a bacterium known to cause peptic ulcer and gastric cancer. The aptamer HPA-2 has shown potential for the detection of H. pylori, demonstrating high specificity within a limit of detection of 88 cfu/mL [128]. In addition to the mentioned applications, aptamers can also be used to monitor changes in microbiota and detecting shifts in the microbial community, allowing for the identification of potential health risks or the monitoring of the effectiveness of treatments [129].
The use of aptamers in gut microbiota research is still in its early stages and there is significant potential for future applications. Aptamers have the potential to be used as a diagnostic tool for gut microbiota-related disease, such as inflammatory bowel disease and colorectal cancer, but also for gut microbiota-related diseases, such as antibiotic-resistant infections [123]. Moreover, aptamers can be integrated with other technologies, such as high-throughput sequencing and mass spectrometry, to provide a more comprehensive analysis of the gut microbiota [130]. This approach has the potential to provide a more detailed understanding of the gut microbiota and its role in human health and disease.
2.3. Final Remarks
The high affinity displayed by aptamer over their targets, and its facility in conjugating with labelling molecules makes them a suitable tool, acting as recognition elements, for biomarker discovery and disease diagnosis, especially in imaging and biosensor devices [8,9,11].
In the field of both in vivo and in vitro bioimaging, aptamers have advantages over antibodies, owing to their good solubility and low molecular weight when compared with antibodies, which allows its blood clearance and easy tissue penetration abilities [11,12].
Biosensors that use aptamers as recognition elements are called aptasensors and generally rely on the conformational changes in the aptamers after binding to its target. They have been applied in point-of-care miniaturized devices to detect environmental contaminants, cancer biomarkers or drugs, as example [85,131,132]. Additionally, they can also be used as a theragnostic tool, allowing to detect and treat cancer [133]. In terms of diagnosis, regardless the field of pathology, the main advantage in the application of the sensing strategy relies on early detection. Early detection allows, by itself a higher rate of success in overall survival. Moreover, since most diseases lack proper early diagnosis due to a lack of specific biomarkers, biosensors also allow the possibility of multiplexing analysis, helping eliminate inaccurate diagnoses [134], or even diagnosing diseases with higher feasibility [135]. Ultimately, this translates to proper and personalized therapy with fewer side effects and lower overall costs.
3. Conclusions
Aptamers, short synthetic single-stranded RNA or DNA molecules, have emerged as powerful tools with a wide range of applications in various fields, including cancer therapeutics, immune diseases, microbiota research, and diagnosis.
Within cancer therapeutics, aptamers have shown tremendous promise. They can act as antagonists or agonists, blocking or stimulating specific targets, making them valuable for drug delivery systems. Aptamers offer a more targeted and potentially less toxic alternative to traditional cancer treatments like chemotherapy. Their application spans various cancer types, with numerous aptamers having been identified to target specific markers associated with breast, colorectal, lung, liver, renal, prostate, and other cancers. The versatility of aptamers allows for their combination with other therapeutic agents, potentially enhancing treatment responses and reducing the likelihood of resistance.
Aptamers have also been found to have applications in the treatment of immune diseases, where they can be employed for diagnosis and therapy. They can detect immune markers, facilitating the diagnosis of conditions such as HIV or autoimmune diseases. Aptamers can also modulate immune responses, influence immune checkpoint inhibitors, or stimulate immune cells, contributing to more effective immunotherapies. Furthermore, aptamers have potential in drug delivery strategies, optimizing drug targeting, and minimizing side effects.
In microbiota research and infection control, aptamers play a pivotal role in identifying and quantifying specific microbial species or strains. Their high specificity and sensitivity allow for the rapid detection of pathogens or beneficial microorganisms in complex microbial communities. Aptamers can also target microbial metabolites and signal molecules, aiding in understanding the intricate interactions between the microbiome and host health. Additionally, they have applications in therapy, where they can target and modulate specific microorganisms or deliver therapeutic molecules precisely to infection sites, reducing side effects and improving treatment efficacy. Aptamers have the potential to revolutionize the study and management of microbiota-related diseases and infections.
Finally, aptamers are valuable tools for disease diagnosis, offering high affinity, ease of conjugation with labeling molecules, and versatility in biosensors and bioimaging. They outperform antibodies in various diagnostic applications, such as biosensors for detecting environmental contaminants, cancer biomarkers, or drugs. Aptasensors based on aptamer-conformational changes have been deployed in point-of-care devices, providing rapid and accurate detection, even in resource-limited settings. These aptamer-based diagnostic tools hold great promise for improving early disease detection and personalized medicine.
Overall, aptamers have transcended the confines of the laboratory and have emerged as indispensable components in various scientific disciplines. Their versatility, specificity, and applicability continue to expand, offering innovative solutions for tackling complex challenges in cancer research, immunology, microbiota analysis, and disease diagnosis. As research in aptamers continues to evolve, we can anticipate even more breakthroughs and advancements with profound implications for healthcare, diagnostics, and therapeutic interventions.
Author Contributions
A.P.S.: Data research, original draft preparation; A.C.R.: Data research, original draft preparation; C.A.: Data research, original draft preparation; M.C.C.G.C.: Data research, original draft preparation; P.P.P.: Data research, original draft preparation; R.V.: Data research, original draft preparation; R.F.: Reviewing and editing; P.B. (Pedro Barata): Reviewing; Á.G.: Editing; S.R.: Research; D.M.-M.: Research; P.B. (Pilar Baylina): Conceptualization, research, reviewing, and editing. A.C.P.: Conceptualization, reviewing, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
Pilar Baylina (P.B.) acknowledges on the behalf of the authors the support of Fundação para a Ciência e Tecnologia (FCT), Portuguese Government, under the Strategic Project Reference: UID/BIM/04293/2013. Pilar Baylina (P.B.) and R.F. was also supported by FEDER/02/SAICT/2020/072560. Ana Cláudia Pereira (A.C.P.) acknowledges the support of Fundação para a Ciência e Tecnologia (FCT), Portuguese Government, under the Strategic Project Reference: 2022.09032.PTDC. André P. Sousa acknowledges FCT for the PhD grant 2022.12441.BD. Ana C. Rocha acknowledges FCT for the PhD grant 2021.06521.BD. Patrick P. Pais acknowledges FCT for the PhD grant 2021.09498.BD. Susana Ramalho acknowledges LaBMI for the PhD grant LABMI/BI/2021/01 (NEURO4COVID).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data presented in this study are openly available in PubMed, NCBI. The DOI for each study is presented on the references.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kumar Kulabhusan, P.A.-O.; Hussain, B.; Yuce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef] [PubMed]
- Byun, J. Recent Progress and Opportunities for Nucleic Acid Aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Sousa, D.A.; Carneiro, M.; Ferreira, D.; Moreira, F.T.C.; Sales, M.G.F.; Rodrigues, L.R. Recent Advances in the Selection of Cancer-Specific Aptamers for the Development of Biosensors. Curr. Med. Chem. 2022, 29, 5850–5880. [Google Scholar] [CrossRef]
- Agnello, L.; Camorani, S.; Fedele, M.; Cerchia, L. Aptamers and antibodies: Rivals or allies in cancer targeted therapy? Explor. Target. Antitumor Ther. 2021, 2, 107–121. [Google Scholar] [CrossRef]
- Thevendran, R.; Citartan, M. Assays to Estimate the Binding Affinity of Aptamers. Talanta 2022, 238, 122971. [Google Scholar] [CrossRef]
- Ozer, A.; Pagano, J.M.; Lis, J.T. New Technologies Provide Quantum Changes in the Scale, Speed, and Success of SELEX Methods and Aptamer Characterization. Mol. Ther. Nucleic Acids 2014, 3, e183. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef]
- Radom, F.; Jurek, P.M.; Mazurek, M.P.; Otlewski, J.; Jelen, F. Aptamers: Molecules of great potential. Biotechnol. Adv. 2013, 31, 1260–1274. [Google Scholar] [CrossRef]
- Santosh, B.; Yadava, P.K. Nucleic acid aptamers: Research tools in disease diagnostics and therapeutics. Biomed. Res. Int. 2014, 2014, 540451. [Google Scholar] [CrossRef]
- Sun, H.; Zu, Y. A Highlight of Recent Advances in Aptamer Technology and Its Application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef]
- Gijs, M.; Penner, G.; Blackler, G.B.; Impens, N.R.; Baatout, S.; Luxen, A.; Aerts, A.M. Improved Aptamers for the Diagnosis and Potential Treatment of HER2-Positive Cancer. Pharmaceuticals 2016, 9, 29. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, I.S.; Park, S.A.; Kim, Y.; Lee, J.E.; Noh, D.Y.; Kim, K.T.; Ryu, S.H.; Suh, P.G. Periostin-binding DNA aptamer inhibits breast cancer growth and metastasis. Mol. Ther. 2013, 21, 1004–1013. [Google Scholar] [CrossRef]
- Pinto-Diez, C.; Ferreras-Martin, R.; Carrion-Marchante, R.; Klett-Mingo, J.I.; Garcia-Hernandez, M.; Perez-Morgado, M.I.; Sacristan, S.; Barragan, M.; Seijo-Vila, M.; Tundidor, I.; et al. An optimized MNK1b aptamer, apMNKQ2, and its potential use as a therapeutic agent in breast cancer. Mol. Ther. Nucleic Acids 2022, 30, 553–568. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fu, X.; Huang, J.; Zeng, P.; Huang, Y.; Chen, X.; Liang, C. Advances in Screening and Development of Therapeutic Aptamers Against Cancer Cells. Front. Cell Dev. Biol. 2021, 9, 662791. [Google Scholar] [CrossRef]
- Esposito, C.L.; Quintavalle, C.; Ingenito, F.; Rotoli, D.; Roscigno, G.; Nuzzo, S.; Thomas, R.; Catuogno, S.; de Franciscis, V.; Condorelli, G. Identification of a novel RNA aptamer that selectively targets breast cancer exosomes. Mol. Ther. Nucleic Acids 2021, 23, 982–994. [Google Scholar] [CrossRef]
- Aldag, J.; Persson, T.; Hartmann, R.K. 2’-Fluoro-Pyrimidine-Modified RNA Aptamers Specific for Lipopolysaccharide Binding Protein (LBP). Int. J. Mol. Sci. 2018, 19, 3883. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef]
- Viraka Nellore, B.P.; Kanchanapally, R.; Pramanik, A.; Sinha, S.S.; Chavva, S.R.; Hamme, A., 2nd; Ray, P.C. Aptamer-conjugated graphene oxide membranes for highly efficient capture and accurate identification of multiple types of circulating tumor cells. Bioconjug Chem. 2015, 26, 235–242. [Google Scholar] [CrossRef]
- Sae-Lim, S.; Soontornworajit, B.; Rotkrua, P. Inhibition of Colorectal Cancer Cell Proliferation by Regulating Platelet-Derived Growth Factor B Signaling with a DNA Aptamer. Asian Pac. J. Cancer Prev. 2019, 20, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Ahmadyousefi, Y.; Malih, S.; Mirzaee, Y.; Saidijam, M. Nucleic acid aptamers in diagnosis of colorectal cancer. Biochimie 2019, 156, 1–11. [Google Scholar] [CrossRef]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef]
- Lai, W.Y.; Huang, B.T.; Wang, J.W.; Lin, P.Y.; Yang, P.C. A Novel PD-L1-targeting Antagonistic DNA Aptamer With Antitumor Effects. Mol. Ther. Nucleic Acids 2016, 5, e397. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.T.; Liang, C.H.; Yu, J.H.; Huang, K.C.; Tung, C.H.; Wu, J.E.; Wu, Y.Y.; Chang, C.H.; Hong, T.M.; Chen, Y.L. A DNA Aptamer Targeting Galectin-1 as a Novel Immunotherapeutic Strategy for Lung Cancer. Mol. Ther. Nucleic Acids 2019, 18, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Esposito, V.; Russo, A.; Vellecco, V.; Bucci, M.; Russo, G.; Mayol, L.; Virgilio, A.; Galeone, A. Thrombin binding aptamer analogues containing inversion of polarity sites endowed with antiproliferative and anti-motility properties against Calu-6 cells. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2645–2650. [Google Scholar] [CrossRef]
- Wang, H.; Qin, M.; Liu, R.; Ding, X.; Chen, I.S.Y.; Jiang, Y. Characterization of A Bifunctional Synthetic RNA Aptamer and A Truncated Form for Ability to Inhibit Growth of Non-Small Cell Lung Cancer. Sci. Rep. 2019, 9, 18836. [Google Scholar] [CrossRef]
- Trinh, T.L.; Zhu, G.; Xiao, X.; Puszyk, W.; Sefah, K.; Wu, Q.; Tan, W.; Liu, C. A Synthetic Aptamer-Drug Adduct for Targeted Liver Cancer Therapy. PLoS ONE 2015, 10, e0136673. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef]
- Wang, T.; Philippovich, S.; Mao, J.; Veedu, R.N. Efficient Epidermal Growth Factor Receptor Targeting Oligonucleotide as a Potential Molecule for Targeted Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 4700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Z.; Xie, L.; Zhang, Y.; Deng, T.; Li, J.; Liu, J.; Xiong, W.; Zhang, L.; Zhang, L.; et al. Molecular Recognition and In-Vitro-Targeted Inhibition of Renal Cell Carcinoma Using a DNA Aptamer. Mol. Ther. Nucleic Acids 2018, 12, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N., Jr.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Speransky, S.; Serafini, P.; Caroli, J.; Bicciato, S.; Lippman, M.E.; Bishopric, N.H. A novel RNA aptamer identifies plasma membrane ATP synthase beta subunit as an early marker and therapeutic target in aggressive cancer. Breast Cancer Res. Treat. 2019, 176, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Hernandez, L.I.; Thomas, G.S.; Long, M.E.; Rockey, W.M.; Howell, C.A.; Chen, Y.; Hernandez, F.J.; Liu, X.Y.; Wilson, M.E.; et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol. Ther. 2014, 22, 1910–1922. [Google Scholar] [CrossRef]
- Jiao, Y.; Xu, P.; Luan, S.; Wang, X.; Gao, Y.; Zhao, C.; Fu, P. Molecular imaging and treatment of PSMA-positive prostate cancer with (99m)Tc radiolabeled aptamer-siRNA chimeras. Nucl. Med. Biol. 2022, 104–105, 28–37. [Google Scholar] [CrossRef]
- Singh, S.S. Preclinical pharmacokinetics: An approach towards safer and efficacious drugs. Curr. Drug Metab. 2006, 7, 165–182. [Google Scholar] [CrossRef]
- Marangoni, K.; Neves, A.F.; Rocha, R.M.; Faria, P.R.; Alves, P.T.; Souza, A.G.; Fujimura, P.T.; Santos, F.A.; Araujo, T.G.; Ward, L.S.; et al. Prostate-specific RNA aptamer: Promising nucleic acid antibody-like cancer detection. Sci. Rep. 2015, 5, 12090. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Salipur, F.R.; Shams, M.; Forsthoefel, M.K.; Bates, P.J. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015, 9, 1392–1405. [Google Scholar] [CrossRef]
- Kim, E.; Jung, Y.; Choi, H.; Yang, J.; Suh, J.S.; Huh, Y.M.; Kim, K.; Haam, S. Prostate cancer cell death produced by the co-delivery of Bcl-xL shRNA and doxorubicin using an aptamer-conjugated polyplex. Biomaterials 2010, 31, 4592–4599. [Google Scholar] [CrossRef]
- Affinito, A.; Quintavalle, C.; Esposito, C.L.; Roscigno, G.; Vilardo, C.; Nuzzo, S.; Ricci-Vitiani, L.; De Luca, G.; Pallini, R.; Kichkailo, A.S.; et al. The Discovery of RNA Aptamers that Selectively Bind Glioblastoma Stem Cells. Mol. Ther. Nucleic Acids 2019, 18, 99–109. [Google Scholar] [CrossRef]
- Vayrynen, O.; Piippo, M.; Jamsa, H.; Vaisanen, T.; de Almeida, C.E.B.; Salo, T.; Missailidis, S.; Risteli, M. Effects of ionizing radiation and HPSE1 inhibition on the invasion of oral tongue carcinoma cells on human extracellular matrices in vitro. Exp. Cell Res. 2018, 371, 151–161. [Google Scholar] [CrossRef]
- Cesur, O.; Nicol, C.; Groves, H.; Mankouri, J.; Blair, G.E.; Stonehouse, N.J. The Subcellular Localisation of the Human Papillomavirus (HPV) 16 E7 Protein in Cervical Cancer Cells and Its Perturbation by RNA Aptamers. Viruses 2015, 7, 3443–3461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Li, P.C.; Guo, J.; Huo, F.; Yang, J.; Jia, R.; Wang, J.; Huang, Q.; Theodorescu, D.; et al. Development of Novel Aptamer-Based Targeted Chemotherapy for Bladder Cancer. Cancer Res. 2022, 82, 1128–1139. [Google Scholar] [CrossRef] [PubMed]
- Mahlknecht, G.; Maron, R.; Mancini, M.; Schechter, B.; Sela, M.; Yarden, Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc. Natl. Acad. Sci. USA 2013, 110, 8170–8175. [Google Scholar] [CrossRef]
- Soundararajan, S.; Wang, L.; Sridharan, V.; Chen, W.; Courtenay-Luck, N.; Jones, D.; Spicer, E.K.; Fernandes, D.J. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol. Pharmacol. 2009, 76, 984–991. [Google Scholar] [CrossRef]
- Kotula, J.W.; Sun, J.; Li, M.; Pratico, E.D.; Fereshteh, M.P.; Ahrens, D.P.; Sullenger, B.A.; Kovacs, J.J. Targeted disruption of beta-arrestin 2-mediated signaling pathways by aptamer chimeras leads to inhibition of leukemic cell growth. PLoS ONE 2014, 9, e93441. [Google Scholar] [CrossRef]
- Ul-Haq, A.; Jin, M.L.; Jeong, K.W.; Kim, H.M.; Chun, K.H. Isolation of MLL1 Inhibitory RNA Aptamers. Biomol. Ther. 2019, 27, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Nozari, A.; Berezovski, M.V. Aptamers for CD Antigens: From Cell Profiling to Activity Modulation. Mol. Ther. Nucleic Acids 2017, 6, 29–44. [Google Scholar] [CrossRef]
- Kryza, D.; Debordeaux, F.; Azema, L.; Hassan, A.; Paurelle, O.; Schulz, J.; Savona-Baron, C.; Charignon, E.; Bonazza, P.; Taleb, J.; et al. Ex Vivo and In Vivo Imaging and Biodistribution of Aptamers Targeting the Human Matrix MetalloProtease-9 in Melanomas. PLoS ONE 2016, 11, e0149387. [Google Scholar] [CrossRef]
- Ray, P.; Cheek, M.A.; Sharaf, M.L.; Li, N.; Ellington, A.D.; Sullenger, B.A.; Shaw, B.R.; White, R.R. Aptamer-mediated delivery of chemotherapy to pancreatic cancer cells. Nucleic Acid. Ther. 2012, 22, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Huang, K.W.; Reebye, V.; Mintz, P.; Tien, Y.W.; Lai, H.S.; Saetrom, P.; Reccia, I.; Swiderski, P.; Armstrong, B.; et al. Targeted Delivery of C/EBPalpha -saRNA by Pancreatic Ductal Adenocarcinoma-specific RNA Aptamers Inhibits Tumor Growth In Vivo. Mol. Ther. 2016, 24, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Huang, K.W.; Andrikakou, P.; Vasconcelos, D.; Swiderski, P.; Reebye, V.; Sodergren, M.; Habib, N.; Rossi, J.J. Targeted Delivery of C/EBPalpha-saRNA by RNA Aptamers Shows Anti-tumor Effects in a Mouse Model of Advanced PDAC. Mol. Ther. Nucleic Acids 2019, 18, 142–154. [Google Scholar] [CrossRef]
- Liu, A.D.; Zhou, J.; Bi, X.Y.; Hou, G.Q.; Li, S.S.; Chen, Q.; Xu, H.; Cao, X. Aptamer-SH2 superbinder-based targeted therapy for pancreatic ductal adenocarcinoma. Clin. Transl. Med. 2021, 11, e337. [Google Scholar] [CrossRef]
- Dua, P.; Kang, H.S.; Hong, S.M.; Tsao, M.S.; Kim, S.; Lee, D.K. Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein. Cancer Res. 2013, 73, 1934–1945. [Google Scholar] [CrossRef] [PubMed]
- Dua, P.; Sajeesh, S.; Kim, S.; Lee, D.K. ALPPL2 Aptamer-Mediated Targeted Delivery of 5-Fluoro-2’-Deoxyuridine to Pancreatic Cancer. Nucleic Acid. Ther. 2015, 25, 180–187. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, H.S.; Jung, D.E.; Kim, J.M.; Song, S.Y. The DNA aptamer binds stemness-enriched cancer cells in pancreatic cancer. J. Mol. Recognit. 2017, 30, e2591. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Lai, L.A.; Brentnall, T.A.; Dawson, D.W.; Kelly, K.A.; Chen, R.; Pan, S. X-aptamers targeting Thy-1 membrane glycoprotein in pancreatic ductal adenocarcinoma. Biochimie 2021, 181, 25–33. [Google Scholar] [CrossRef]
- Porciani, D.; Tedeschi, L.; Marchetti, L.; Citti, L.; Piazza, V.; Beltram, F.; Signore, G. Aptamer-Mediated Codelivery of Doxorubicin and NF-kappaB Decoy Enhances Chemosensitivity of Pancreatic Tumor Cells. Mol. Ther. Nucleic Acids 2015, 4, e235. [Google Scholar] [CrossRef]
- Citro, A.; Pellegrini, S.; Dugnani, E.; Eulberg, D.; Klussmann, S.; Piemonti, L. CCL2/MCP-1 and CXCL12/SDF-1 blockade by L-aptamers improve pancreatic islet engraftment and survival in mouse. Am. J. Transpl. 2019, 19, 3131–3138. [Google Scholar] [CrossRef]
- Ray, P.; Sullenger, B.A.; White, R.R. Further characterization of the target of a potential aptamer biomarker for pancreatic cancer: Cyclophilin B and its posttranslational modifications. Nucleic Acid. Ther. 2013, 23, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Sung, H.J.; Kim, S.; Kim, E.O.; Lee, J.W.; Moon, J.Y.; Choi, K.; Jung, J.E.; Lee, Y.; Koh, S.S.; et al. An RNA aptamer that specifically binds pancreatic adenocarcinoma up-regulated factor inhibits migration and growth of pancreatic cancer cells. Cancer Lett. 2011, 313, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Leslie, M.; Kameyama, H.; Volk, D.E.; Tanaka, T. Aptamer Therapeutics in Cancer: Current and Future. Cancers 2018, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Compte, M.; Harwood, S.L.; Munoz, I.G.; Navarro, R.; Zonca, M.; Perez-Chacon, G.; Erce-Llamazares, A.; Merino, N.; Tapia-Galisteo, A.; Cuesta, A.M.; et al. A tumor-targeted trimeric 4-1BB-agonistic antibody induces potent anti-tumor immunity without systemic toxicity. Nat. Commun. 2018, 9, 4809. [Google Scholar] [CrossRef] [PubMed]
- Dollins, C.M.; Nair, S.; Boczkowski, D.; Lee, J.; Layzer, J.M.; Gilboa, E.; Sullenger, B.A. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 2008, 15, 675–682. [Google Scholar] [CrossRef]
- Cerchia, L.; Esposito, C.L.; Camorani, S.; Rienzo, A.; Stasio, L.; Insabato, L.; Affuso, A.; de Franciscis, V. Targeting Axl with an high-affinity inhibitory aptamer. Mol. Ther. 2012, 20, 2291–2303. [Google Scholar] [CrossRef]
- Dinis Ano Bom, A.P.; da Costa Neves, P.C.; Bonacossa de Almeida, C.E.; Silva, D.; Missailidis, S. Aptamers as Delivery Agents of siRNA and Chimeric Formulations for the Treatment of Cancer. Pharmaceutics 2019, 11, 684. [Google Scholar] [CrossRef]
- Bai, C.; Gao, S.; Hu, S.; Liu, X.; Li, H.; Dong, J.; Huang, A.; Zhu, L.; Zhou, P.; Li, S.; et al. Self-Assembled Multivalent Aptamer Nanoparticles with Potential CAR-like Characteristics Could Activate T Cells and Inhibit Melanoma Growth. Mol. Ther. Oncolytics 2020, 17, 9–20. [Google Scholar] [CrossRef]
- Macdonald, J.; Henri, J.; Goodman, L.; Xiang, D.; Duan, W.; Shigdar, S. Development of a Bifunctional Aptamer Targeting the Transferrin Receptor and Epithelial Cell Adhesion Molecule (EpCAM) for the Treatment of Brain Cancer Metastases. ACS Chem. Neurosci. 2017, 8, 777–784. [Google Scholar] [CrossRef]
- Ma, Y.; Ai, G.; Zhang, C.; Zhao, M.; Dong, X.; Han, Z.; Wang, Z.; Zhang, M.; Liu, Y.; Gao, W.; et al. Novel Linear Peptides with High Affinity to alphavbeta3 Integrin for Precise Tumor Identification. Theranostics 2017, 7, 1511–1523. [Google Scholar] [CrossRef]
- Fechter, P.; Cruz Da Silva, E.; Mercier, M.C.; Noulet, F.; Etienne-Seloum, N.; Guenot, D.; Lehmann, M.; Vauchelles, R.; Martin, S.; Lelong-Rebel, I.; et al. RNA Aptamers Targeting Integrin alpha5beta1 as Probes for Cyto- and Histofluorescence in Glioblastoma. Mol. Ther. Nucleic Acids 2019, 17, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Lee, S.H.; Hwang, Y.; Yoo, H.; Jung, H.; Kim, S.H.; Mok, H. Multivalent Aptamer-RNA Conjugates for Simple and Efficient Delivery of Doxorubicin/siRNA into Multidrug-Resistant Cells. Macromol. Biosci. 2017, 17, 1600343. [Google Scholar] [CrossRef]
- Wang, C.Y.; Lin, B.L.; Chen, C.H. An aptamer targeting shared tumor-specific peptide antigen of MAGE-A3 in multiple cancers. Int. J. Cancer 2016, 138, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Lin, B.L.; Chen, C.H. Targeted drug delivery using an aptamer against shared tumor-specific peptide antigen of MAGE-A3. Cancer Biol. Ther. 2021, 22, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Liu, Z.; Parveen, N.; Rehman, U.; Aziz, A.; Sheikh, A.; Abourehab, M.A.S.; Guo, W.; Huang, J.; Wang, Z.; Kesharwani, P. Unravelling the enigma of siRNA and aptamer mediated therapies against pancreatic cancer. Mol. Cancer 2023, 22, 8. [Google Scholar] [CrossRef]
- Zhang, X.H.; Wang, W.; Chen, X. Selection and identification of an ssDNA aptamer to NB4 cell. J. Clin. Lab. Anal. 2021, 35, e23718. [Google Scholar] [CrossRef]
- Gao, X.; Teng, X.; Dai, Y.; Li, J. Rolling Circle Amplification-Assisted Flow Cytometry Approach for Simultaneous Profiling of Exosomal Surface Proteins. ACS Sens. 2021, 6, 3611–3620. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, Y.; Shin, D.S.; Silangcruz, J.; Tuleuova, N.; Revzin, A. Aptamer-containing surfaces for selective capture of CD4 expressing cells. Langmuir 2012, 28, 12544–12549. [Google Scholar] [CrossRef]
- Cheng, E.L.; Kacherovsky, N.; Pun, S.H. Aptamer-Based Traceless Multiplexed Cell Isolation Systems. ACS Appl. Mater. Interfaces 2022, 14, 44136–44146. [Google Scholar] [CrossRef]
- Song, Z.; Mao, J.; Barrero, R.A.; Wang, P.; Zhang, F.; Wang, T. Development of a CD63 Aptamer for Efficient Cancer Immunochemistry and Immunoaffinity-Based Exosome Isolation. Molecules 2020, 25, 5585. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Satheesan, S.; Li, H.; Weinberg, M.S.; Morris, K.V.; Burnett, J.C.; Rossi, J.J. Cell-specific RNA aptamer against human CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity. Chem. Biol. 2015, 22, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Nunes, J.; Lifante, J.; Shen, Y.; Ximendes, E.C.; Jaque, D.; Iglesias-de la Cruz, M.C.; Cruz, C. Biological studies of an ICG-tagged aptamer as drug delivery system for malignant melanoma. Eur. J. Pharm. Biopharm. 2020, 154, 228–235. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhao, J.; Li, J.; Liao, X.; Chen, F. Advances of aptamers screened by Cell-SELEX in selection procedure, cancer diagnostics and therapeutics. Anal. Biochem. 2020, 598, 113620. [Google Scholar] [CrossRef]
- Wang, C.W.; Chung, W.H.; Cheng, Y.F.; Ying, N.W.; Peck, K.; Chen, Y.T.; Hung, S.I. A new nucleic acid-based agent inhibits cytotoxic T lymphocyte-mediated immune disorders. J. Allergy Clin. Immunol. 2013, 132, 713–722. [Google Scholar] [CrossRef]
- Ott, L.E.; Carson, S. Immunological tools: Engaging students in the use and analysis of flow cytometry and enzyme-linked immunosorbent assay (ELISA). Biochem. Mol. Biol. Educ. 2014, 42, 382–397. [Google Scholar] [CrossRef] [PubMed]
- Bognár, Z.; Gyurcsányi, R.E. Aptamers against Immunoglobulins: Design, Selection and Bioanalytical Applications. Int. J. Mol. Sci. 2020, 21, 5748. [Google Scholar] [CrossRef]
- Camorani, S.; Passariello, M.; Agnello, L.; Esposito, S.; Collina, F.; Cantile, M.; Di Bonito, M.; Ulasov, I.V.; Fedele, M.; Zannetti, A.; et al. Aptamer targeted therapy potentiates immune checkpoint blockade in triple-negative breast cancer. J. Exp. Clin. Cancer Res. 2020, 39, 180. [Google Scholar] [CrossRef]
- Pastor, F.; Berraondo, P.; Etxeberria, I.; Frederick, J.; Sahin, U.; Gilboa, E.; Melero, I. An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 2018, 17, 751–767. [Google Scholar] [CrossRef]
- Liu, C.G.; Wang, Y.; Liu, P.; Yao, Q.L.; Zhou, Y.Y.; Li, C.F.; Zhao, Q.; Liu, G.H.; Zhang, X.L. Aptamer-T Cell Targeted Therapy for Tumor Treatment Using Sugar Metabolism and Click Chemistry. ACS Chem. Biol. 2020, 15, 1554–1565. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Yao, F.; An, Y.; Li, X.; Yang, X. Novel Nanotherapeutics for Cancer Immunotherapy by PD-L1-Aptamer-Functionalized and Fexofenadine-Loaded Albumin Nanoparticles. Molecules 2023, 11, 2556. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Qi, Y.; Wang, G.; Li, L.; Jin, Z.; Tian, J.; Du, Y. PD-L1 Aptamer-Functionalized Metal-Organic Framework Nanoparticles for Robust Photo-Immunotherapy against Cancer with Enhanced Safety. Angew. Chem. Int. Ed. Engl. 2023, 26, e202214750. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, X.; Yeganeh, P.N.; Lee, D.J.; Valle-Garcia, D.; Meza-Sosa, K.F.; Junqueira, C.; Su, J.; Luo, H.R.; Hide, W.; et al. Immunotherapy for breast cancer using EpCAM aptamer tumor-targeted gene knockdown. Proc. Natl. Acad. Sci. USA 2021, 118, e2022830118. [Google Scholar] [CrossRef]
- Shigdar, S.; Schrand, B.; Giangrande, P.H.; de Franciscis, V. Aptamers: Cutting edge of cancer therapies. Mol. Ther. 2021, 29, 2396–2411. [Google Scholar] [CrossRef] [PubMed]
- Camorani, S.; Granata, I.; Collina, F.; Leonetti, F.; Cantile, M.; Botti, G.; Fedele, M.; Guarracino, M.R.; Cerchia, L. Novel Aptamers Selected on Living Cells for Specific Recognition of Triple-Negative Breast Cancer. iScience 2020, 23, 100979. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Marisetty, A.; Schrand, B.; Gabrusiewicz, K.; Hashimoto, Y.; Ott, M.; Grami, Z.; Kong, L.Y.; Ling, X.; Caruso, H.; et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Investig. 2019, 129, 137–149. [Google Scholar] [CrossRef]
- Song, W.; Li, H.; Liu, H.; Wu, Z.; Qiang, W.; Xu, D. Fabrication of streptavidin functionalized silver nanoparticle decorated graphene and its application in disposable electrochemical sensor for immunoglobulin E. Electrochem. Commun. 2013, 31, 16–19. [Google Scholar] [CrossRef]
- Li, H.; Wang, C.; Wu, Z.; Lu, L.; Qiu, L.; Zhou, H.; Shen, G.; Yu, R. An electronic channel switching-based aptasensor for ultrasensitive protein detection. Anal. Chim. Acta 2013, 758, 130–137. [Google Scholar] [CrossRef]
- Maehashi, K.; Matsumoto, K.; Takamura, Y.; Tamiya, E. Aptamer-Based Label-Free Immunosensors Using Carbon Nanotube Field-Effect Transistors. Electroanalysis 2009, 21, 1285–1290. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, J.P.; Han, S.J.; Sim, S.J. Aptamer biosensor for lable-free detection of human immunoglobulin E based on surface plasmon resonance. Sens. Actuators B: Chem. 2009, 139, 471–475. [Google Scholar] [CrossRef]
- Liu, Y.-M.; Yang, J.-J.; Cao, J.-T.; Zhang, J.-J.; Chen, Y.-H.; Ren, S.-W. An electrochemiluminescence aptasensor based on CdSe/ZnS functionalized MoS2 and enzymatic biocatalytic precipitation for sensitive detection of immunoglobulin E. Sens. Actuators B: Chem. 2016, 232, 538–544. [Google Scholar] [CrossRef]
- Momeni, M.; Mashayekhi, K.; Navashenaq, J.G.; Sankian, M. Identification of G-quadruplex anti-Interleukin-2 aptamer with high specificity through SELEX stringency. Heliyon 2022, 8, e09721. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yuan, C.; Wang, W.; Yu, Z.; Hao, R.; Zhang, Y.; Guan, M.; Li, N.; Yang, H. Aptasensor-enabled quantitative analysis of nano-sized extracellular vesicles by flow cytometry. Analyst 2020, 145, 7551–7558. [Google Scholar] [CrossRef]
- Phung, N.L.; Walter, J.G.; Jonczyk, R.; Seiler, L.K.; Scheper, T.; Blume, C. Development of an Aptamer-Based Lateral Flow Assay for the Detection of C-Reactive Protein Using Microarray Technology as a Prescreening Platform. ACS Comb. Sci. 2020, 22, 617–629. [Google Scholar] [CrossRef]
- Tan, X.; Jia, F.; Wang, P.; Zhang, K. Nucleic acid-based drug delivery strategies. J. Control Release 2020, 323, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G. Applications in Which Aptamers Are Needed or Wanted in Diagnostics and Therapeutics. Pharmaceuticals 2022, 15, 693. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Meng, X.; de Vos, W.M.; Wu, H.; Fang, X.; Maiti, A.K. Implications of Gut Microbiota in Complex Human Diseases. Int. J. Mol. Sci. 2021, 22, 12661. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Khan, I.; Bai, Y.; Zha, L.; Ullah, N.; Ullah, H.; Shah, S.R.H.; Sun, H.; Zhang, C. Mechanism of the Gut Microbiota Colonization Resistance and Enteric Pathogen Infection. Front. Cell Infect. Microbiol. 2021, 11, 716299. [Google Scholar] [CrossRef]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Czepiel, J.; Dróżdż, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultańska, D.; Garlicki, A.; Biesiada, G. Clostridium difficile infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhou, J.; Peng, Y.; Xie, Y.; Xiao, Y. Aptamers: A prospective tool for infectious diseases diagnosis. J. Clin. Lab. Anal. 2022, 36, e24725. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wang, L.; Lu, W.; Zhao, J.; Zhang, H.; Chen, W. Selection, Characterization and Interaction Studies of a DNA Aptamer for the Detection of Bifidobacterium bifidum. Int. J. Mol. Sci. 2017, 18, 883. [Google Scholar] [CrossRef]
- Song, S.; Wang, X.; Xu, K.; Ning, L.; Yang, X. Rapid identification and quantitation of the viable cells of Lactobacillus casei in fermented dairy products using an aptamer-based strategy powered by a novel cell-SELEX protocol. J. Dairy. Sci. 2019, 102, 10814–10824. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Zhang, Y.; Krämer, M.; Kissmann, A.K.; Amann, V.; Raber, H.F.; Weil, T.; Stieger, K.R.; Knippschild, U.; Henkel, M.; et al. A Polyclonal Aptamer Library for the Specific Binding of the Gut Bacterium Roseburia intestinalis in Mixtures with Other Gut Microbiome Bacteria and Human Stool Samples. Int. J. Mol. Sci. 2022, 23, 7744. [Google Scholar] [CrossRef]
- Bhardwaj, J.; Chaudhary, N.; Kim, H.; Jang, J. Subtyping of influenza A H1N1 virus using a label-free electrochemical biosensor based on the DNA aptamer targeting the stem region of HA protein. Anal. Chim. Acta 2019, 1064, 94–103. [Google Scholar] [CrossRef]
- Bai, C.; Lu, Z.; Jiang, H.; Yang, Z.; Liu, X.; Ding, H.; Li, H.; Dong, J.; Huang, A.; Fang, T.; et al. Aptamer selection and application in multivalent binding-based electrical impedance detection of inactivated H1N1 virus. Biosens. Bioelectron. 2018, 110, 162–167. [Google Scholar] [CrossRef]
- Chen, H.L.; Hsiao, W.H.; Lee, H.C.; Wu, S.C.; Cheng, J.W. Selection and Characterization of DNA Aptamers Targeting All Four Serotypes of Dengue Viruses. PLoS ONE 2015, 10, e0131240. [Google Scholar] [CrossRef]
- Morais, L.M.; Chaves, T.S.; Medeiros, M.A.; Pereira, K.A.B.; Jurgilas, P.B.; Barbosa de Lima, S.M.; Missailidis, S.; Bispo de Filippis, A.M. Selection and Characterization of Single-Stranded DNA Aptamers of Diagnostic Potential against the Whole Zika Virus. Viruses 2022, 14, 1867. [Google Scholar] [CrossRef] [PubMed]
- Afrasiabi, S.; Pourhajibagher, M.; Raoofian, R.; Tabarzad, M.; Bahador, A. Therapeutic applications of nucleic acid aptamers in microbial infections. J. Biomed. Sci. 2020, 27, 6. [Google Scholar] [CrossRef]
- Wandtke, T.; Woźniak, J.; Kopiński, P. Aptamers in diagnostics and treatment of viral infections. Viruses 2015, 7, 751–780. [Google Scholar] [CrossRef]
- Li, J.; Gu, J.; Zhang, H.; Liu, R.; Zhang, W.; Mohammed-Elsabagh, M.; Xia, J.; Morrison, D.; Zakaria, S.; Chang, D.; et al. A Highly Specific DNA Aptamer for RNase H2 from Clostridium difficile. ACS Appl. Mater. Interfaces 2021, 13, 9464–9471. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Peng, Z.; Ning, Y.; Chen, Y.; Zhou, Q.; Deng, L. Highly specific and cost-efficient detection of Salmonella Paratyphi A combining aptamers with single-walled carbon nanotubes. Sensors 2013, 13, 6865–6881. [Google Scholar] [CrossRef] [PubMed]
- Amraee, M.; Oloomi, M.; Yavari, A.; Bouzari, S. DNA aptamer identification and characterization for E. coli O157 detection using cell based SELEX method. Anal. Biochem. 2017, 536, 36–44. [Google Scholar] [CrossRef]
- Wu, H.; Gu, L.; Ma, X.; Tian, X.; Fan, S.; Qin, M.; Lu, J.; Lyu, M.; Wang, S. Rapid Detection of Helicobacter pylori by the Naked Eye Using DNA Aptamers. ACS Omega 2021, 6, 3771–3779. [Google Scholar] [CrossRef]
- Guo, K.; Song, Z.; Wang, G.; Tang, C. Detecting Redox Potentials Using Porous Boron Nitride/ATP-DNA Aptamer/Methylene Blue Biosensor to Monitor Microbial Activities. Micromachines 2022, 13, 83. [Google Scholar] [CrossRef]
- Wang, K.; Wang, M.; Ma, T.; Li, W.; Zhang, H. Review on the Selection of Aptamers and Application in Paper-Based Sensors. Biosensors 2022, 13, 39. [Google Scholar] [CrossRef]
- Sousa, M.P.; Piloto, A.M.L.; Pereira, A.C.; Schmitt, F.C.; Fernandes, R.; Moreira, F.T.C. New Quantum-Dot-Based Fluorescent Immunosensor for Cancer Biomarker Detection. Chemosensors 2022, 10, 518. [Google Scholar] [CrossRef]
- Pereira, A.C.; Moreira, F.T.C.; Rodrigues, L.R.; Sales, M.G.F. Paper-based aptasensor for colorimetric detection of osteopontin. Anal. Chim. Acta 2022, 1198, 339557. [Google Scholar] [CrossRef] [PubMed]
- Razlansari, M.; Jafarinejad, S.; Rahdar, A.; Shirvaliloo, M.; Arshad, R.; Fathi-Karkan, S.; Mirinejad, S.; Sargazi, S.; Sheervalilou, R.; Ajalli, N.; et al. Development and classification of RNA aptamers for therapeutic purposes: An updated review with emphasis on cancer. Mol. Cell Biochem. 2023, 478, 1573–1598. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Parolo, C.; Idili, A.; Gomis, R.R.; Rodrigues, L.; Sales, G.; Merkoçi, A. Paper-based biosensors for Cancer Diagnosis. Trends Chem. 2022, 4, 554–567. [Google Scholar] [CrossRef]
- Sousa, A.P.; Cunha, D.M.; Franco, C.; Teixeira, C.; Gojon, F.; Baylina, P.; Fernandes, R. Which Role Plays 2-Hydroxybutyric Acid on Insulin Resistance? Metabolites. 2021, 11, 835. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).