Abstract
Pseudomonas aeruginosa (PAO1) is an opportunistic pathogen, lethal in immunocompromised individuals. The clinical management of PAO1 infections still depends deeply on antibiotic therapy. However, this therapy has been alarmingly overpowered by growing bacterial resistance mechanisms over the years. One of these bacterial mechanisms is quorum sensing (QS). QS is involved in the production of biofilm, rhamnolipids and pyocyanin, among other factors. The present study aimed to study the effect of the mutations in the genes of rubredoxin (Rub A1 and Rub A2) and glutaredoxin (GLRx) in the production of virulence traits and susceptibility of PAO1 to the antibiotic ciprofloxacin (CIP) and to infection by a phage cocktail. Rub A1, Rub A2, and GLRx showed a decrease in the expression of genes lasI, lasR, mvfR, and rpsL when compared to the wild type, PAO1. Rub A1 and Rub A2 also showed a decrease in the expression of the gene pqsA, while the mutant GLRx showed an increase of over 200% in expression compared to PAO1. The biofilm produced by the mutants Rub A1, Rub A2, and GLRx increased more than 1.5 times in comparison to PAO1, with statistical significance (p < 0.0001). In the viability assay, the mutant strain Rub A2 was the most susceptible to ciprofloxacin in both concentrations tested (p < 0.0001). The production of proteases increased in the mutant strains when compared to PAO1 (p < 0.05). However, there was a decrease in the production of rhamnolipids and pyocyanins in the mutant strains. In the phage assay, we could perceive a reduction in the growth of the mutant strains when compared to PAO1. Additionally, after the addition of the phages, all the strains showed susceptibility to the phage assay (p < 0.0001), observed in the decrease in the absorbance values. These results may highlight the relevance of the genes Rub A1, Rub A2, and GLRX in the proliferation and treatment of infections with PAO1. Overall, this study gives preliminary insights into how gene expression may be helpful in strategies to overcome antibiotic resistance.
1. Introduction
Pseudomonas aeruginosa (PAO1) is a Gram-negative opportunistic bacteria associated with acute and chronic infections in humans; it is not only linked to hospital-acquired infections but also to individuals with conditions such as immunodeficiency virus infection, cancer, severe burns, immunologic genetic diseases, and diabetes mellitus, as well as other underlying health concerns [1,2].
Over the course of recent years, there has been a notable escalation in the prevalence of P. aeruginosa strains that are resistant to a range of drug substances, including trimethoprim, chloramphenicol, cephalosporins, macrolides, tetracyclines, and fluoroquinolones [2]. Inside the quinolones, the most effective antibiotic reported for P. aeruginosa is ciprofloxacin (CIP). Ciprofloxacin has a bactericidal effect by inhibiting the bacterial DNA topoisomerase and gyrase. Worldwide reports have been describing a decrease in the overall susceptibility of P. aeruginosa and CIP remains within the small group of antibiotics administered to treat P. aeruginosa infections via oral [3]. The increase in the drug-resistant strains of P. aeruginosa has garnered significant attention from the global medical community, highlighting not only the need to perceive the mechanisms behind the resistance mechanisms to conventional drugs but also to discover new strategies to deal with these pathogens [2].
One important bacterial resistance mechanism is the formation of biofilms. Biofilms are structures composed of extracellular components secreted by bacterial cells that lower the penetration of antimicrobial drugs in bacteria cells [4,5,6,7,8,9]. Therefore, biofilms increase the bacterial survival rate in adverse environments [7]. In a study, 63 cases of carbapenems-resistant P. aeruginosa were evaluated on their capacity to form biofilms as a predictive factor for mortality. In the described study, Jeong et al. found that an increase of just one percent in the biofilm production in these strains could increase the risk for mortality in patients suffering from bacteremia 1.10 times [10].
To overcome bacterial resistance and pathogenicity, new strategies have been studied, with three of them being the inhibition or modulation of the quorum sensing (QS), the use of QS inhibitors [11] or gene mutations [1], and the use of phage therapy [4,11].
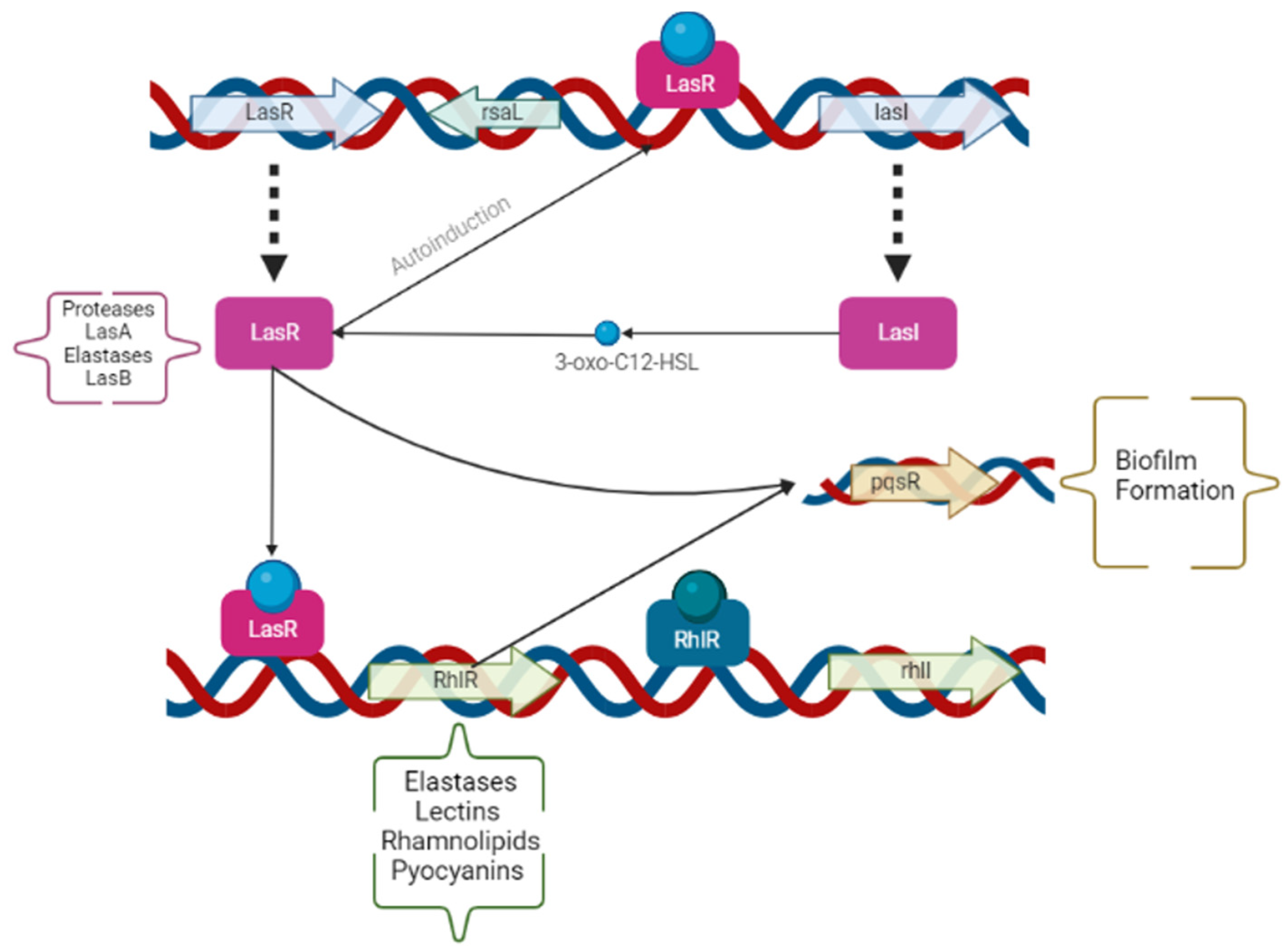
The opportunist pathogen PAO1 carries the capacity to coordinate the whole bacterial population by cell-to-cell communication, which is also called QS. The expression of QS is often increased proportionally to the increase in the bacterial population. Therefore, the modulation of QS benefits bacterial survival and adaption to stressful environments, due to the expression of PAO1 virulence traits and biofilm formation. These traits are regulated by two acyl-homoserine lactoses, LasI/LasR, and PQS-MvfR [6,7,8,9]. The N-acyl homoserine lactones are signal molecules that control de production of proteases, hemolysins, chitinases, lipases, siderophores, extracellular r DNA, and extracellular polysaccharides [2,4,8]. Modifications on either lasR (protease and elastase) or lasI (autoinducer) in the strains of PAO1 lower the expression of the QS system genes, the virulence traits, and the biofilm formation. The virulence traits of PAO1 can be observed in Figure 1. Therefore, the modulation of the QS is known as anti-virulence therapy [8]. A study, conducted by Ahmed et al., showed a reduction of 65% in proteases, 22% in elastases, and 32% in pyocyanins by applying QS inhibitors in PAO1 cells [12].
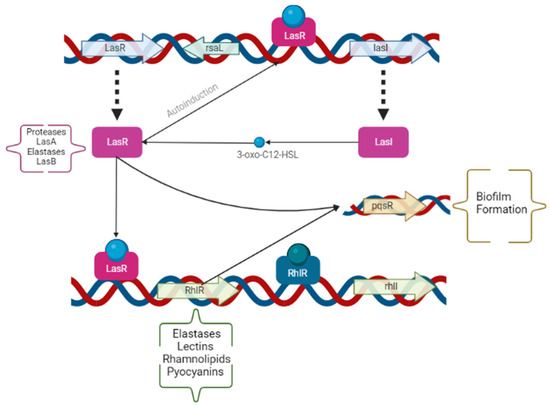
Figure 1.
Operons and genes involved on the expression of the different Pseudomonas aeruginosa virulence factors in the quorum sensing system. The genes and operons have been summarized by arrows and boxes, and the virulent traits influenced by each are described alongside (in parentheses). Created by the authors using BioRender.com (accessed on 11 August 2022).
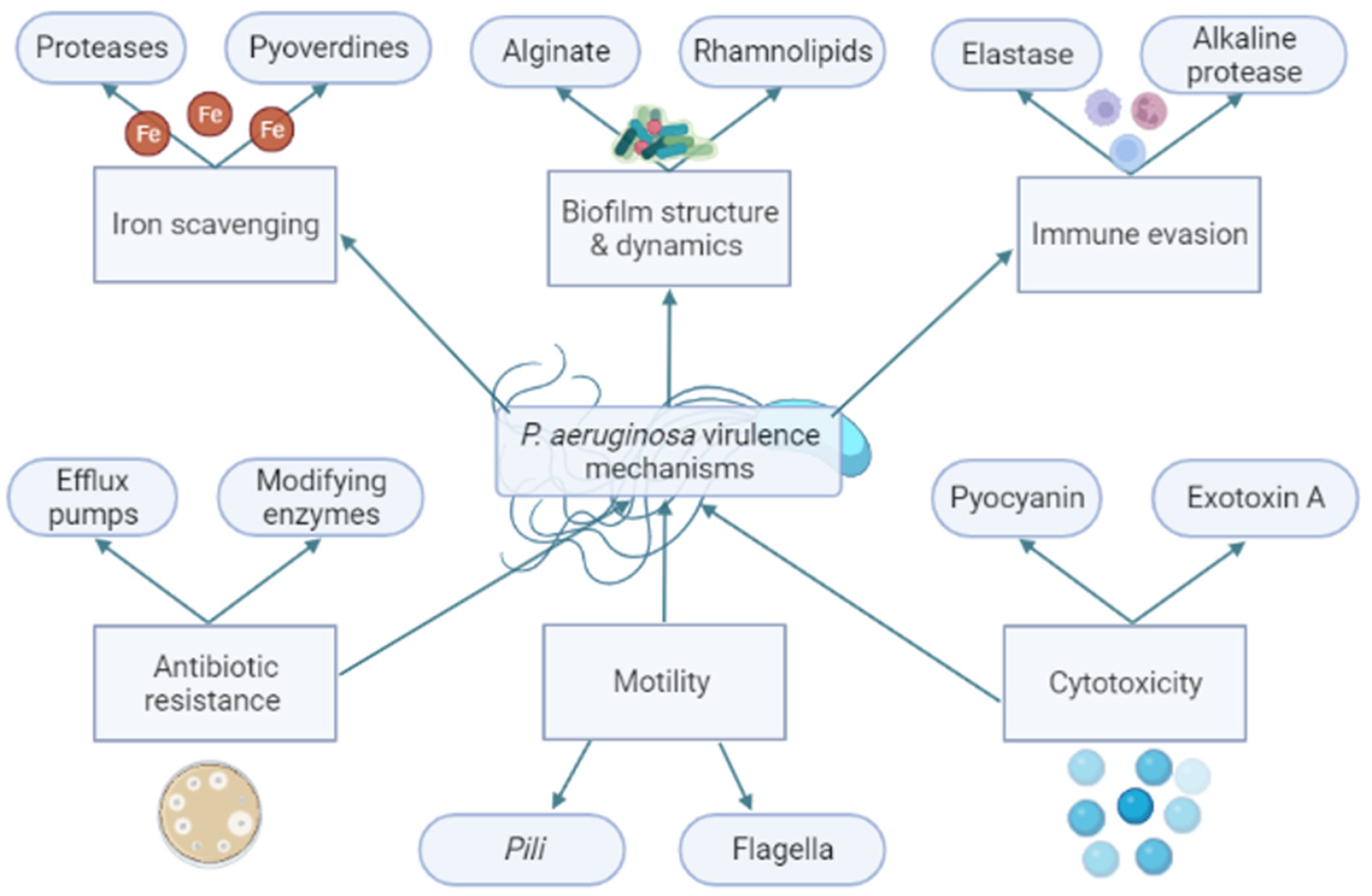
The virulence traits of PAO1 can be divided into functional groups based on the bacteria function in the cell, as seen in Figure 2. One of these groups is the proteases. Proteases are composed of a signal peptide, a pro-peptide, and a mature protease domain. They are firstly expressed and then processed twice to reach their mature form by cleavage processes during the translocation on the cytoplasmatic membrane and secondly in the periplasm [13]. Mature proteases promote a proteolytic effect in tissues, leading to proinflammatory cytokines production and enhancing the inflammatory host response [14]. Ahmed et al. achieved the inhibition of the QS using isolated plant compounds, such as trans-cinnamaldehyde (CA) and salicylic acid (SA). In their study, both compounds achieved a downregulation of las and rhl within the QS, and the CA achieved a reduction of 13-fold in las and 7-fold in rhl. Additionally, the compounds reduced the production of extracellular virulence factors such as proteases by 65%, elastases by 22%, and pyocyanins by 32% [12]. Pyocyanins are another produced PAO1 virulence trait. Pyocyanins are a blue phenazine pigment with antioxidant capacity produced by 90 to 95% of PAO1 strains [15]. Therefore, pyocyanins confer a protective mechanism against the reactive oxygen species (ROS) produced by the immune system of the host by the inactivation of catalases [1].
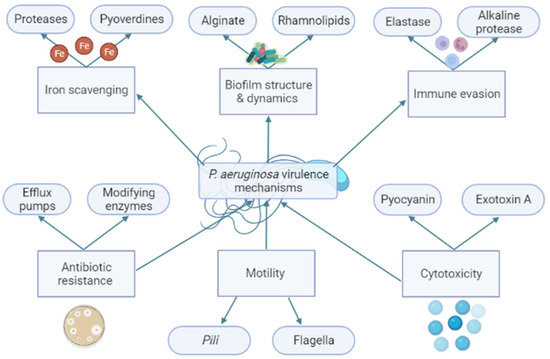
Figure 2.
Brief schematic enumeration of Pseudomonas aeruginosa virulence mechanisms divided by their bacterial function. The bacterial virulence mechanisms are involved in the survival and pathogenesis of P. aeruginosa. For example: Alginate and rhamnolipids are constituents of the structure of biofilm. Biofilm is a structure like a barrier that can, for example, protect the bacteria against antibiotics. The reduction of alginate and rhamnolipids may affect the capacity to produce a functional biofilm by P. aeruginosa. Created by the authors using BioRender.com (accessed on 11 August 2022).
Rhamnolipids are another example of a PAO1 virulence trait. Rhamnolipids are made up of glycolipids, biosurfactants, and phenazines, which are components of the biofilm matrix and have extracellular redox activity. Additionally, rhamnolipids can cause the lysis of polymorphonuclear cells and macrophages [14]. Yang et al. perceived that the compound paenol, a phenolic compound, could significantly downregulate the transcription of the QS-related genes in PA01, lasI/R, rhlI/R, and pqs/mvfR. In their study, paenol could significantly inhibit rhamnolipid production by 66.27% after 24 h [16].
The resistance of P. aeruginosa strains can also be affected by interfering with the specific gene of each metabolic pathway. For example, rubA2, and rubA1 are two genes encoded by the cluster rubB that are related to the expression of the rubredoxins reductases (Rdxs). Rdxs are redox-active iron–sulfur proteins crucial for the reduction of oxygen species produced by the host, and, consequently, for the survival of the bacteria during infection [17]. In P. aeruginosa, the Rdxs are necessary for the growth on alkanes. The Rdxs are found upregulated in several highly virulent mutant strains. The Rdxs are considered essential for survival and proliferation after the phagocytosis process of the host defenses. Additionally, the mutant strain in Rdxs have a very impaired QS and exhibited lower virulence in animal infection models [18].
Another example is the glutaredoxin (GLRX) gene. The GLRX gene codifies small ubiquitous redox enzymes; these enzymes catalyze the reduction of thiol-dissulfide exchange reactions dependent on glutathione. A study performed with a grxD mutant demonstrated reduced virulence in Drosophila melanogaster [19]. Additionally, GLRX has a protective effect against antibiotic drugs [20].
To control bacterial infections, an alternative method against antibiotic drug-resistant bacteria is phage therapy. Phages are viruses that infect specific strains of bacteria. This specificity contrasts with antibiotics, which display a more general specter of bacterial strains [21]. Phage therapy uses lytic phages, phages whose infection results in the lyse of bacteria, that are able to penetrate difficult environments such as biofilms. Phages are usually used in cocktails, where more than one phage type is used. Phages encode a polysaccharide hydrolase that has the ability to adsorb, invade, and shatter the bacterial host [21]. Phage therapy is useful in many sectors, including agriculture, to control plant bacterial infections [22], multi-drug resistant microorganisms in humans and animals, and in research, where phages have been studied for the treatment of bacterial infections in in vitro and in vivo models [23]. For example, Arumugam et al. evaluated the antibacterial capacity of two lytic phages for multidrug-resistant P. aeruginosa in bacteremia mice models. They achieved the recovery of the healthy state of every specimen with the phage concentrations of 1:10 and 1:100 [24].
In this study, we aimed to perceive the role of three mutations, Rub A1, Rub A2, and GLRx, in QS bacterial modulation by comparing the QS gene expression, production of virulence traits, and viability of mutants to PAO1. Moreover, we also assessed the effects of these mutations on bacteria susceptibility to the antibiotic ciprofloxacin (CIP) and bacterial infection by a phage cocktail.
2. Materials and Methods
2.1. Chemicals
The specifications for the preparation of all solutions and chemicals utilized in this study are consolidated in Table 1.

Table 1.
Brief description of the solutions and reagents used, and respective sources and preparation specifications used in this study.
2.2. Strains and Maintenance
The tested strains used in this study were purchased from the PAO1 transposon mutant library [25]. These mutant strains encompass Rub A1, Rub A2, GLRX, and the wild-type PAO1. Before experiments, fresh cultures of all strains were grown overnight in TSB, followed by a final optical density adjustment to 0.1 at a wavelength of 600 nm. The final concentrations of ciprofloxacin used were 0.125 and 0.25 µg/mL.
2.3. Quorum Sensing Operons and Gene Expression
In this study, the expression of regulatory genes involved in QS, namely the operons lasI, lasR, which are responsible for the expression of virulence factors protease Las and elastase Las, and the genes rhlI, rhlR, pqsA, and mvfRI, were quantified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), performed according to Malgaonkar [26]. The primers selected for this assay can be observed in Table 2 [11]. Briefly, the total RNA isolated from each bacterial strain was obtained using the Lab-Aid 824s DNA Extraction Kit (Zeesan, China). The concentration and purity of the extracted RNA were verified by using a µDrop™ Plate and a Multiskan SkyHigh Microplate Spectrophotometer (Thermo Scientific™, Waltham, MA, USA). The RT-qPCR was performed using the one-step NZYSpeedy RT-qPCR Green Master Mix Kit (NZYtech-Genes and Enzymes) in a qTOWER3 Real-Time PCR Thermal Cycler (Analytik jena, Jena, Germany) under the following conditions: incubation at 95 °C for 3 min and 40 cycles of 95 °C/10 s and 60 °C/40 s. Relative quantification was determined by the qPCR by delta-delta Ct Method 2−ΔΔCT, using the ribosomal gene rpsL as an internal control.

Table 2.
List of primers designed for reverse transcription-quantitative polymerase chain reaction for the quorum sensing genes of the four strains of Pseudomonas aeruginosa in this study.
2.4. Viability Assay of Rub A1, Rub A2, GLRX and PAO1 under CIP Concentrations
The cell viability was assessed by using a vital dye: erythrosine (EB). EB dyes only stain the dead bacteria pink, leaving the viable bacteria uncolored. Therefore, a more intense color correlates to higher cell death in the suspension, which is easily quantified through spectrophotometry [27].
The bacterial suspensions with and without antibiotics were centrifuged for 10 min at 4000 rpm/1073× g using a Gyrozen 1248R Multi-Purpose (High-Speed) Refrigerated Centrifuge. Then, the pellet was resuspended with 3 mL of fresh PBS. Afterward, 1 mL was separated for the control of living cells and 1 mL for the control of dead cells (cells were heated at 70 °C for 30 min). Then, we prepared five concentrations of dead cells (0%, 25%, 50%, 75%, and 100%) to construct a standard curve that could quantify the viable cells in each strain of PAO1 (wild and mutants). Following this, 300 μL of EB was added and the tubes were laid for 10 to 15 min at room temperature (RT). After that time, the suspensions were centrifuged at 8000 rpm/4293× g for 10 min. Then, the supernatant was removed, and the pellet was resuspended in 300 μL of PBS. Afterward, 100 μL of each standard was added to a microplate with 96 wells, in triplicate, and the absorbance was measured at 530 nm. The percentage of viable cells on the different strains was calculated with a standard curve. After the treatment with the two concentrations of CIP (0.125 and 0.25 µg/mL) overnight, the number of viable cells of each strain was calculated using the respective standard curve. The samples of each strain were prepared using the same method [27].
2.5. Biofilm Formation of Rub A1, Rub A2, GLRX and PAO1 under Normal Conditions
To observe the biofilms of the PAO1 strains, the methodology employed was similar to the one described elsewhere, with some changes [1]. Briefly, the cultures were grown overnight and were inoculated on a new medium of TSB to grow for a further 24 h at 37 °C. After that period, the cultures were diluted in the proportion 1:100 in a fresh medium of TSB. Then, they were inoculated in four to eight wells of a 96-well microplate, with 100 μL of culture per well. The microplates were incubated from 4 to 24 h at 37 °C. After incubation, the microplates were vigorously shaken to release all fluid and were washed carefully in water; this process was repeated two times for each microplate. Then, we added 125 μL of the solution of 0.1% of CV to each well, and the microplates were incubated at RT for 10 to 15 min. Afterward, the microplates were rinsed three to four times with water by submersion, and the excess water was removed by vigorous shakes against a block of paper. Then, the microplates were left to dry upside down. After they were dried, we added 125 μL of AC 30% per well, and the microplates were left at RT for a further 10 to 15 min. Next, 125 μL from each well was transferred to new microplates, and the absorbance of each well was measured at 550 nm. The taring was determined with 30% AC.
2.6. Evaluation of the Production of Proteases LasA in the Strains Rub A1, Rub A2, GLRX and PAO1 under CIP Concentrations
The quantification of the proteases LasA began with the centrifugation and collection of the culture supernatant after 24 h of growth and 24 h of contact with CIP in a concentration of 0.125 and 0.25 µg/mL. Then, to 100 μL of the recovered supernatant, we added 500 μL of 0.8% AZO. These suspensions were incubated at 25 °C for 3 h. After the 3 h incubation, the reaction was stopped by the addition of 500 μL of HCL 1.5 M in contact with an ice bath for 30 min. Afterward, the suspensions were centrifuged at 10,000× g for 10 min, and to half of the supernatant we added 1 N of NaOH. Finally, the concentration of proteins in each suspension was measured at 440 nm and the values were normalized using the values of a standard straight of BSA measured at 600 nm wavelength [12].
2.7. Rhamnolipids Production in the Strains Rub A1, Rub A2, GLRX and PAO1 under CIP Concentrations
To achieve the determination of the rhamnolipids, we also used the supernatants of each strain’s tested culture, as described in the protease methodology with the two concentrations of CIP mentioned. The pH of these supernatants was adjusted to 2.5 ± 0.2. Then, the suspensions were extracted using five volumes of CHCl3. Next, 4 mL of each suspension was added to 200 µL of 1 g/L of MB and 4.9 mL of distilled water. Then, the mixture was mixed vigorously and the absorbance was measured at 638 nm [26].
2.8. Pyocyanin Comparison between the Strains Rub A1, Rub A2, GLRX and PAO1 under CIP Concentrations
The pyocyanins in RubA1, RubA2, GLRX, and PAO1 were measured before and after the exposure to CIP in concentrations of 0.125 and 0.25 µg/mL. From each bacterial culture, 7.5 mL of suspension supernatant was filtered. We added 4.5 mL of CHCl3 to the suspensions, and the mixture was vortexed until the color changed to a greenish blue. Then, the samples were centrifuged at 4000 RPM/1073× g for 10 min, and 3 mL of the resulting supernatant was transferred to a new tube containing 1.5 mL of 0.2 M HCl and shaken until it turned soft pink. Then, the pink suspension was transferred to a microplate and the absorbance was measured at 520 nm. To obtain the concentration of pyocyanin in μg/mL, the absorbances were multiplied by the factor 17.072 [12].
2.9. Bacteriophage Assay
To perform the phage assay, we needed fresh cultures of Rub A1, Rub A2, GLRX, and PAO1 grown overnight and adjusted to 0.2 of optical density using the spectrophotometer. The cultures were inoculated in a 96-well microplate in a 1:1 ratio with TSB (control) and 1:1 with a phage cocktail (BCP БAKTEPͶOΦAГ®, Atlanta, GA, USA) that was phage-specific for PAO1, and the concentration of phages used was 1.3 × 107 phages per mL. The microplate was incubated at 37 °C, and the measurements at 600 nm were obtained at 0, 2, 4, 6, and 8 h post-inoculation.
2.10. Statistics, Data Treatment and Analysis
The results from the measurements of the cell viability, biofilm production, proteases, rhamnolipids, and bacteriophage assay were statistically obtained through one-way and two-way ANOVA with multiple comparisons analysis with 95% confidence. The rest of the results are shown as a comparison between the mutated strains and the wild-type PAO1 through graphs and heatmaps.
3. Results
3.1. QS Gene Expression
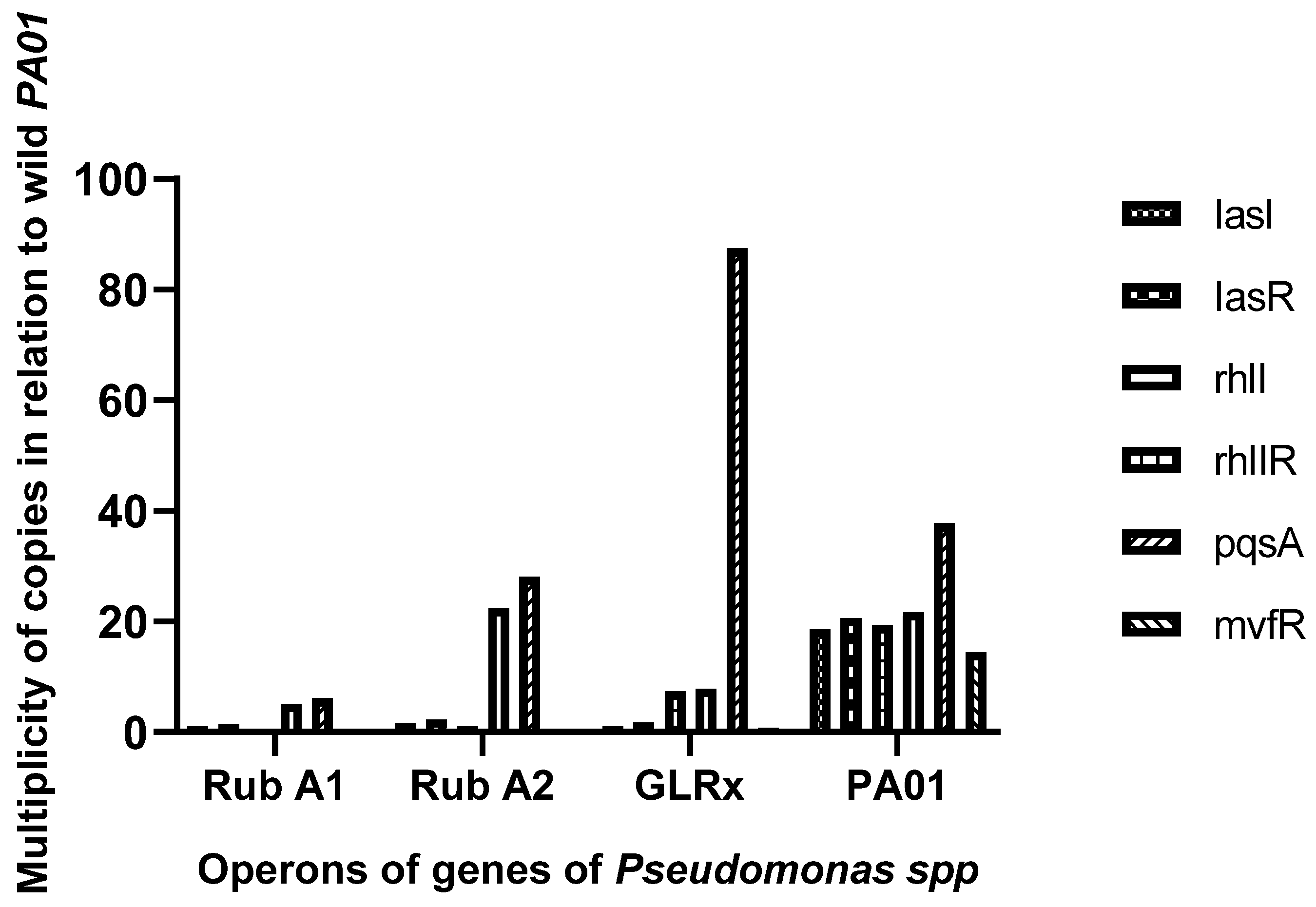
The activity of specific genes and operons related to QS, including lasI, lasR, rhlI, rhlR, pqsA, and mvfRI, was measured using the 2−ΔΔCT method and compared to a reference gene, rpsL (see Figure 3). The pqsA gene exhibited significantly higher expression in the strain harboring the mutated grlx gene compared to the wild-type PAO1 strain. On the other hand, the other genes showed lower expression in the mutated strains overall. Additionally, Rub A1 demonstrated a major decrease in pqsA and rhII when compared to the strain Rub A2.
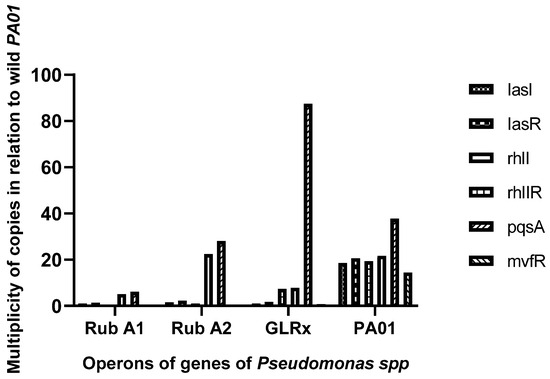
Figure 3.
Graphic representation of the assessment of the expression of the operon (lasI, lasR) and genes (rhlI, rhlR, pqsA, and mvfRI) assay, in three mutant strains of PAO1 (rubredoxins: strains Rub A1 and Rub A2; a glutaredoxin: strain GLRX) and one wild type (PAO1).
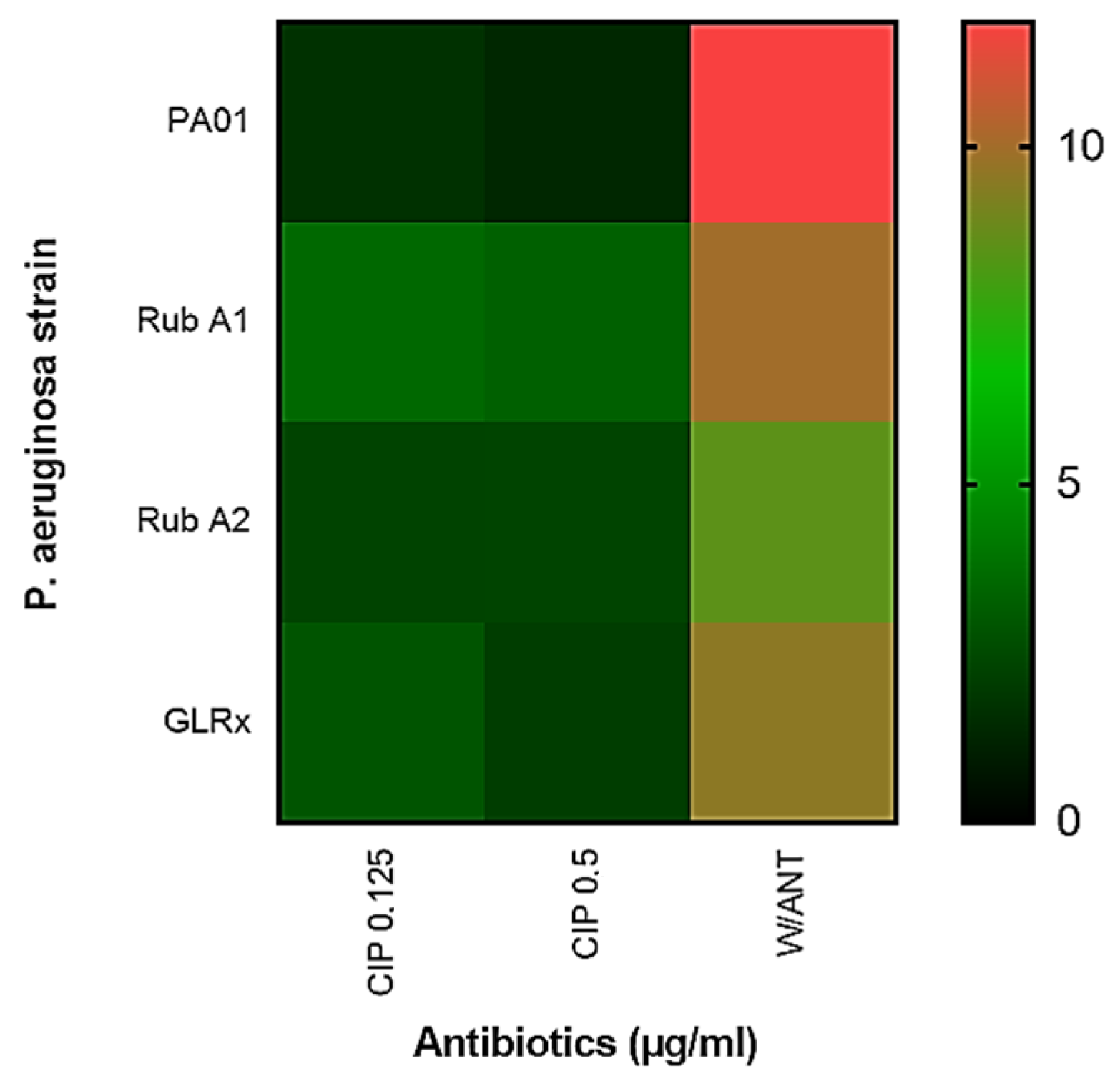
3.2. Viability Assay of Rub A1, Rub A2, GLRX and PAO1 under CIP Concentrations
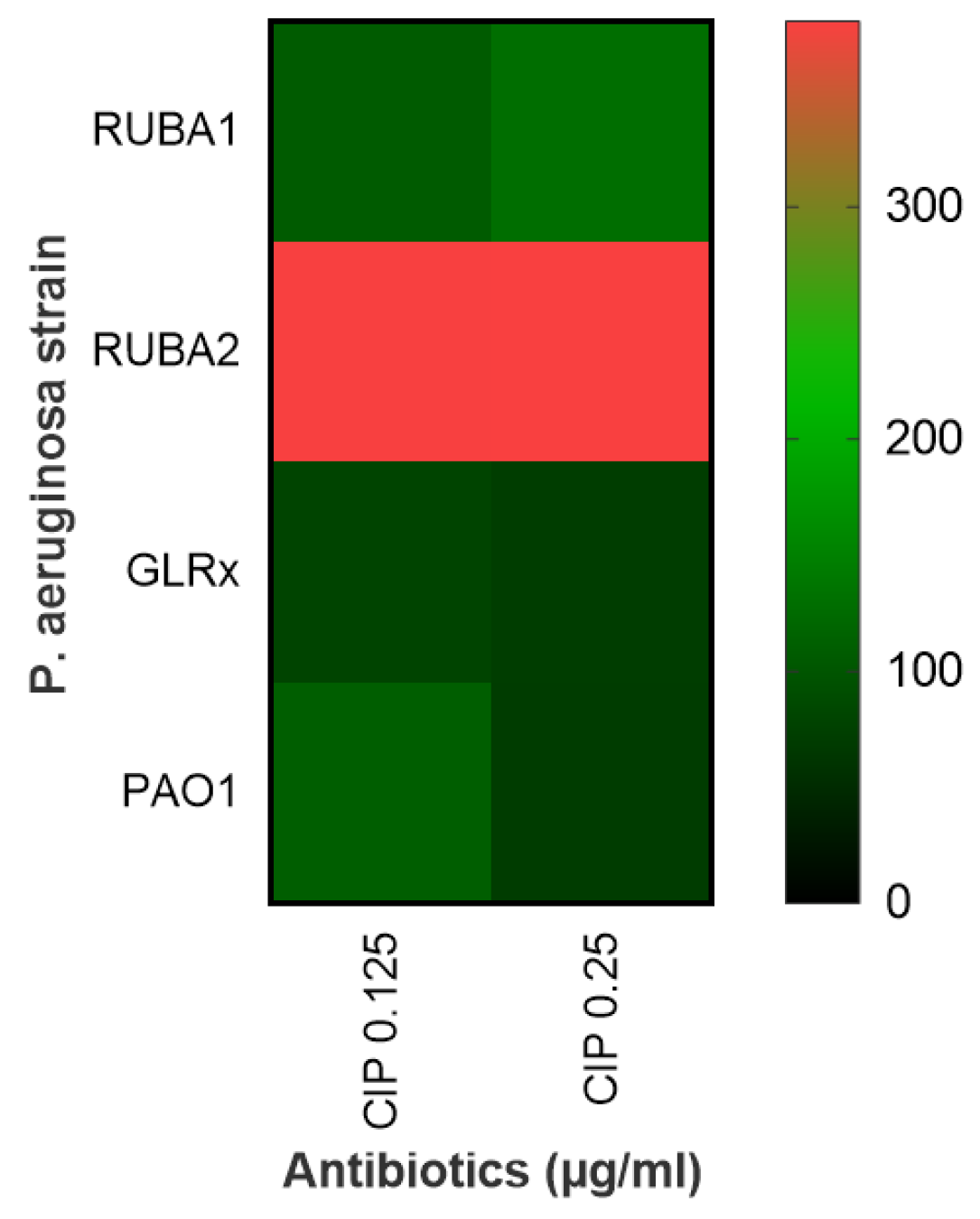
The viability assay involved assessing the percentage of dead cells using both concentrations of CIP and subsequently contrasting these results with those from the wild-type PAO1 strain. The increase in the number of dead cells was notably higher within the mutant strain Rub A2, as visually depicted in Figure 4. This observation underscored the impact of the mutation on cell viability under the influence of different CIP concentrations.
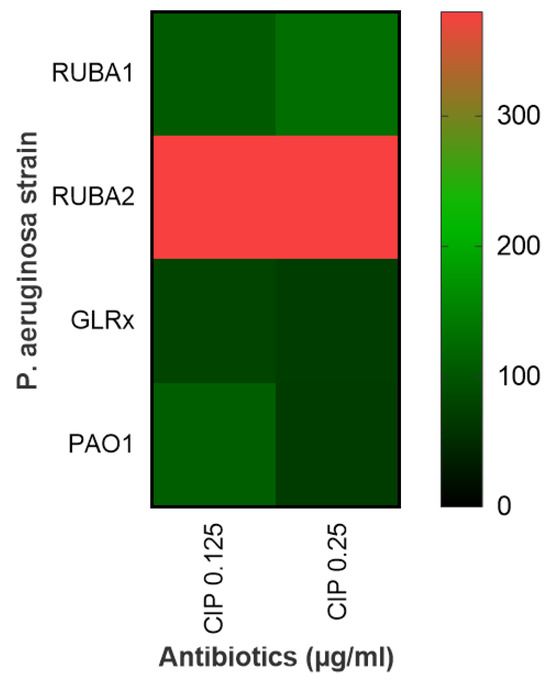
Figure 4.
Heatmap showing the effect of ciprofloxacin (CIP) on cell viability at two different concentrations (0.125 and 0.25 µg/mL) for Pseudomonas aeruginosa wild type (PAO1) in three mutant strains of PAO1 (rubredoxins: strains Rub A1 and Rub A2; a glutaredoxin: strain GLRX). The scale reflects cell death, with a higher death rate for higher values (from green to red).
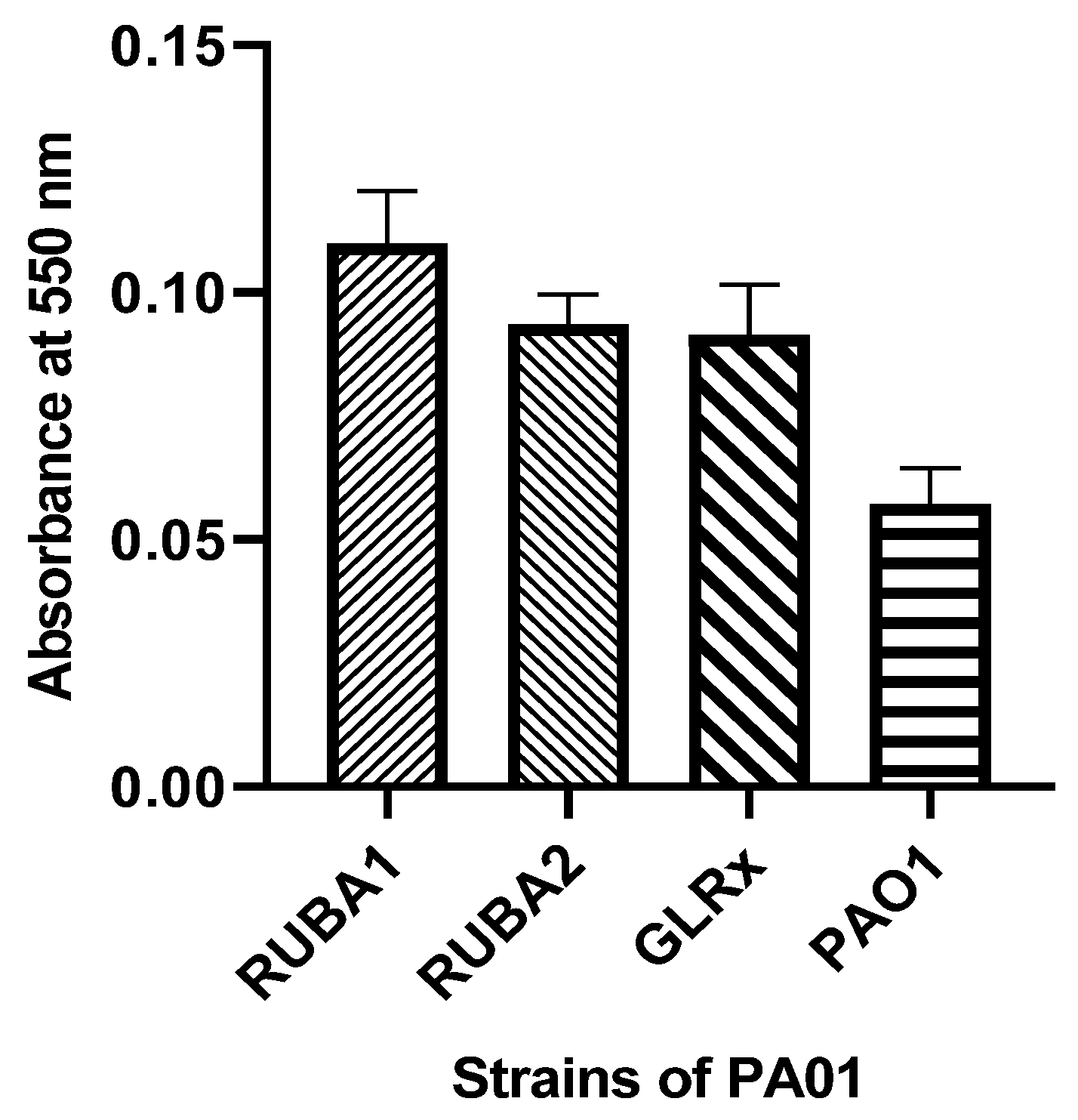
3.3. Biofilm Formation of Rub A1, Rub A2, GLRX and PAO1 under Normal Conditions
Within the biofilm assay, as illustrated in Figure 5, it is evident that the mutated strains exhibited a notable increase in biofilm production when contrasted with the biofilm production of the wild-type PAO1 strain.
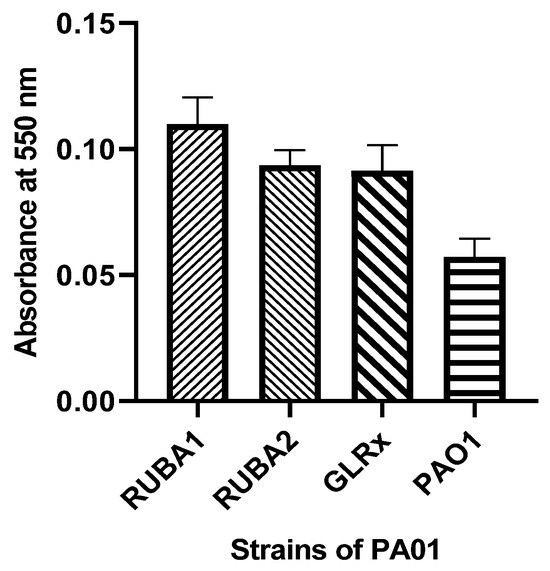
Figure 5.
Biofilm production in Pseudomonas aeruginosa wild type (PAO1) and three mutants of PAO1: two of them mutant in rubredoxins (strains named as Rub A1 and Rub A2) and one in glutaredoxin (strain named as GLRX). The results were measured at 550 nm.
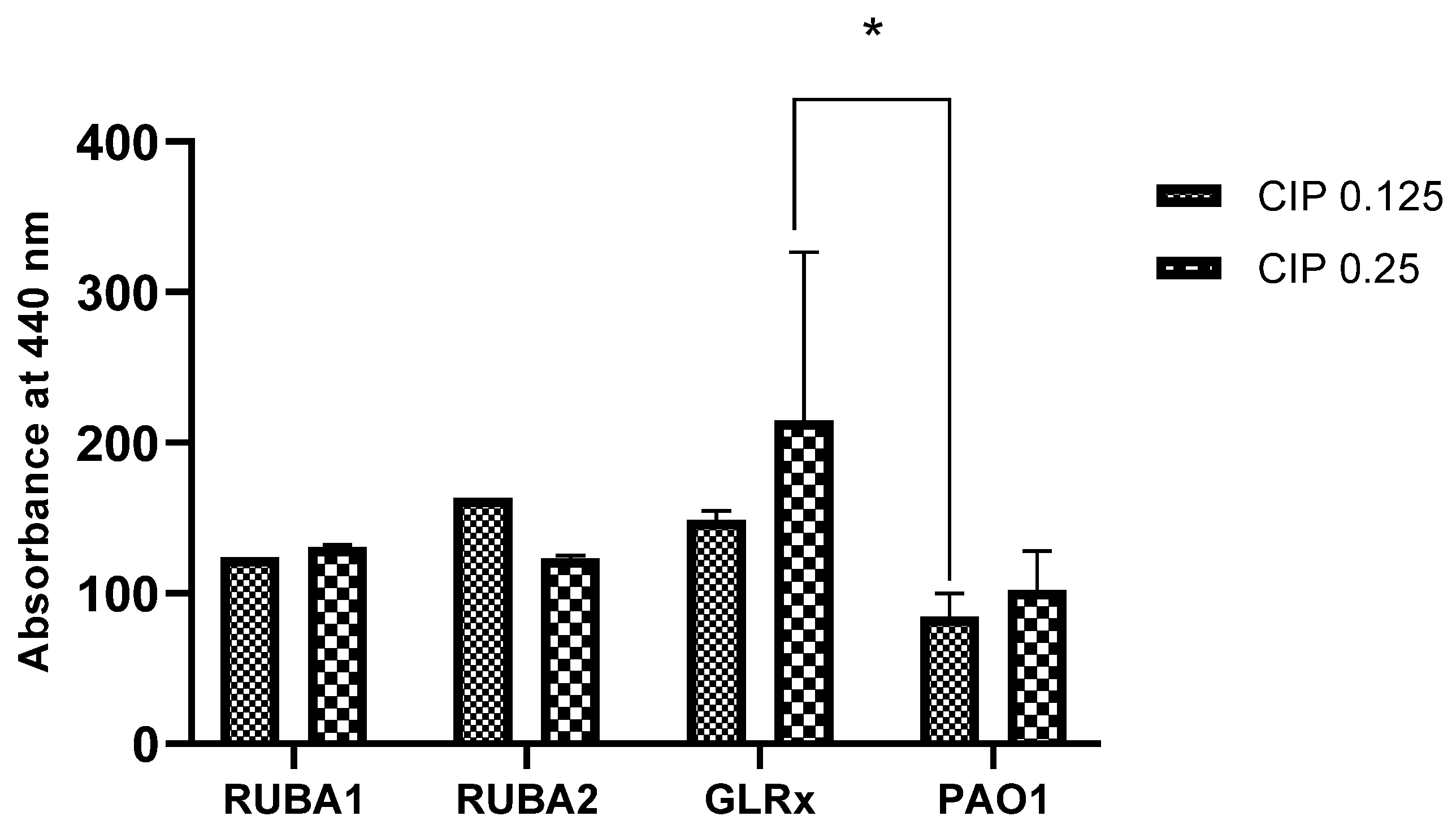
3.4. Evaluation of the Production of Proteases LasA in the Strains Rub A1, Rub A2, GLRX and PAO1 at Two CIP Concentrations
Following the assay of biofilm production, we aimed to evaluate the effect of ciprofloxacin on LasA protease production. The results, presented in Figure 6, show how LasA protease production varied among strains and CIP concentrations. The enzyme production was higher for all mutants in comparison to the wild type, for both CIP concentrations. Moreover, the GLRx mutant showed increased LasA protease production when compared to PAO1, for CIP at 0.25 µg/mL, with statistical significance (p < 0.05).
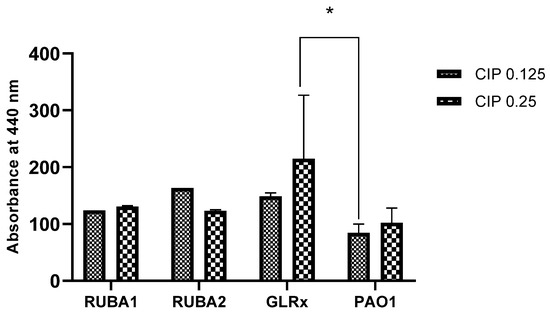
Figure 6.
Production of proteases in Pseudomonas aeruginosa wild type (PAO1) and three mutant strains of PAO1 (rubredoxins: strains Rub A1 and Rub A2; a glutaredoxin: strain GLRX). The results were measured at 440 nm under two concentrations of ciprofloxacin (CIP)— 0.125 and 0.25 µg/mL. * p < 0.05.
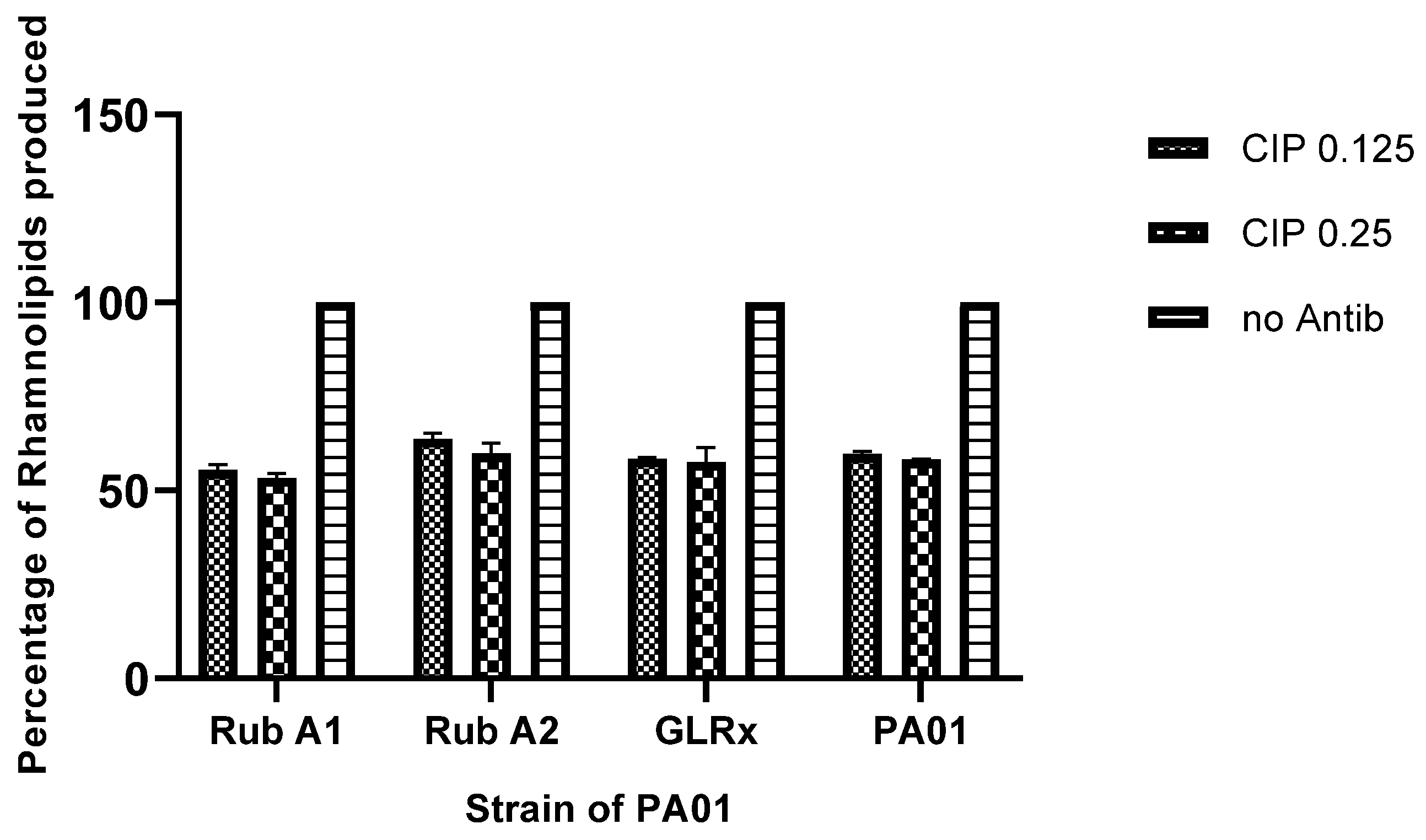
3.5. Rhamnolipids Production in the Strains Rub A1, Rub A2, GLRX and PAO1 under CIP Concentrations
Following treatment with the CIP antibiotic, the production of rhamnolipids was assessed. The results exhibited a decrease in rhamnolipid production by around 50% across all the tested strains, as shown in Figure 7. Moreover, the variation of CIP concentration provided no variation in the rhamnolipid production in all tested strains.
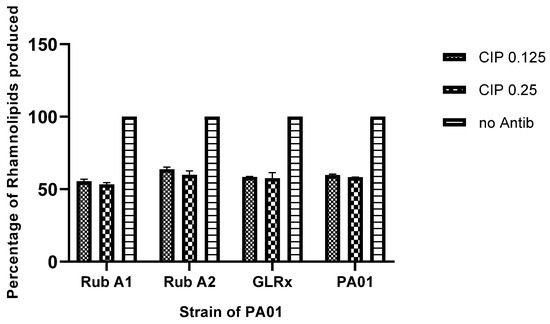
Figure 7.
The comparison of rhamnolipids produced in Pseudomonas aeruginosa wild type (PAO1) and three mutant strains of PAO1 (rubredoxins: strains Rub A1 and Rub A2; a glutaredoxin: strain GLRX) under two concentrations of ciproflaxin (CIP)—0.125 and 0.25 µg/mL. Results compared by percentage to the same strain without antibiotic.
3.6. Pyocyanin Comparison between the Strains Rub A1, Rub A2, GLRX and PAO1 under CIP Concentrations
The synthesis of pyocyanin in each distinct PAO1 strain was also assessed and is shown, in Figure 8, as a heatmap, at both CIP concentrations as well as without the antibiotic. In this heatmap is shown that, without antibiotics, PAO1 exhibits higher pyocyanin production when compared to any of the mutant strains. However, in the presence of CIP, results show that all strains produce less pyocyanin, with no statistical significance between them.
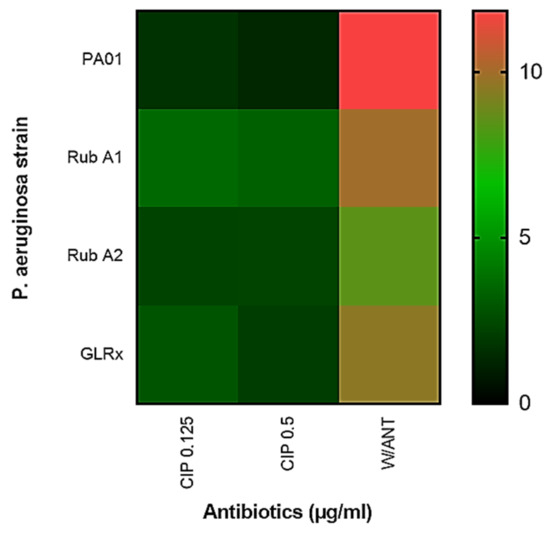
Figure 8.
Comparison of the pyocyanin produced in Pseudomonas aeruginosa wild type (PAO1) to the three mutant strains of PAO1 (rubredoxins: strains Rub A1 and Rub A2; a glutaredoxin: strain GLRX) with and without the use of the antibiotic ciprofloxacin (CIP) at the concentrations of 0.125 and 0.25 µg/mL.
3.7. Phage Assay
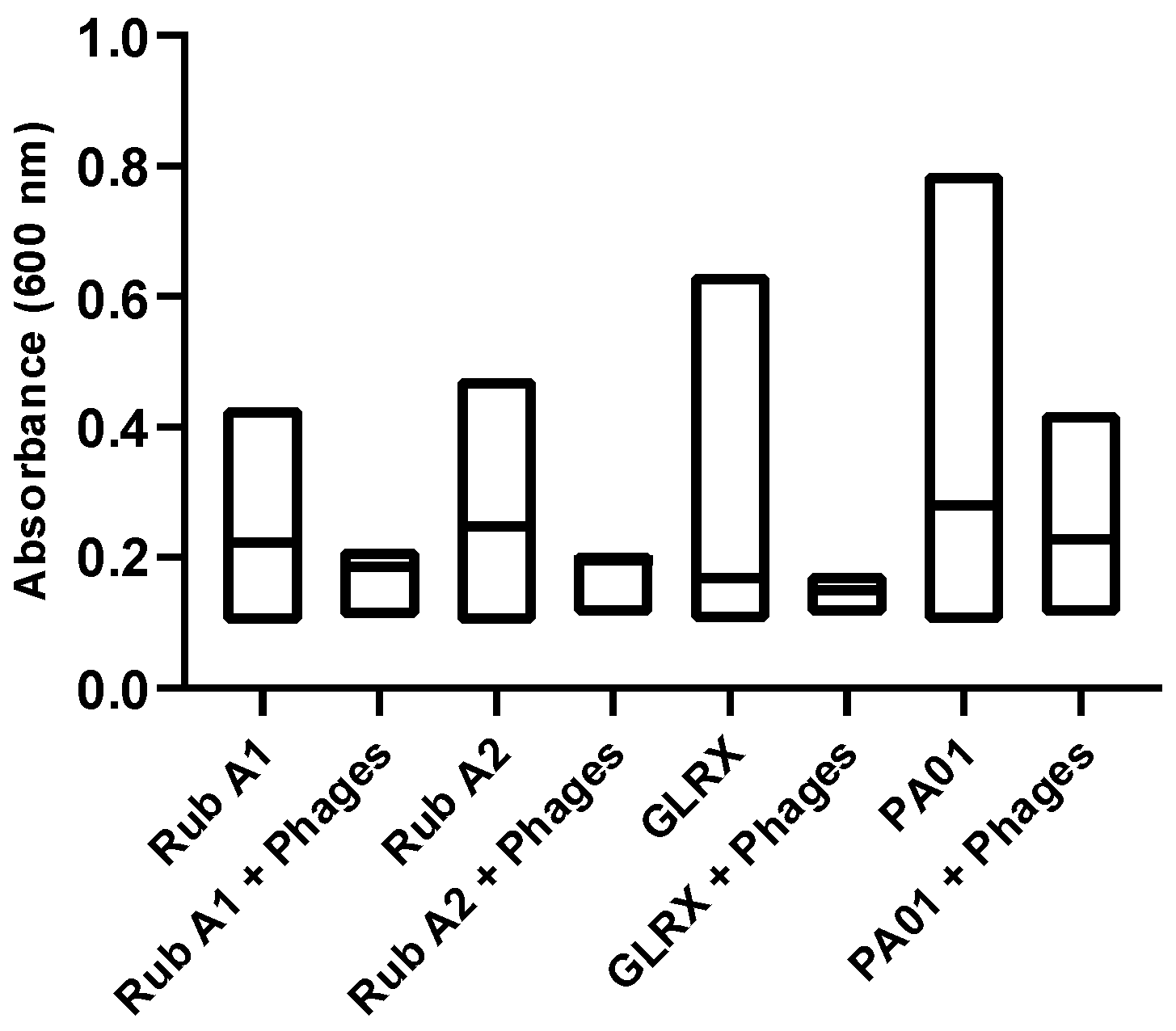
Next, the susceptibility of PAO1 strains to a specific phage cocktail was assessed, to evaluate the significance of each studied mutation. For that, we analyzed the growth curves of each strain both in the absence and presence of phage inoculation. This quantitative assessment, as illustrated in Figure 9, involved monitoring the changes in optical density at a wavelength of 600 nm using a spectrophotometer. The results unveiled a prevailing trend—a substantial portion of the strains exhibited resistance to phage infection, shedding light on the potential role of these mutations in influencing the bacterium’s interaction with the phage cocktail.
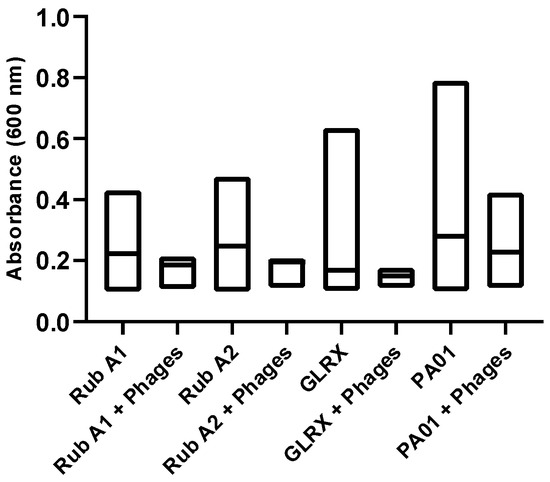
Figure 9.
The effect of the phage cocktail mix was tested for Pseudomonas aeruginosa wild type (PAO1) and three mutant strains of PAO1 (rubredoxins: strains Rub A1 and Rub A2; a glutaredoxin: strain GLRX) and measured by spectrophotometry at 600 nm. The distribution of the values in absorbance, at 600 nm, was compared between the strains with and without the inoculation of the phage cocktail.
4. Discussion
The study aimed to investigate the impact of three mutations (Rub A1, Rub A2, and GLRx) on bacterial quorum sensing modulation, including gene expression, virulence trait production, viability compared to PAO1, susceptibility to ciprofloxacin, and susceptibility to a phage cocktail for bacterial infections. For that, we started by assessing the expression of the genes lasI, lasR, pqsA, mvfR, and rpsL in the three mutant strains and comparing to the wild strain.
Considering the role of the mutated genes, the silencing of each of them in each of the mutants studied should implicate a decrease in some of the production of virulence traits, such as protease LasA, which are regulated by the gene lasR [28], and rhamnolipid and pyocyanin production, that is regulated by the gene rhlR [29]. The results in Figure 3 agree with the literature, showing an overall decrease in gene expression, in particular for genes LasR and rhlR, both associated with antioxidant protection, for all mutants in comparison with the wild type. Worthy of mentioning is that although the 2-ΔΔCT method is considered the gold standard of gene quantification [30], the interactions between the genes were not investigated in this study. For this, high-density DNA microarrays could be a useful methodology to understand the dynamic of the gene interactions and responses to the different environment conditions applied [31].
A representation in a heatmap for the viability of each tested strain in each concentration of CIP used can be observed in Figure 4. In this assay, the mutant strains Rub A2 and GLRX were significantly the most susceptible (p < 0.0001) and the most resistant (p < 0.05), respectively, to the CIP’s lower concentration when compared to the wild strain. The mutant Rub A1 did not show a significant difference in the viability cell numbers in the lower concentration of CIP when compared to the wild strain, only in the higher CIP concentration (p < 0.001). Rub A2 also showed vulnerability to the higher CIP concentration (p < 0.0001). GLRX did not show significant results when compared to the wild strain at higher concentration of CIP. The inhibition of catalase and rubredoxin reductase may be related to the increase in bacterial membrane lipid peroxidation, and this fact may have contributed to an increase in the susceptibility of the mutant Rub A2 to the CIP concentrations tested [32]. The non-significant result observed in the other mutant strain, Rub A1, may be related to the fact that, although Rub A1 and Rub A2 share 80% of the amino acid sequence, the differences in the substitutions can change the molecular surface and consequently modify the electron acceptors of each Rdxs [17]. GLRX function in P. aeruginosa is still unclear, although Saninjuk et al. found recently that this gene is involved in the maturation of [Fe-S] clusters [19]. Since iron can act as a cofactor for many enzymes, our suggestion for the result obtained is that the iron metabolism might be affected [33] and consequently the lack of a cofactor may have changed the structure of the main target of CIP, bacterial DNA topoisomerase and gyrase [3], leading to the resistance observed in the GLRX mutant. Further studies are necessary to assess this suggestion.
Biofilm production, shown in Figure 5, was significantly higher in mutant strains—Rub A1 (p < 0.0001), Rub A2 (p < 0.0001), and GLRX (p < 0.0001)—when compared to the wild strain. Although the biofilm increase for strain GLRX was expected according to the bibliography [34], the observed increase for mutants Rub A1 and Rub A2 were not foreseen, as the suppression in correspondent gene expression is associated with lower biofilm production [2,9,26]. The PQS is constituted by different genes: pqsABCDE, phnAB and pqsH [35]. The expression of pqsA affects the PQS and, consequently, the biofilm production [36]. Our study conducted a comparison between the expression of the gene pqsA in three mutant PAO1 versus the wild-type PAO1. Storz, et al. identified the pqsD enzyme as a key enzyme whose inhibition could repress the PQS and the biofilm production in P. aeruginosa [37]. Therefore, the possibility of the interaction between the other genes of PQS and biofilm production may be a hypothesis for the results observed.
In the protease production, shown in Figure 6, only the mutant strain GLRX showed a significant increase in proteases when compared to the wild strain for the higher CIP concentration (p < 0.05). These results were contradictory to those in a previous study, where the expression of the QS genes was decreased [28]. Once again, a possible explanation could be related with the iron metabolism. GLRX has been described before to have a synergy with iron metabolism [19], with this inorganic compound having a repressive effect on the production of LasA [38]. More studies are needed to explore this hypothesis.
Rhamnolipid production decreased when compared to the same strains without the antibiotic inoculation, as expected by the decrease observed in the production of QS genes previously [29]. There was no significant difference observed between the two concentrations of CIP used in this study in the rhamnolipid production in all the strains tested. Rhamnolipids are essential for the swarming motility of PAO1 and are involved in biofilm formation and bacteria protection from phagocytosis by macrophages. Additionally, these glycolipids can also act as hemolysins, making this virulence trait determinant in the pathogenesis of the bacteria. Therefore, more studies could be helpful to perceive the mechanism underlying the inhibition of these genes as a protective mechanism associated with host phagocytosis in virulent P. aeruginosa strains [39].
In Figure 8, a heatmap of the results regarding the production of pyocyanins by the PAO1 strains shows that all mutant strains had a lower production of pyocyanins when compared to the wild strain, in the absence of the antibiotic. With the inoculation of antibiotics, there was a lower production of pyocyanins in all strains tested due to the bactericidal effect of the CIP [40]. Pyocyanins are inducers of host oxidative stress, increasing the intracellular levels of reactive oxygen species. It has also been suggested by some authors that pyocyanins induce biofilm formation by promoting the interactions between PAO1 cells [41].
Regarding the results for the phage assay, in Figure 9, the mutant strains show significantly lower absorbance values than the wild strain of PAO1 (p < 0.0001), indicating that these three genes may be playing an important role or/and may affect the proliferation of this bacterium. Secondly, the phage cocktail used was significantly effective in all the PAO1 strains tested in this study, with lower growth density when compared to the control (p < 0.0001). Finally, the distribution of the results with the phage inoculation was lower than the mean of the control of these strains; otherwise, the distribution of the results obtained between the phages and the wild strain was higher and lower than the mean of the control of the growth of this bacteria. Therefore, the genes Rub A1, Rub A2, and GLRX may be involved in the ability of this strain to resist phage infection. More studies are needed to understand how this phage cocktail affects biofilm formation.
Finally, the mutation on genes Rdxs and GLRX influenced the expression of QS, the resistance to the antibiotic CIP, biofilm production, and other virulence factors quantified in this study. The results highlight the relevance of further studying the interaction of these genes as possible therapeutic aims for the treatment of P. aeruginosa infections.
5. Conclusions and Future Perspectives
This study aimed to investigate the impact of mutations in Rub A1, Rub A2, and GLRx genes on Pseudomonas aeruginosa (PAO1) by assessing gene expression, virulence trait production, susceptibility to ciprofloxacin (CIP), and susceptibility to a phage cocktail. The mutants exhibited altered gene expression profiles, with a notable decrease in genes associated with quorum sensing (QS). The study found that mutants generally displayed increased biofilm production, with unexpected results for Rub A1 and Rub A2 mutants, suggesting potential gene interactions influencing biofilms. Moreover, protease production significantly increased only in the GLRX mutant for higher CIP concentrations, potentially being linked to iron metabolism. Rhamnolipid production decreased as expected, while pyocyanin production was lower in all mutants and further reduced by CIP. Additionally, the mutants showed increased susceptibility to the phage cocktail, indicating the relevance of these genes in PAO1 resistance to phage infections. These findings underscore the potential therapeutic significance of exploring these genes in combating P. aeruginosa infections.
Author Contributions
S.S.: Research, software, original draft preparation; C.S.: Investigation, software, original draft preparation; M.C.D.: Investigation; M.V.: Investigation; S.L.: Investigation; R.F.: Investigation, review and editing; A.C.R.: Investigation; P.J.P.: Investigation; M.O.: Investigation; J.M.: Investigation; G.N.: Investigation; C.L.: Investigation; Á.G.: Investigation; J.M.M.: Investigation; D.M.-M.: Investigation; A.C.P.: Conceptualization, investigation, review and editing, project administration. P.B.: Conceptualization, investigation, review and editing, funding and resources, project administration. All authors have read and agreed to the published version of the manuscript.
Funding
Pilar Baylina (PB) acknowledges on the behalf of the authors the support of Fundação para a Ciência e Tecnologia (FCT), Portuguese Government, under the Strategic Project Reference: UID/BIM/04293/2013. PB was also supported by FEDER/02/SAICT/2020/072560. Marco Oliveira was funded by LaBMI, grant number PORTIC/IPP4COVID/BI/2021/01. Gonçalo Novais was funded by LaBMI, grant number PORTIC/LABMI/BII/2021/01. Patrick J. Pais was funded by FCT, grant number 2021.09498.BD. Ana Catarina Rocha was funded by FCT, grant number 2021.06521.BD.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Van Laar, T.A.; Esani, S.; Birges, T.J.; Hazen, B.; Thomas, J.M.; Rawata, M. Pseudomonas aeruginosa gshA Mutant Is Defective in Biofil Formation, Swarming, and Pyocyanin Production. Am. Soc. Microbiol. MSpere 2018, 3, e00155-18. [Google Scholar] [CrossRef]
- Tam, M.; Thi, T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Thai, T.; Salisbury, B.H.; Zito, P.M. Ciprofloxacin. StatPearls Publishing. 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK535454/ (accessed on 23 June 2022).
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef]
- Lee, K.; Yoon, S.S. Pseudomonas aeruginosa Biofilm, a Programmed Bacterial Life for Fitness. J. Microbiol. Biotechnol. 2017, 27, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Kostylev, M.; Kim, D.Y.; Smalley, N.E.; Salukhe, I.; Peter Greenberg, E.; Dandekar, A.A. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc. Natl. Acad. Sci. USA 2019, 116, 7027–7032. [Google Scholar] [CrossRef] [PubMed]
- Prateeksha; Bajpai, R.; Yusuf, M.A.; Upreti, D.K.; Gupta, V.K.; Singh, B.N. Endolichenic fungus, Aspergillus quandricinctus of Usnea longissima inhibits quorum sensing and biofilm formation of Pseudomonas aeruginosa PAO1. Microb. Pathog. 2020, 140, 103933. [Google Scholar] [CrossRef]
- Singh, B.N.; Prateeksha; Upreti, D.K.; Singh, B.R.; Defoirdt, T.; Gupta, V.K.; De Souza, A.O.; Singh, H.B.; Barreira, J.C.M.; Ferreira, I.C.F.R.; et al. Bactericidal, quorum quenching and anti-biofilm nanofactories: A new niche for nanotechnologists. Crit. Rev. Biotechnol. 2017, 37, 525–540. [Google Scholar] [CrossRef]
- Williams, P.; Cámara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- Jeong, S.J.; Yoon, S.S.; Bae, I.K.; Jeong, S.H.; Kim, J.M.; Lee, K. Risk factors for mortality in patients with bloodstream infections caused by carbapenem-resistant Pseudomonas aeruginosa: Clinical impact of bacterial virulence and strains on outcome. Diagn. Microbiol. Infect. Dis. 2014, 80, 130–135. [Google Scholar] [CrossRef]
- Malešević, M.; Di Lorenzo, F.; Filipić, B.; Stanisavljević, N.; Novović, K.; Senerovic, L.; Polović, N.; Molinaro, A.; Kojić, M.; Jovčić, B. Pseudomonas aeruginosa quorum sensing inhibition by clinical isolate Delftia tsuruhatensis 11304: Involvement of N-octadecanoylhomoserine lactones. Sci. Rep. 2019, 9, 16465. [Google Scholar] [CrossRef]
- Ahmed, S.A.K.S.; Rudden, M.; Smyth, T.J.; Dooley, J.S.G.; Marchant, R.; Banat, I.M. Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl. Microbiol. Biotechnol. 2019, 103, 3521–3535. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Li, X.-H.; Kim, S.-K.; Lee, J.-H. Post-secretional activation of Protease IV by quorum sensing in Pseudomonas aeruginosa. Sci. Rep. 2017, 7, 4416. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, M.; Jakobsen, T.H.; Kühl, M.; Bjarnsholt, T. The structure—Function relationship of Pseudomonas aeruginosa in infections and its influence on the microenvironment. FEMS Microbiol. Rev. 2022, 46, fuac018. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, A.A.; Kamer, A.M.A.; Al-Monofy, K.B.; Al-Madboly, L.A. Pseudomonas aeruginosa’s greenish-blue pigment pyocyanin: Its production and biological activities. Microb. Cell Factories 2023, 22, 110. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Hao, S.; Zhao, L.; Shi, F.; Ye, G.; Zou, Y.; Song, X.; Li, L.; Yin, Z.; He, X.; et al. Paeonol Attenuates Quorum-Sensing Regulated Virulence and Biofilm Formation in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 692474. [Google Scholar] [CrossRef]
- Hagelueken, G.; Wiehlmann, L.; Adams, T.M.; Kolmar, H.; Heinz, D.W.; Tu, B. Crystal structure of the electron transfer complex rubredoxin—Rubredoxin reductase of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2007, 104, 12276–12281. [Google Scholar] [CrossRef]
- Wiehlmann, L.; Urbanke, C.; Adams, T.M.; Munder, A.; Tümmler, B. The ancient rubredoxin system: An efficient defense mechanism of pseudomonas aeruginosa during infections. Eur. Respir. J. 2014, 44, 484. [Google Scholar]
- Saninjuk, K.; Romsang, A.; Duang-Nkern, J.; Wongsaroj, L.; Leesukon, P.; Dubbs, J.M.; Vattanaviboon, P.; Mongkolsuk, S. Monothiol Glutaredoxin Is Essential for Oxidative Stress Protection and Virulence in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2023, 89, e0171422. [Google Scholar] [CrossRef]
- Napper, K.R.; Leeper, T.C.; Khattri, R.; Morris, D.; Davis, C. Drug Interactions with Glutaredoxin Orthologues. Bachelor’s Thesis, The Honors College, University of Akron, Akron, OH, USA, 2015. [Google Scholar]
- Chegini, Z.; Khoshbayan, A.; Moghadam, M.T.; Farahani, I.; Jazireian, P. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 45. [Google Scholar] [CrossRef]
- Liu, R.; Han, G.; Li, Z.; Cun, S.; Hao, B.; Zhang, J.; Liu, X. Bacteriophage therapy in aquaculture: Current status and future challenges. Folia Microbiol. 2022, 67, 573–590. [Google Scholar] [CrossRef]
- Uyttebroek, S.; Chen, B.; Onsea, J.; Ruythooren, F.; Debaveye, Y.; Devolder, D.; Spriet, I.; Depypere, M.; Wagemans, J.; Lavigne, R.; et al. Safety and efficacy of phage therapy in difficult-to-treat infections: A systematic review. Lancet 2022, 22, e208–e220. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, S.N.; Manohar, P.; Sukumaran, S.; Sadagopan, S.; Loh, B.; Leptihn, S.; Nachimuthu, R. Antibacterial efficacy of lytic phages against multidrug-resistant Pseudomonas aeruginosa infections in bacteraemia mice models. BMC Microbiol. 2022, 22, 187. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.A.; Alwood, A.; Thaipisuttikul, I.; Spencer, D.; Haugen, E.; Ernst, S.; Will, O.; Kaul, R.; Raymond, C.; Levy, R.; et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 14339–14344. [Google Scholar] [CrossRef] [PubMed]
- Malgaonkar, A.; Nair, M. Quorum sensing in Pseudomonas aeruginosa mediated by RhlR is regulated by a small RNA PhrD. Sci. Rep. 2019, 9, 11. [Google Scholar] [CrossRef]
- Franke, J.D.; Braverman, A.L.; Cunningham, A.M.; Eberhard, E.E.; Perry, G.A. Erythrosin B: A versatile colorimetric and fluorescent vital dye for bacteria. BioTechniques 2019, 68, 7–13. [Google Scholar] [CrossRef]
- Gambello, M.J.; Kaye, S.; Iglewski, B.H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 1993, 61, 1180–1184. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Koch, A.K.; Fiechter, A.; Reiser, J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1994, 176, 2044–2054. [Google Scholar] [CrossRef]
- Adamski, M.G.; Gumann, P.; Baird, A.E. A Method for Quantitative Analysis of Standard and High-Throughput qPCR Expression Data Based on Input Sample Quantity. PLoS ONE 2014, 9, e103917. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Mathee, K. Comparative transcriptome analyses of Pseudomonas aeruginosa. Hum. Genom. 2009, 3, 349. [Google Scholar] [CrossRef]
- Shome, S.; Talukdar, A.D.; Nath, R.; Tewari, S. Curcumin-ZnO nanocomposite mediated inhibition of Pseudomonas aeruginosa biofilm and its mechanism of action. J. Drug Deliv. Sci. Technol. 2023, 81, 104301. [Google Scholar] [CrossRef]
- Rao, V.A. Iron Chelators with Topoisomerase-Inhibitory Activity and Their Anticancer Applications. Antioxid. Redox Signal. 2013, 18, 930–955. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Turner, K.E.; Kirienko, N.V. PqsA Promotes Pyoverdine Production via Biofilm Formation. Pathogens 2018, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front. Cell. Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef]
- Shahab, M.; Danial, M.; Khan, T.; Liang, C.; Duan, X.; Wang, D.; Gao, H.; Zheng, G. In Silico Identification of Lead Compounds for Pseudomonas Aeruginosa PqsA Enzyme: Computational Study to Block Biofilm Formation. Biomedicines 2023, 11, 961. [Google Scholar] [CrossRef] [PubMed]
- Storz, M.P.; Maurer, C.K.; Zimmer, C.; Wagner, N.; Brengel, C.; de Jong, J.C.; Lucas, S.; Müsken, M.; Häussler, S.; Steinbach, A.; et al. Validation of PqsD as an Anti-biofilm Target in Pseudomonas aeruginosa by Development of Small-Molecule Inhibitors. J. Am. Chem. Soc. 2012, 134, 16143–16146. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.C.; Ohman, D.E. Efficient production and processing of elastase and LasA by Pseudomonas aeruginosa require zinc and calcium ions. J. Bacteriol. 1992, 174, 4140–4147. [Google Scholar] [CrossRef]
- Wittgens, A.; Kovacic, F.; Müller, M.M.; Gerlitzki, M.; Santiago-schübel, B.; Hofmann, D.; Tiso, T.; Blank, L.M.; Henkel, M.; Hausmann, R.; et al. Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Appl. Microbiol. Biotechnol. 2017, 101, 2865–2878. [Google Scholar] [CrossRef]
- Kart, D.; Reçber, T.; Nemutlu, E.; Sagiroglu, M. Sub-Inhibitory Concentrations of Ciprofloxacin Alone and Combinations with Plant-Derived Compounds against P. aeruginosa Biofilms and Their Effects on the Metabolomic Profile of P. aeruginosa Biofilms. Antibiotics 2021, 10, 414. [Google Scholar] [CrossRef]
- Alatraktchi, F.A.; Svendsen, W.E.; Molin, S. Electrochemical Detection of Pyocyanin as a Biomarker for Pseudomonas aeruginosa: A Focused Review. Sensors 2020, 20, 5218. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).