RMAU-Net: Breast Tumor Segmentation Network Based on Residual Depthwise Separable Convolution and Multiscale Channel Attention Gates
Abstract
1. Introduction
- (1)
- The feature extraction module RDw block was designed, which is simple in structure and can capture more global breast tumor feature information.
- (2)
- It proposes a multi-scale channel attention gate module to better localize irregular breast tumors by portraying low-level features in both spatial and channel dimensions.
- (3)
- It uses the Patch Merging operation for downsampling so that breast ultrasound image information will not be lost.
- (4)
- Experiments were conducted on two breast ultrasound datasets, Dataset B and BUSI, and the results show that the method in this paper has superior segmentation performance and better generalization.
2. Related Work
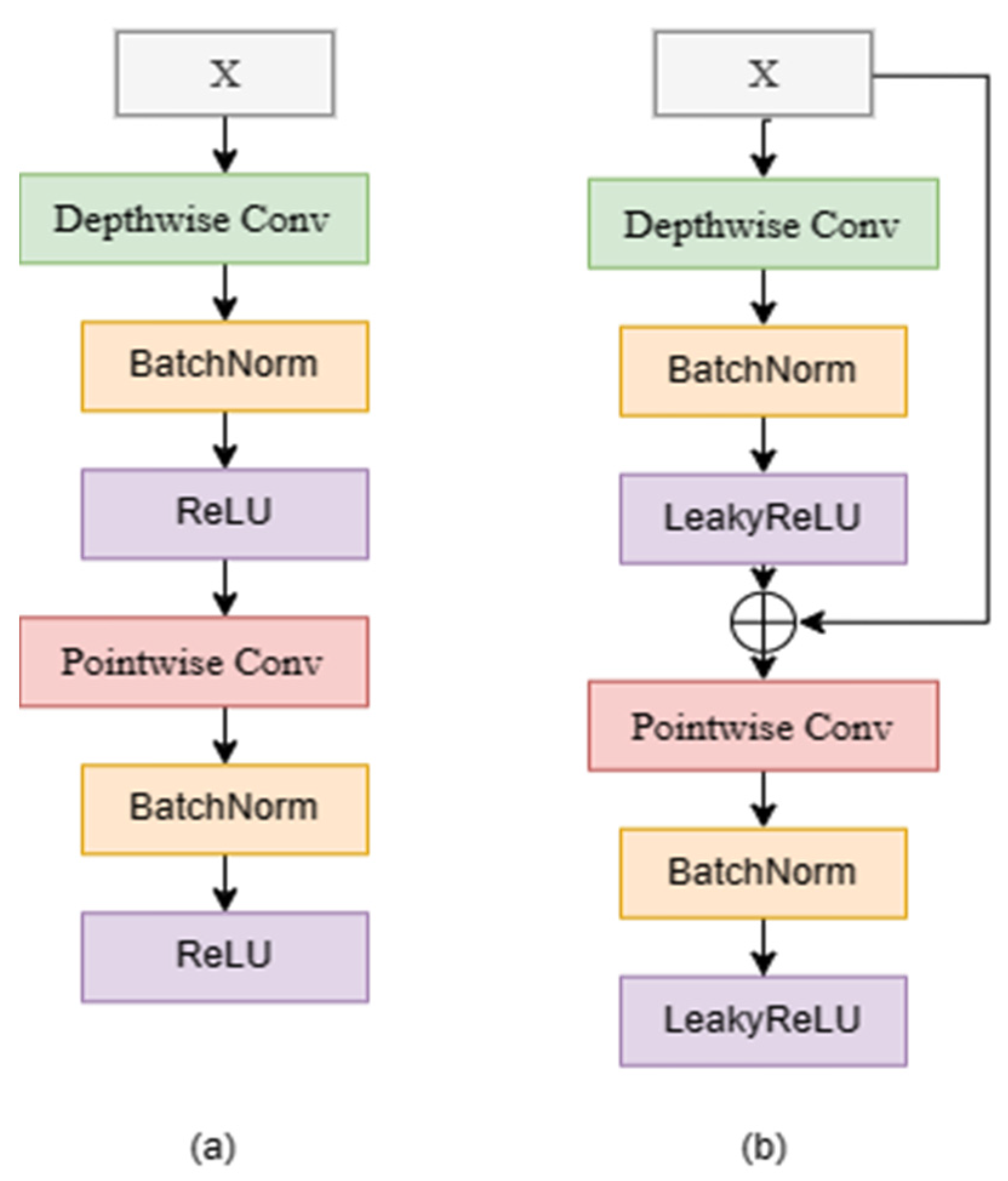
2.1. Depthwise Separable Convolution
2.2. Skip Connection
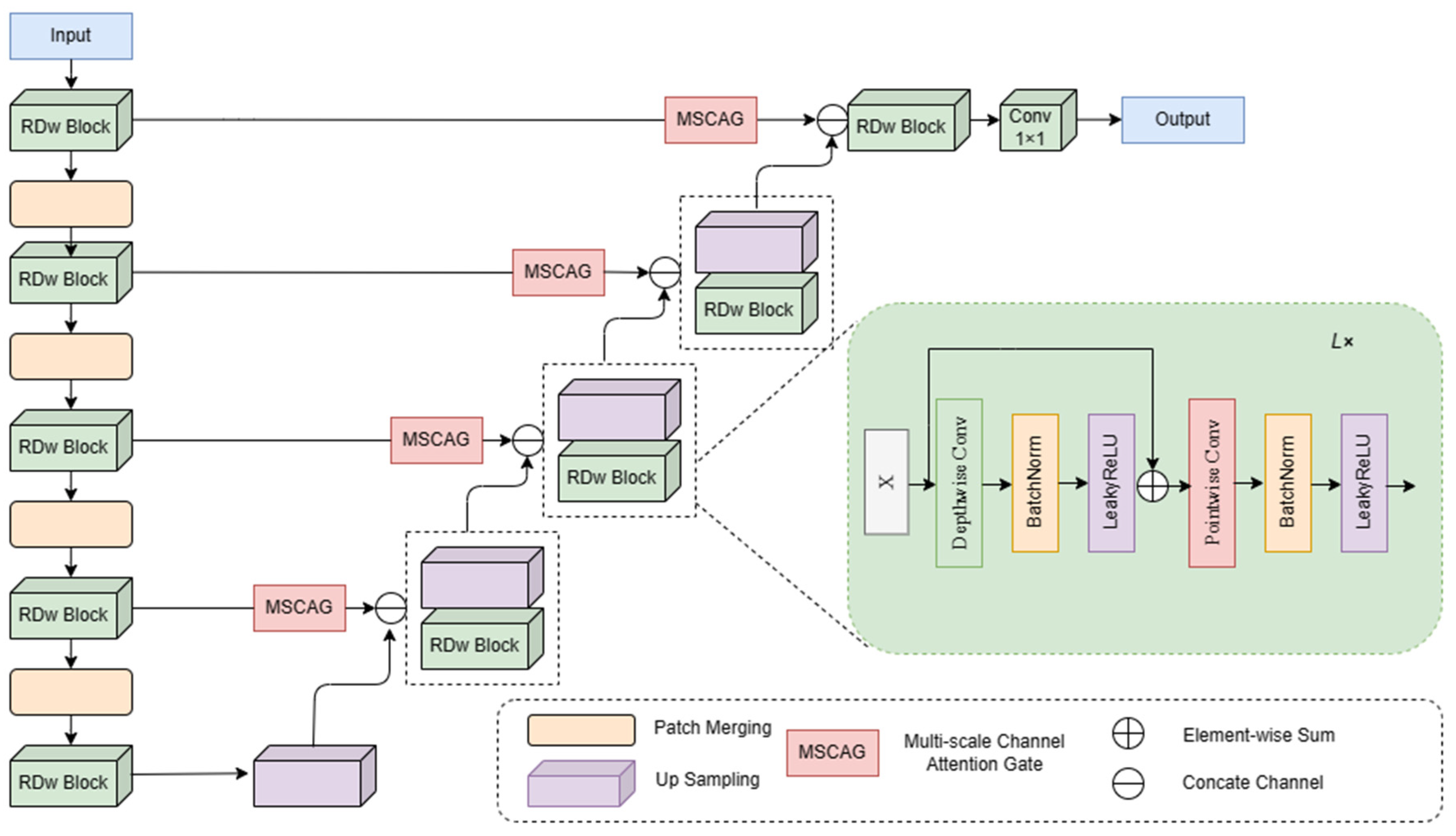
3. Method
3.1. RDw Block
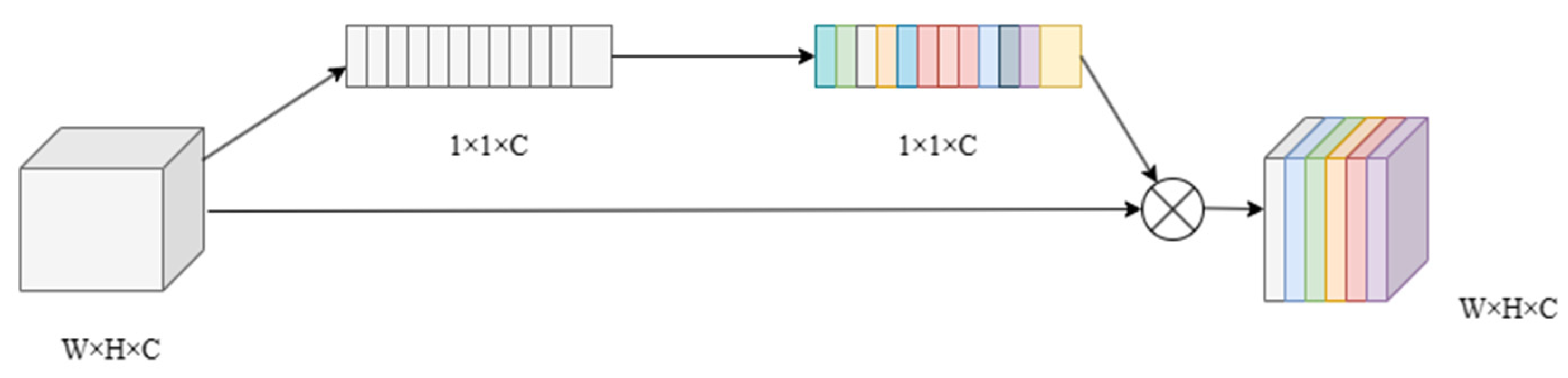
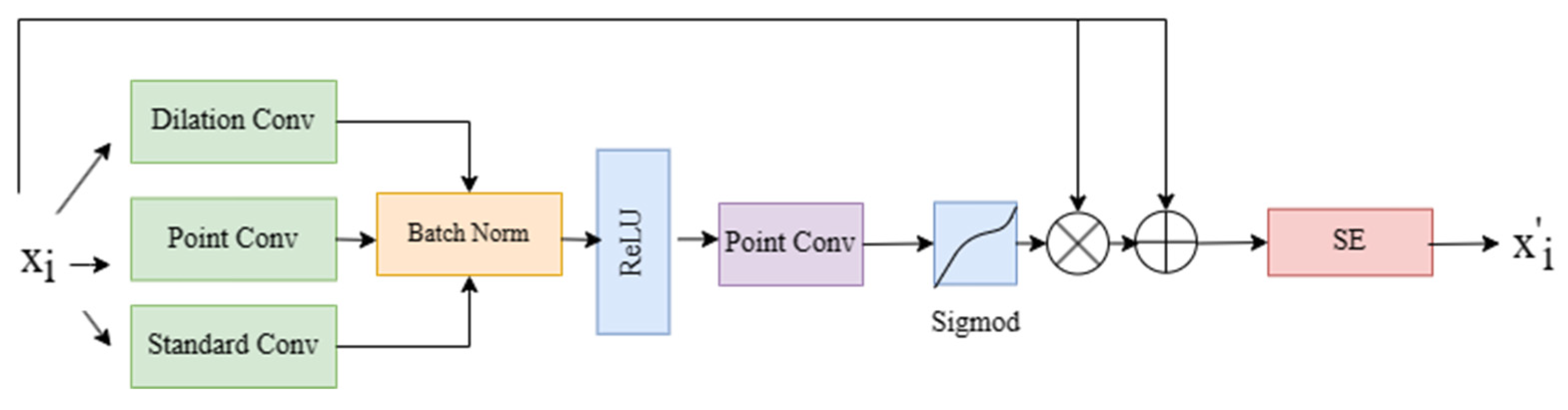
3.2. Multi-Scale Channel Attention Gate
3.3. Patch Merging
3.4. Loss Function
4. Experiment and Analysis
4.1. Dataset and Preprocessing
4.2. Experimental Settings
| Algorithm 1. The detailed training process of the RMAU-Net |
| Input: Augmented training sample S = {X1, X2, …Xn}, where X ∈ R256×256×3 1. Begin 2. Randomly initialize the model parameters 3. While ε have not converged do 4. For epoch = 0, 1, …, 300 do 5. The parameters were retrained on the target dataset 6. The Adam optimizer was used to update the weights, as expressed by α (learning rate), (the first corrected bias), (the second corrected bias) 7. The BDice loss function was used to update the weights, as expressed by , where “BDice” means BCE Loss and Dice Loss 8. Apply the cosine annealing strategy to adjust α 9. Continuously update the weighs using the S 10. 11. End for 12. End while 13. End Output: the best weight parameter |
4.3. Evaluation Indicators
4.4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Matsumoto, R.A.E.K.; Catani, J.H.; Campoy, M.L.; Oliveira, A.M.; Barros, N.D. Radiological findings of breast involvement in benign and malignant systemic diseases. Radiol. Bras. 2018, 51, 328–333. [Google Scholar] [CrossRef][Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Sahiner, B.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Wei, J.; Zhou, C.; Lu, Y. Computer-aided detection of clustered microcalcifications in digital breast tomosynthesis: A 3D approach. Med. Phys. 2012, 39, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.D.; Shan, J.; Ju, W.; Guo, Y.; Zhang, L. Automated breast cancer detection and classification using ultrasound images: A survey. Pattern Recognit. 2010, 43, 299–317. [Google Scholar]
- Noble, J.A.; Boukerroui, D. Ultrasound image segmentation: A survey. IEEE Trans. Med. Imaging 2006, 25, 987–1010. [Google Scholar] [PubMed]
- Wells, P.N.T.; Halliwell, M. Speckle in ultrasonic imaging. Ultrasonics 1981, 19, 225–229. [Google Scholar] [CrossRef]
- Xiao, G.; Brady, M.; Noble, J.A.; Zhang, Y. Segmentation of ultrasound B-mode images with intensity inhomogeneity correction. IEEE Trans. Med. Imaging 2002, 21, 48–57. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, L.; Cao, X.; Tong, Y. Breast tumors recognition based on edge feature extraction using support vector machine. Biomed. Signal Process. Control 2020, 58, 101825. [Google Scholar]
- Inoue, K.; Yamanaka, C.; Kawasaki, A.; Koshimizu, K.; Sasaki, T.; Doi, T. Computer aided detection of breast cancer on ultrasound imaging using deep learning. Ultrasound Med. Biol. 2017, 43, S19. [Google Scholar] [CrossRef][Green Version]
- Drukker, K.; Giger, M.L.; Horsch, K.; Kupinski, M.A.; Vyborny, C.J.; Mendelson, E.B. Computerized lesion detection on breast ultrasound. Med. Phys. 2002, 29, 1438–1446. [Google Scholar] [CrossRef]
- Horsch, K.; Giger, M.L.; Venta, L.A.; Vyborny, C.J. Automatic segmentation of breast lesions on ultrasound. Med. Phys. 2001, 28, 1652–1659. [Google Scholar] [CrossRef] [PubMed]
- Moon, W.K.; Lo, C.M.; Chen, R.T.; Shen, Y.W.; Chang, J.M.; Huang, C.S.; Chen, J.; Hsu, W.; Chang, R.F. Tumor detection in automated breast ultrasound images using quantitative tissue clustering. Med. Phys. 2014, 41, 042901. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Cheng, H.D.; Wang, Y. A novel segmentation method for breast ultrasound images based on neutrosophic l-means clustering. Med. Phys. 2012, 39, 5669–5682. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; De, A.; Bhattacharjee, A.K. MRI Skull Bone Lesion Segmentation Using Distance Based Watershed Segmentation. In Proceedings of the 3rd International Conference on Frontiers of Intelligent Computing: Theory and Applications (FICTA), Bhubaneswar, Odisha, India, 14–15 November 2014. [Google Scholar]
- Zhou, Z.; Wu, W.; Wu, S.; Tsui, P.H.; Lin, C.C.; Zhang, L.; Wang, T. Semi-automatic breast ultrasound image segmentation based on mean shift and graph cuts. Ultrason. Imaging 2014, 36, 256–276. [Google Scholar] [CrossRef]
- Huang, Q.H.; Lee, S.Y.; Liu, L.Z.; Lu, M.H.; Jin, L.W.; Li, A.H. A robust graph-based segmentation method for breast tumors in ultrasound images. Ultrasonics 2012, 52, 266–275. [Google Scholar] [CrossRef]
- Sohail, A.; Arif, F. Supervised and unsupervised algorithms for bioinformatics and data science. Prog. Biophys. Mol. Biol. 2020, 151, 14–22. [Google Scholar] [CrossRef]
- Sohail, A.; Younas, M.; Bhatti, Y.; Li, Z.; Tunç, S.; Abid, M. Analysis of trabecular bone mechanics using machine learning. Evol. Bioinform. 2019, 15, 1176934318825084. [Google Scholar] [CrossRef]
- Al-Utaibi, K.A.; Idrees, M.; Sohail, A.; Arif, F.; Nutini, A.; Sait, S.M. Artificial intelligence to link environmental endocrine disruptors (EEDs) with bone diseases. Int. J. Model. Simul. Sci. Comput. 2022, 13, 2250019. [Google Scholar] [CrossRef]
- Hu, G.; Du, Z. Adaptive kernel-based fuzzy c-means clustering with spatial constraints for image segmentation. Int. J. Pattern Recognit. Artif. Intell. 2019, 33, 1954003. [Google Scholar] [CrossRef]
- Xu, M.; Huang, K.; Chen, Q.; Qi, X. Mssa-net: Multi-scale self-attention network for breast ultrasound image segmentation. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021. [Google Scholar]
- Mannem, R.; Ca, V.; Ghosh, P.K. A SegNet based image enhancement technique for air-tissue boundary segmentation in real-time magnetic resonance imaging video. In Proceedings of the 2019 National Conference on Communications (NCC), Bangalore, India, 20–23 February 2019. [Google Scholar]
- Ben-Cohen, A.; Klang, E.; Raskin, S.P.; Soffer, S.; Ben-Haim, S.; Konen, E.; Amitai, M.M.; Greenspan, H. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng. Appl. Artif. Intell. 2019, 78, 186–194. [Google Scholar] [CrossRef]
- Amiri, M.; Brooks, R.; Rivaz, H. Fine tuning u-net for ultrasound image segmentation: Which layers? In Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data; Springer: Cham, Switzerland, 2019; pp. 235–242. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted res-unet for high-quality retina vessel segmentation. In Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018. [Google Scholar]
- Ibtehaz, N.; Sohel Rahman, M.M. Rethinking the U-Net architecture for multimodal biomedical image segmentation. arXiv 2019, arXiv:1902.04049. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.-W.; Wu, J. Unet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020. [Google Scholar]
- Valanarasu, J.M.J.; Patel, V.M. Unext: Mlp-based rapid medical image segmentation network. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Valanarasu, J.M.J.; Oza, P.; Hacihaliloglu, I.; Patel, V.M. Medical transformer: Gated axial-attention for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021. [Google Scholar]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. Transbts: Multimodal brain tumor segmentation using transformer. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021. [Google Scholar]
- Shareef, B.; Xian, M.; Vakanski, A. Stan: Small tumor-aware network for breast ultrasound image segmentation. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020. [Google Scholar]
- Lei, B.; Huang, S.; Li, R.; Bian, C.; Li, H.; Chou, Y.H.; Cheng, J.Z. Segmentation of breast anatomy for automated whole breast ultrasound images with boundary regularized convolutional encoder–decoder network. Neurocomputing 2018, 321, 178–186. [Google Scholar] [CrossRef]
- Xue, C.; Zhu, L.; Fu, H.; Hu, X.; Li, X.; Zhang, H.; Heng, P.A. Global guidance network for breast lesion segmentation in ultrasound images. Med. Image Anal. 2021, 70, 101989. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Lin, M.; Dou, H.; Lin, Z.; Ying, Q.; Jia, X.; Xu, W.; Mei, Z.; Yang, X.; Dong, Y.; et al. Boundary-rendering network for breast lesion segmentation in ultrasound images. Med. Image Anal. 2022, 80, 102478. [Google Scholar] [CrossRef]
- Tong, Y.; Liu, Y.; Zhao, M.; Meng, L.; Zhang, J. Improved U-net MALF model for lesion segmentation in breast ultrasound images. Biomed. Signal Process. Control 2021, 68, 102721. [Google Scholar] [CrossRef]
- Zhuang, Z.; Li, N.; Joseph Raj, A.N.; Mahesh, V.G.; Qiu, S. An RDAU-NET model for lesion segmentation in breast ultrasound images. PLoS ONE 2019, 14, e0221535. [Google Scholar] [CrossRef]
- Cho, S.W.; Baek, N.R.; Park, K.R. Deep Learning-based Multi-stage segmentation method using ultrasound images for breast cancer diagnosis. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 10273–10292. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Dar, M.F.; Ganivada, A. EfficientU-Net: A Novel Deep Learning Method for Breast Tumor Segmentation and Classification in Ultrasound Images. Neural Process. Lett. 2023, 1–24. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021. [Google Scholar]
- Yap, M.H.; Goyal, M.; Osman, F.; Martí, R.; Denton, E.; Juette, A.; Zwiggelaar, R. Breast ultrasound region of interest detection and lesion localisation. Artificial Intelligence in Medicine 2020, 107, 101880. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar] [CrossRef] [PubMed]

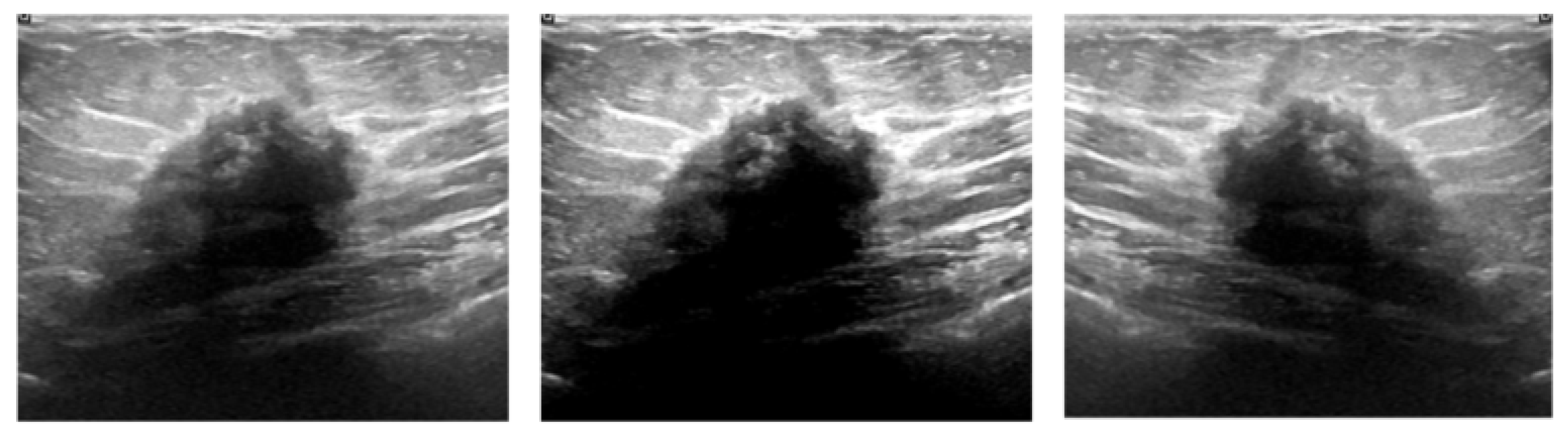
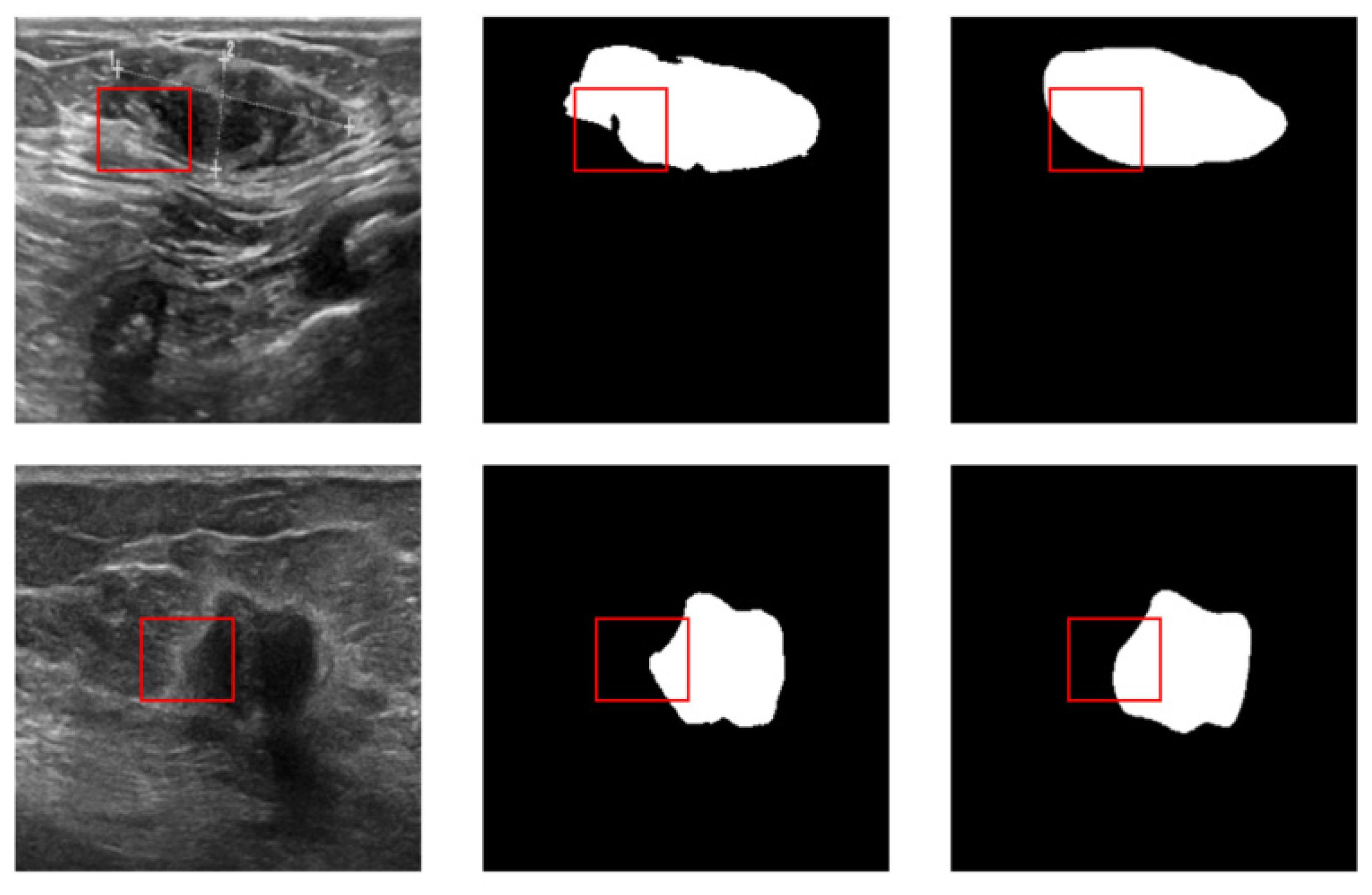
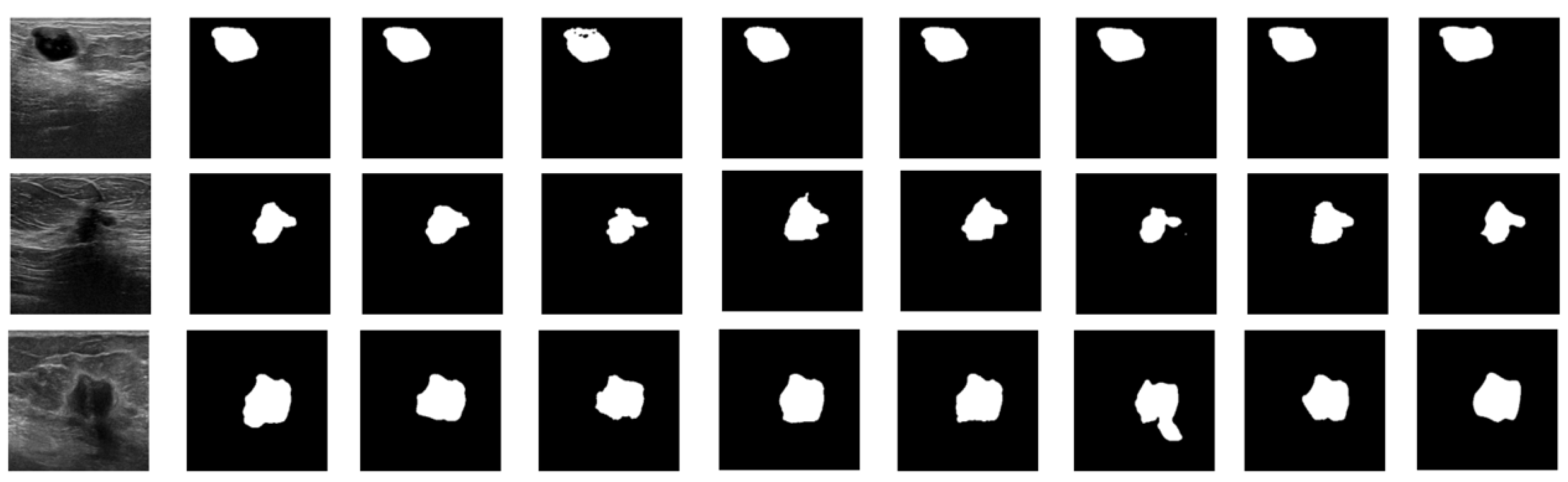

| Equipment | Benign | Malignant | Total | |
|---|---|---|---|---|
| Dataset B | Siemens ACUSON Sequoia C512 | 110 | 53 | 163 |
| BUSI | LOGIQ E9 and LOGIQ E9 Agile | 437 | 210 | 647 |
| Model | Dice | IoU | Recall | Precision | Accuracy |
|---|---|---|---|---|---|
| U-Net [24] | 86.28 | 75.89 | 87.15 | 85.75 | 99.15 |
| U-Net++ [25] | 83.84 | 72.20 | 85.17 | 82.79 | 98.99 |
| SegNet [23] | 83.51 | 71.86 | 80.33 | 87.49 | 99.05 |
| Attention U-Net [26] | 84.27 | 72.86 | 84.46 | 84.70 | 99.05 |
| UNeXt [30] | 81.09 | 68.57 | 80.09 | 81.66 | 98.81 |
| ResU-Net [27] | 84.37 | 73.71 | 85.35 | 83.98 | 99.04 |
| Ours | 87.12 | 77.61 | 86.04 | 88.55 | 99.22 |
| Model | Dice | IoU | Recall | Precision | Accuracy |
|---|---|---|---|---|---|
| U-Net [24] | 75.65 | 62.33 | 71.97 | 81.90 | 95.95 |
| U-Net++ [25] | 75.03 | 61.59 | 69.50 | 85.15 | 96.04 |
| SegNet [23] | 77.21 | 64.72 | 72.10 | 86.81 | 96.33 |
| Attention U-Net [26] | 74.65 | 61.52 | 68.85 | 85.60 | 95.99 |
| UNeXt [30] | 72.77 | 58.59 | 67.57 | 82.05 | 95.67 |
| ResU-Net [27] | 75.20 | 61.72 | 69.69 | 84.09 | 95.95 |
| Ours | 79.79 | 66.97 | 79.63 | 84.77 | 96.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, S.; Qiu, Z.; Li, P.; Hong, Y. RMAU-Net: Breast Tumor Segmentation Network Based on Residual Depthwise Separable Convolution and Multiscale Channel Attention Gates. Appl. Sci. 2023, 13, 11362. https://doi.org/10.3390/app132011362
Yuan S, Qiu Z, Li P, Hong Y. RMAU-Net: Breast Tumor Segmentation Network Based on Residual Depthwise Separable Convolution and Multiscale Channel Attention Gates. Applied Sciences. 2023; 13(20):11362. https://doi.org/10.3390/app132011362
Chicago/Turabian StyleYuan, Sheng, Zhao Qiu, Peipei Li, and Yuqi Hong. 2023. "RMAU-Net: Breast Tumor Segmentation Network Based on Residual Depthwise Separable Convolution and Multiscale Channel Attention Gates" Applied Sciences 13, no. 20: 11362. https://doi.org/10.3390/app132011362
APA StyleYuan, S., Qiu, Z., Li, P., & Hong, Y. (2023). RMAU-Net: Breast Tumor Segmentation Network Based on Residual Depthwise Separable Convolution and Multiscale Channel Attention Gates. Applied Sciences, 13(20), 11362. https://doi.org/10.3390/app132011362




