Learning Models for Bone Marrow Edema Detection in Magnetic Resonance Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset and Pre-Processing
2.2. High Intensity Masking (IBM)
2.3. Learning Models
2.4. Experimental Design
3. Results and Discussion
Limitations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maraghelli, D.; Brandi, M.L.; Matucci Cerinic, M.; Peired, A.J.; Colagrande, S. Edema-like marrow signal intensity: A narrative review with a pictorial essay. Skelet. Radiol. 2021, 50, 645–663. [Google Scholar] [CrossRef] [PubMed]
- von Brandis, E.; Zadig, P.K.; Avenarius, D.F.; Flatø, B.; Kristian Knudsen, P.; Lilleby, V.; Nguyen, B.; Rosendahl, K.; Ording Müller, L.S. Whole body magnetic resonance imaging in healthy children and adolescents. bone marrow appearances of the axial skeleton. Eur. J. Radiol. 2022, 154, 110425. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Kramer, J.; Vakil-Adli, A.; Aigner, N.; Breitenseher, M. Painful bone marrow edema of the knee: Differential diagnosis and therapeutic concepts. Orthop. Clin. North Am. 2004, 35, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Choi, S.T.; Lee, G.Y.; Ha, Y.J.; Choi, S.I. Method for diagnosing the bone marrow edema of sacroiliac joint in patients with axial spondyloarthritis using magnetic resonance image analysis based on deep learning. Diagnostics 2021, 11, 1156. [Google Scholar] [CrossRef] [PubMed]
- Tibrewala, R.; Ozhinsky, E.; Shah, R.; Foreman, S.C.; Pedoia, V.; Majumdar, S. Detecting hip osteoarthritic degenerative changes in MRI using deep learning. Osteoarthr. Cartil. 2019, 27, S387–S388. [Google Scholar] [CrossRef]
- Peer, M.; Abboud, S.; Hertz, U.; Amedi, A.; Arzy, S. Intensity-based masking: A tool to improve functional connectivity results of resting-state fMRI. Hum. Brain Mapp. 2016, 37, 2407–2418. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Eweje, F.R.; Bao, B.; Wu, J.; Dalal, D.; hua Liao, W.; He, Y.; Luo, Y.; Lu, S.; Zhang, P.; Peng, X.; et al. Deep Learning for Classification of Bone Lesions on Routine MRI. EBioMedicine 2021, 68, 103402. [Google Scholar] [CrossRef] [PubMed]
- Georgeanu, V.A.; Mămuleanu, M.; Ghiea, S.; Selișteanu, D. Malignant Bone Tumors Diagnosis Using Magnetic Resonance Imaging Based on Deep Learning Algorithms. Medicina 2022, 58, 636. [Google Scholar] [CrossRef] [PubMed]
- Astuto, B.; Flament, I.; Namiri, N.K.; Shah, R.; Bharadwaj, U.; Link, T.M.; Bucknor, M.D.; Pedoia, V.; Majumdar, S. Automatic deep learning-assisted detection and grading of abnormalities in knee MRI studies. Radiol. Artif. Intell. 2021, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kang, J.W.; Lee, D.E.; Son, W.; Lee, S.M.; Park, C.; Kim, M. Deep learning approaches for bone marrow edema detection and interpretation in dual-energy CT. SSRN Electron. J. 2022. [Google Scholar] [CrossRef]
- Levine, M. Assessing Bone Health in children and adolescents. Indian J. Endocrinol. Metab. 2012, 16, 205. [Google Scholar] [CrossRef] [PubMed]
- von Brandis, E.; Jenssen, H.B.; Avenarius, D.F.; Bjørnerud, A.; Flatø, B.; Tomterstad, A.H.; Lilleby, V.; Rosendahl, K.; Sakinis, T.; Zadig, P.K.; et al. Automated segmentation of Magnetic Resonance Bone Marrow Signal: A feasibility study. Pediatr. Radiol. 2022, 52, 1104–1114. [Google Scholar] [CrossRef] [PubMed]
- Chuah, T.K.; Poh, C.L.; Sheah, K. Quantitative texture analysis of MRI images for detection of cartilage-related bone marrow edema. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 5112–5115. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. (IJCV) 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Silva, F.; Pereira, T.; Neves, I.; Morgado, J.; Freitas, C.; Malafaia, M.; Sousa, J.; Fonseca, J.; Negrão, E.; Flor de Lima, B.; et al. Towards Machine Learning-Aided Lung Cancer Clinical Routines: Approaches and Open Challenges. J. Pers. Med. 2022, 12, 480. [Google Scholar] [CrossRef] [PubMed]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning for medical imaging. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019; Volume 32, pp. 3347–3357. [Google Scholar]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar]
- Krogh, A.; Hertz, J. A simple weight decay can improve generalization. Adv. Neural Inf. Process. Syst. 1991, 4, 950–957. [Google Scholar]
- Almajalid, R.; Zhang, M.; Shan, J. Fully automatic knee bone detection and segmentation on three-dimensional MRI. Diagnostics 2022, 12, 123. [Google Scholar] [CrossRef] [PubMed]
- Faisal, A.; Ng, S.C.; Goh, S.L.; George, J.; Supriyanto, E.; Lai, K.W. Multiple LREK active contours for knee meniscus ultrasound image segmentation. IEEE Trans. Med. Imaging 2015, 34, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Faisal, A.; Ng, S.C.; Goh, S.L.; Lai, K.W. Knee cartilage segmentation and thickness computation from ultrasound images. Med. Biol. Eng. Comput. 2017, 56, 657–669. [Google Scholar] [CrossRef]


| # Slices with Bones Segmented | # Patients | |||
|---|---|---|---|---|
| With Edema Regions | Without Edema Regions | Total Slices | Total Patients | |
| Edema Patients | 228 | 85 | 313 | 28 |
| Non-Edema Patients | - | 36 | 36 | 36 |
| Hyperparameter | Range of Values |
|---|---|
| Learning Rate | 0.01, 0.005, 0.001, 0.0001, 0.00001 |
| Batch size | 16, 32, 64, 112 |
| Momentum | 0, 0.5, 0.9 |
| Optimizer | SGD, Adam |
| Training | ROI | Balanced Accuracy | AUC |
|---|---|---|---|
| Scratch | Bone Region | 0.545 | 0.534 |
| HIM | 0.550 | 0.971 | |
| TL (fixed feature extractor) | Bone Region | 0.583 | 0.618 |
| HIM | 0.588 | 0.683 | |
| TL (fine-tuned feature extractor) | Bone Region | 0.568 | 0.550 |
| HIM | 0.852 | 0.964 |
| Predicted Class | |||
|---|---|---|---|
| Non-Edema | Edema | ||
| True class | Non-edema | 8 | 2 |
| Edema | 4 | 38 | |
| Run | Balanded Accuracy without New Non-Edema Slices | Balanded Accuracy with New Non-Edema Slices |
|---|---|---|
| #1 | 0.852 | 0.739 |
| #2 | 0.791 | 0.687 |
| #3 | 0.801 | 0.630 |
| #4 | 0.759 | 0.613 |
| #5 | 0.760 | 0.693 |
| 0.792 ± 0.034 | 0.672 ± 0.045 |
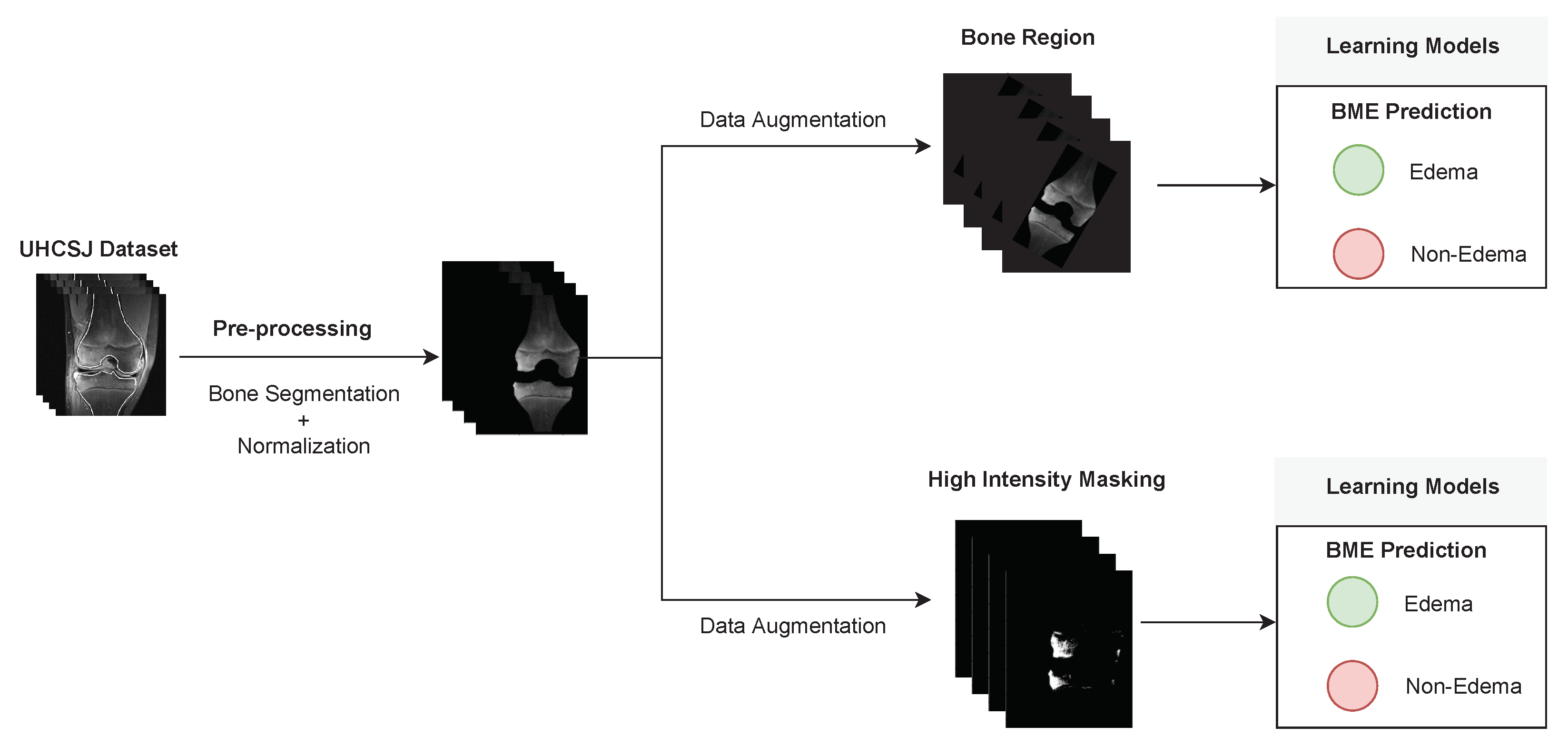
| Patient | Patient ID | Image Label | Image Prediction | Original Image | High Intensity Masking Image |
|---|---|---|---|---|---|
| Edema | 21 | Edema | Non-edema | 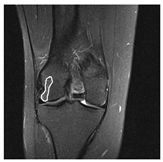 |  |
| Edema | 21 | Non-edema | Edema |  |  |
| Non-edema | 2 | Non-edema | Edema | 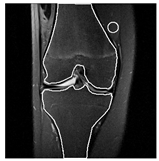 |  |
| Non-edema | 5 | Non-edema | Edema | 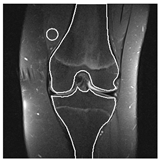 | 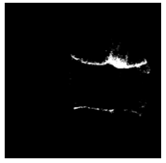 |
| Non-edema | 20 | Non-edema | Edema | 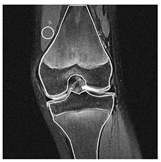 |  |
| Non-edema | 30 | Non-edema | Edema |  | 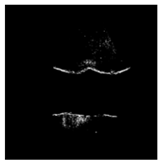 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, G.; Pereira, T.; Silva, F.; Sousa, J.; Carvalho, D.C.; Dias, S.C.; Oliveira, H.P. Learning Models for Bone Marrow Edema Detection in Magnetic Resonance Imaging. Appl. Sci. 2023, 13, 1024. https://doi.org/10.3390/app13021024
Ribeiro G, Pereira T, Silva F, Sousa J, Carvalho DC, Dias SC, Oliveira HP. Learning Models for Bone Marrow Edema Detection in Magnetic Resonance Imaging. Applied Sciences. 2023; 13(2):1024. https://doi.org/10.3390/app13021024
Chicago/Turabian StyleRibeiro, Gonçalo, Tania Pereira, Francisco Silva, Joana Sousa, Diogo Costa Carvalho, Sílvia Costa Dias, and Hélder P. Oliveira. 2023. "Learning Models for Bone Marrow Edema Detection in Magnetic Resonance Imaging" Applied Sciences 13, no. 2: 1024. https://doi.org/10.3390/app13021024
APA StyleRibeiro, G., Pereira, T., Silva, F., Sousa, J., Carvalho, D. C., Dias, S. C., & Oliveira, H. P. (2023). Learning Models for Bone Marrow Edema Detection in Magnetic Resonance Imaging. Applied Sciences, 13(2), 1024. https://doi.org/10.3390/app13021024








