Hypergraph-Clustering Method Based on an Improved Apriori Algorithm
Abstract
:1. Introduction
- (1)
- This paper extends association rules to hypergraphs, then proposes a hypergraph-partitioning high-dimensional data-clustering method based on the improved Apriori algorithm (HDHPA), providing a solution for identifying clustering results of high-dimensional data.
- (2)
- In order to avoid the generation of redundant rules and improve the mining efficiency, the parallel mining of different frequent itemsets is used to obtain hyperedges with corresponding ranks.
- (3)
- The HDHPA method improves clustering accuracy by constructing a hypergraph structure of high-dimensional data. In order to demonstrate the superiority of the method, this paper conducted numerous experiments on multiple datasets.
2. Related Theoretical Foundations
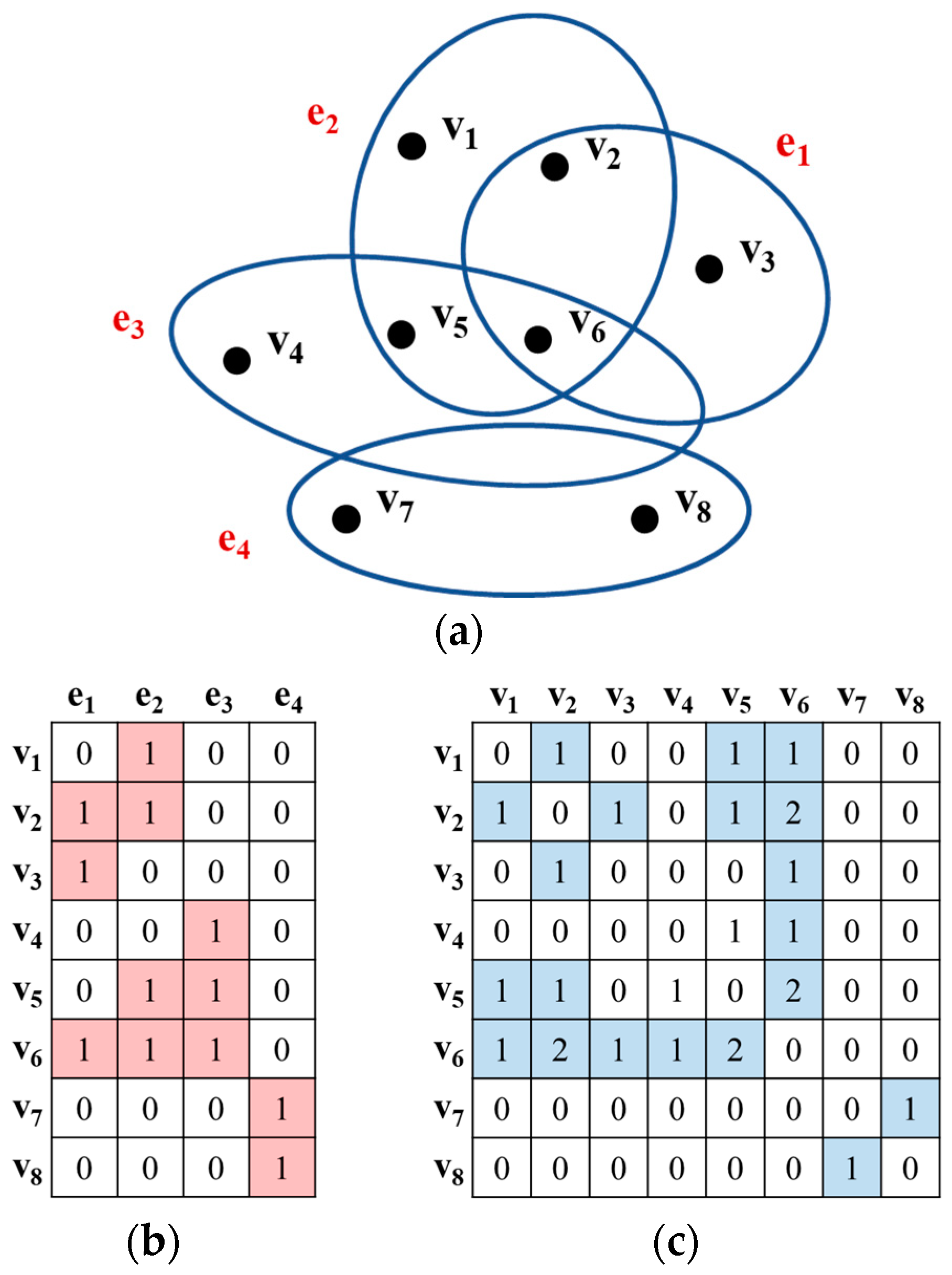
2.1. The Concept of Hypergraph
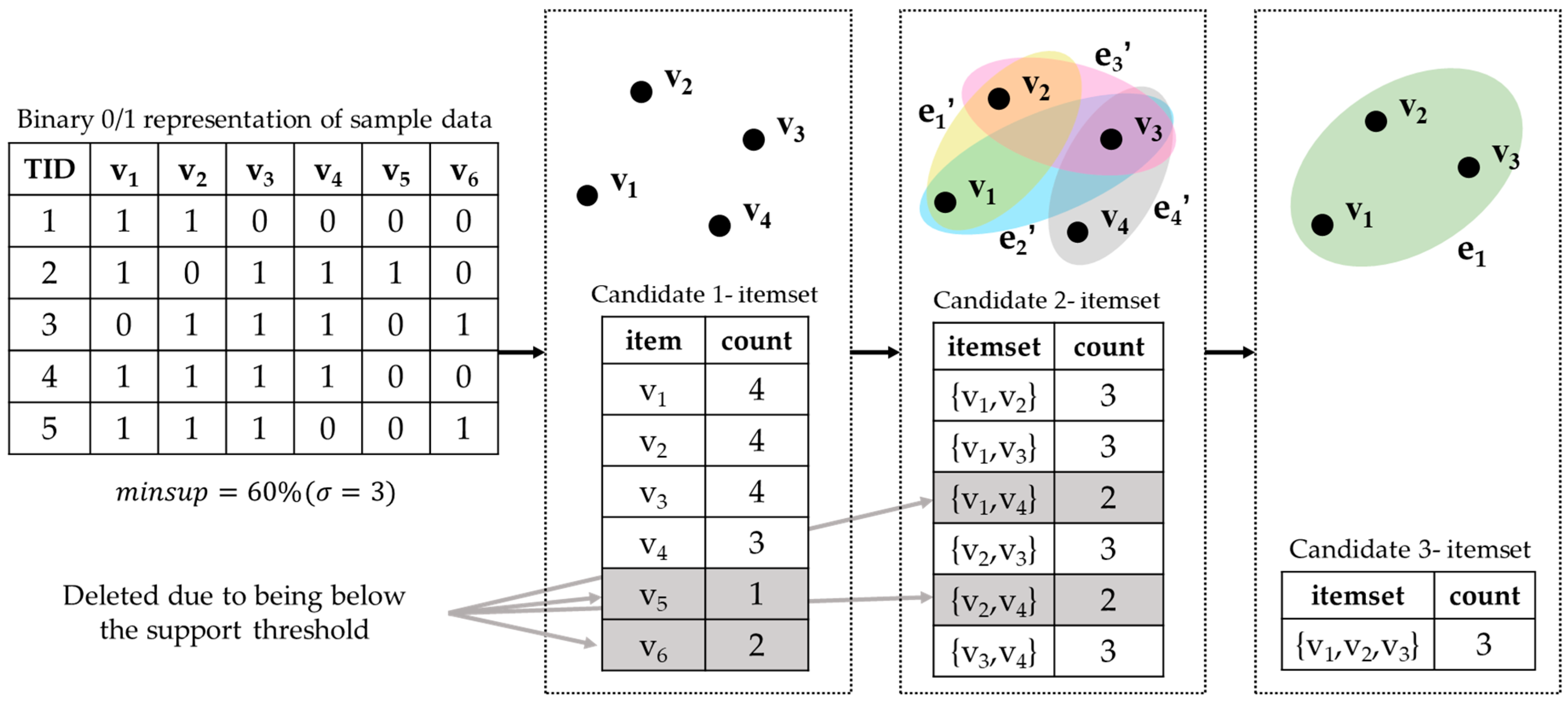
2.2. Apriori of Association Rules
- Scan the dataset L, calculate the support of each item, discard items that do not meet the support threshold, and generate one-dimensional frequent itemsets.
- Iteration: generate two itemsets through the one-dimensional itermsets, discard items that do not meet the support threshold, and generate two-dimensional frequent itemsets.
- Repeat, establish k-dimensional candidate itemsets, discard items that do not meet the support threshold, and generate k-dimensional frequent itemsets.
- Prune each k-itemset; count and retain the association rule itemset that satisfies the set confidence threshold.
2.3. Hypergraph Model Definition with Association Rules
2.4. DSP Algorithm in Hypergraph Partitioning
| Algorithm 1: Partitioning DSP algorithm |
| Input: Hypergraph H and an initial permutation P Output: DSP(H) 1: Apply min-partition () on P to get 2: Set and 3: Repeat 4: for each do 5: if then 6: Apply permutation-reorder () on R to get 7: if then 8: Replace R by in 9: Apply min-partition () on to get 10: Replace R by in 11: end if 12: end if 13: end for 14: Apply min-merge () on to get 15: Set , and 16: until P does not change 17: For each , apply disjoint-partition () on 18: return DSP(H) by Equation (5) |
3. Method Design of HDHPA
3.1. Basic Ideology of HDHPA
3.2. The Stage of Constructing a Hypergraph
| Algorithm 2: HDHPA method for constructing the hypergraph stage |
| Input: A set of all items in dataset L: Output: ,
|
3.3. Hypergraph Partitioning Stage
3.4. Dense Sub-Hypergraph Merging
4. Experiments and Analysis
4.1. Dataset Description
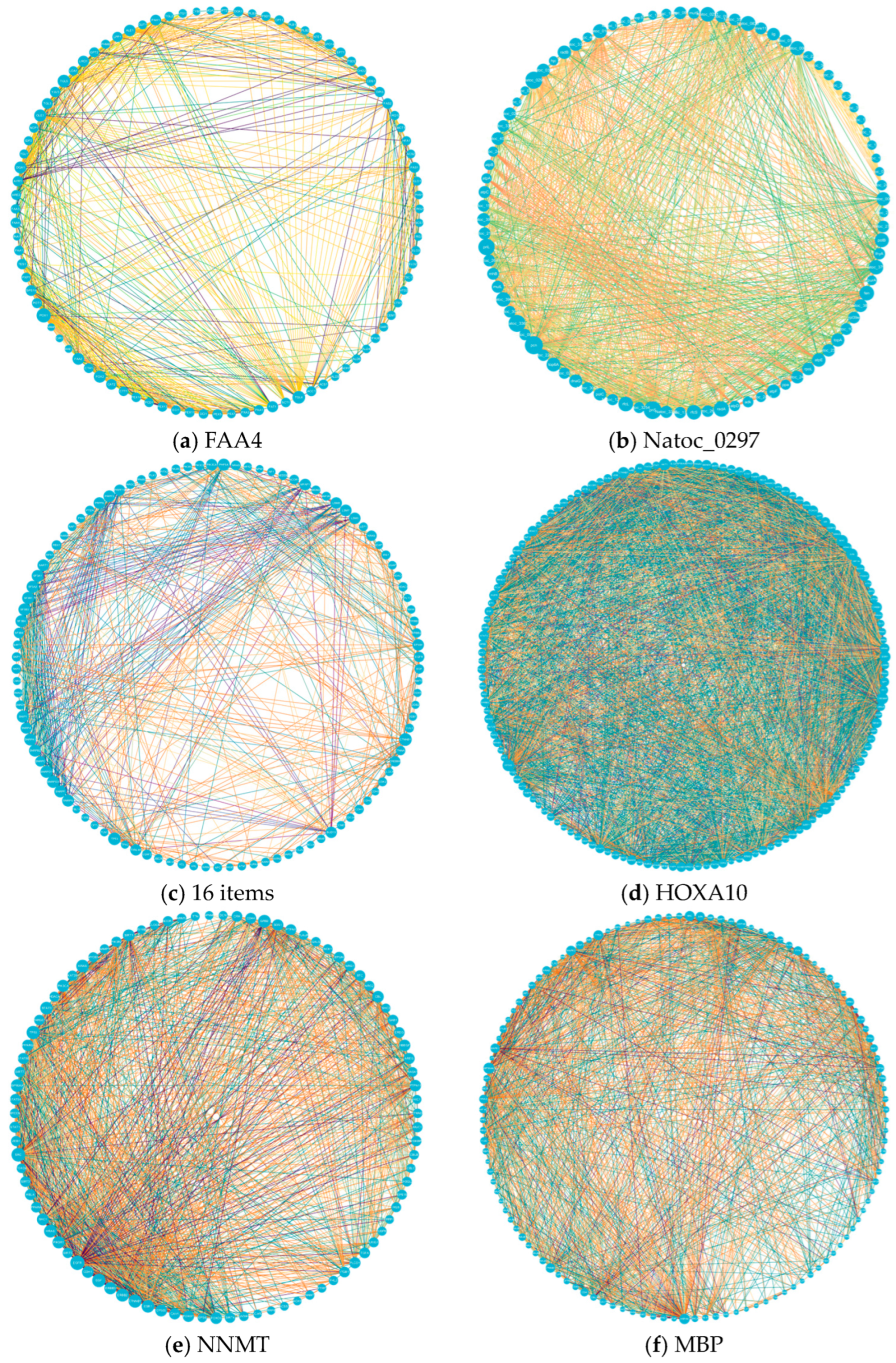
- Protein–protein interaction network: The vertices, attribute values, and edges correspond to proteins, GO (gene ontology [33]) terms, and protein–protein interactions, respectively. The GO terms mainly include molecular functions, cellular components, and biological processes. The experiments selected 6 PPI network datasets from real protein interaction network datasets [34], including FAA4 (from yeast), Natoc_0297 (from Cryptococcal Natrophomonas), 16 items (from Nattococcus), HOXA10 (from chromosome 7), NNMT (from cancer cells), and MBP (from Escherichia coli). Partial PPI networks constructed from the datasets are shown in Figure 3.
- MNIST: A classic handwritten digit recognition dataset, consisting of 60,000 grayscale images divided into 10 classes from “0” to “9”. In this study, 1000 samples were selected from the dataset for experimentation.
4.2. Clustering Evaluation Metrics
4.3. Data Processing
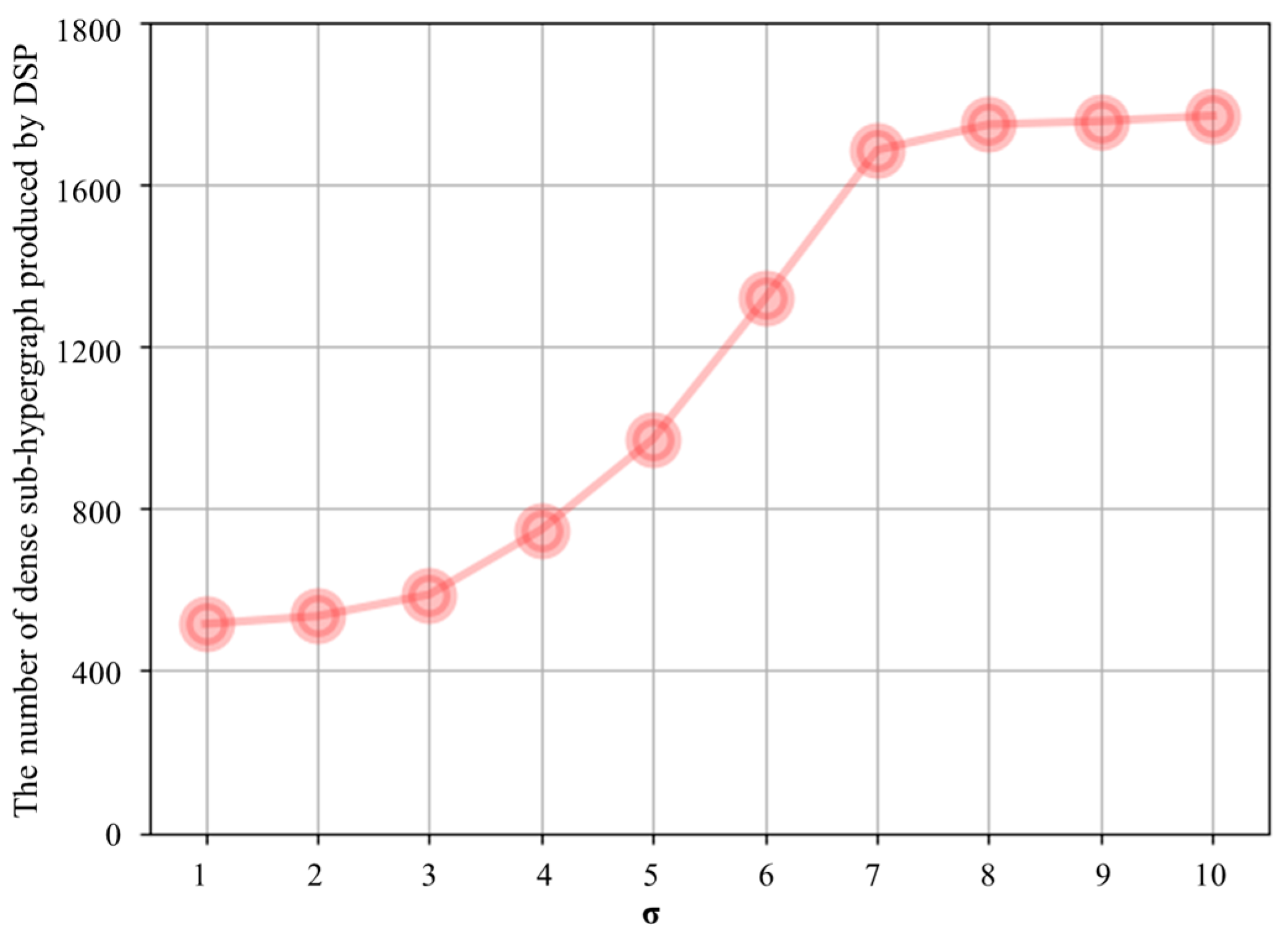
4.4. Parameter Settings
4.5. Experimental Comparison Methods
- k-medoids [39]: A classic clustering method based on representative objects that select objects located at the center of clusters as center points. It is more suitable for processing small datasets with noise and outliers.
- hMETIS [40]: A hypergraph-partitioning method based on the METIS graph-partitioning software package version 5.1.0 that can efficiently partition hypergraphs through k-way multilevel partitioning.
4.6. Experimental Comparison Results
4.6.1. Comparison of Experimental Results on PPI Networks
4.6.2. Comparison of Experimental Results on MNIST
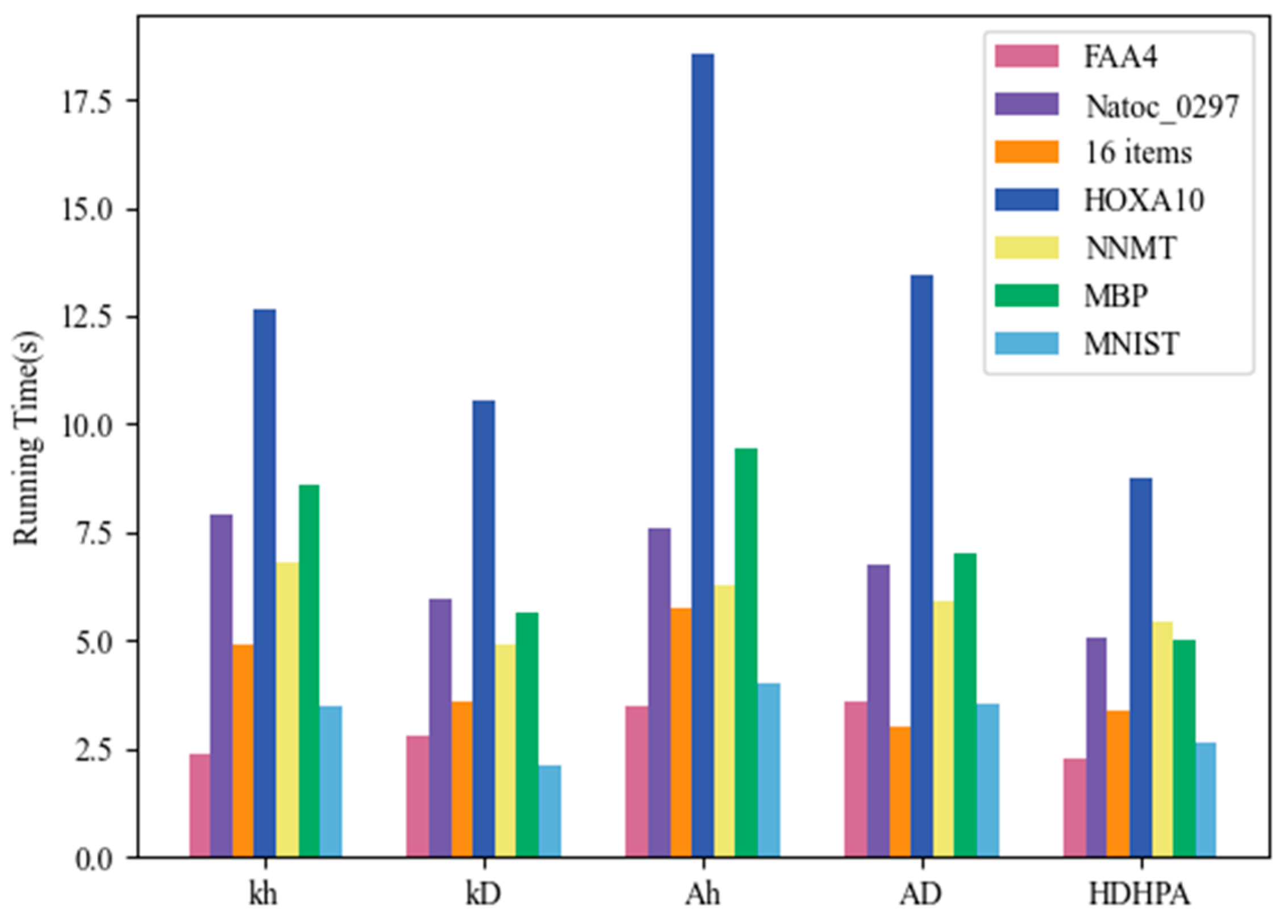
4.6.3. Running Time Comparison
5. Conclusions and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Supplementary Explanation to Figure 3
References
- Guo, X.; Liu, X.; Zhu, E. Adaptive self-paced deep clustering with data augmentation. IEEE Trans. Knowl. Eng. 2019, 32, 1680–1693. [Google Scholar] [CrossRef]
- Mago, N.; Shirwaikar, R.D.; Acharya, U.D.; Hegde, K.G.; Lewis, L.E.S.; Shivakumar, M. Partition and Hierarchical Based Clustering Techniques for Analysis of Neonatal Data. In Proceedings of International Conference on Cognition and Recognition; Springer: Berlin/Heidelberg, Germany, 2017; pp. 345–355. [Google Scholar]
- Von, L.U. A tutorial on spectral clustering. Stat. Comput. 2007, 4, 395–416. [Google Scholar]
- Zeng, J. Analysis of data mining K-means clustering algorithm based on partitioning. Moder. Electron. Technol. 2020, 3, 14–17. [Google Scholar]
- Wang, G.Y. A Preliminary Study on Uncertainty-Oriented Data Clustering. Master’s Thesis, Jilin University, Changchun, China, 2020. [Google Scholar]
- Ackermann, M.R.; Blömer, J.; Kuntze, D.; Sohler, C. Analysis of agglomerative clustering. Algorithmica 2014, 69, 184–215. [Google Scholar] [CrossRef]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabási, A.-L. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef]
- Guo, L.; Cui, Y.; Liang, H.; Zhou, Z. Spectral bisection community detection method for urban road networks. In Proceedings of the 2021 40th Chinese Control Conference (CCC), Shanghai, China, 26–28 July 2021; pp. 806–811. [Google Scholar]
- Newman, M.E.J. Fast algorithm for detecting community structure in networks. Phys. Rev. E 2004, 69, 066133. [Google Scholar] [CrossRef]
- Newman, M.E.J. Spectral methods for community detection and graph partitioning. Phys. Rev. E 2013, 88, 042822. [Google Scholar] [CrossRef]
- Berge, C. Graphs and Hypergraphs; North-Holland: Amsterdam, The Netherlands, 1973. [Google Scholar]
- Brusa, L.; Matias, C. Model-based clustering in simple hypergraphs through a stochastic blockmodel. Comput. Sci. 2022, 10, 05983. [Google Scholar]
- Wang, S.; Li, X.; Liu, D. Hyper-network Model of Architecture for Weapon Equipment System of Systems Based on Granular Computing. J. Syst. Eng. Electron. 2016, 38, 836–843. [Google Scholar]
- Strehl, A.; Ghosh, J.; Cardie, C. Cluster ensembles: A knowledge reuse framework for combining multiple partitions. J. Mach. Learn. Res. 2002, 3, 583–617. [Google Scholar]
- Yang, C.; Liu, D.; Yang, B.; Chi, S.; Jin, D. Research on clustering ensemble methods. Comput. Sci. 2011, 38, 166–170. [Google Scholar]
- Suo, Q.; Guo, J. Hypernetworks: Structure and evolution mechanism. Syst. Eng. Theory Pract. 2017, 37, 720–734. [Google Scholar]
- Tian, L.; Zhang, J.; Zhang, J.; Zhou, W.; Zhou, X. Knowledge graph: Representation, construction, reasoning, and hypergraph theory. J. Comput. Appl. 2021, 41, 2161–2186. [Google Scholar]
- Liu, S.; Huang, X.; Xian, Z.; Zuo, W. Commodity warehouse model based on hypergraph embedding representation. Chin. J. Manag. Sci. 2023, 1–12. [Google Scholar] [CrossRef]
- Wei, L.; Gong, X.; Qian, W.; Zhou, A. Outlier detection in high-dimensional space. J. Softw. 2002, 2, 280–290. [Google Scholar]
- Cui, Y.; Yang, B. Several applications of hypergraphs in data mining. Comput. Sci. 2010, 37, 220–222. [Google Scholar]
- Kadir, M.; Sobhan, S.; Islam, M.Z. Temporal relation extraction using Apriori algorithm. In Proceedings of the 2016 5th International Conference on Informatics, Electronics and Vision (ICIEV), Dhaka, Bangladesh, 13–14 May 2016; pp. 915–920. [Google Scholar]
- Agrawal, R.; Imielinski, T.; Swami, A. Mining Associations between Sets of Items in Massive Databases. In Proceedings of the 1993 ACM SIGMOD International Conference on Management of Data, Washington, DC, USA, 25–28 May 1993; pp. 207–216. [Google Scholar]
- Althuwaynee, O.F.; Aydda, A.; Hwang, I.T.; Lee, Y.-K.; Kim, S.-W.; Park, H.-J.; Lee, M.-S.; Park, Y. Uncertainty reduction of unlabeled features in landslide inventory using machine learning t-SNE clustering and data mining apriori association rule algorithms. Appl. Sci. 2021, 11, 556. [Google Scholar] [CrossRef]
- Esmaeili, H.; Hakami, V.; Bidgoli, B.M.; Shokouhifar, M. Application-specific clustering in wireless sensor networks using combined fuzzy firefly algorithm and random forest. Expert Syst. Appl. 2022, 210, 118365. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, C.; Shi, D.H.; Chen, G.; Li, X. Ranking cliques in higher-order complex networks. Chaos 2023, 33, 073139. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhou, Y.; Mei, K.; Wang, N.; Tang, M.; Cai, G. An Improved Density Peak Clustering Algorithm Based on Chebyshev Inequality and Differential Privacy. Appl. Sci. 2023, 13, 8674. [Google Scholar] [CrossRef]
- Liu, B.; Hsu, W.; Ma, Y. Integrating classification and association rule mining. In Proceedings of the KDD’98: Proceedings of the Fourth International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 27–31 August 1998; pp. 80–86. [Google Scholar]
- Liu, H.; Latecki, L.J.; Yan, S. Dense subgraph partition of positive hypergraphs. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Giannella, C.; Han, J.; Pei, J.; Yan, X.; Yu, P.S. Mining frequent patterns in data streams at multiple time granularities. Next Gener. Data Min. 2006, 35, 61–84. [Google Scholar]
- Hu, J.; He, L.; Mao, Y.; Yang, J. Research on improved algorithm for mining uncertain frequent subgraphs. Comput. Eng. Appl. 2015, 51, 112–116. [Google Scholar]
- Lin, Z. Research on Hierarchical Structure Construction and Maintenance Based on Dense Subgraph Approximation Mode. Ph.D. Thesis, East China Normal University, Shanghai, China, 2022; p. 000745. [Google Scholar]
- Barabási, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nature Rev. Gene. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S. Data Repository. 2020. Available online: https://www.samuel-johnson.org/data (accessed on 1 January 2022).
- Hu, F.; Liu, M.; Zhao, J.; Lei, L. Analysis and application of protein complex hypernetwork characteristics. Complex Syst. Complex. Sci. 2018, 4, 31–38. [Google Scholar]
- Pareek, V.; Tian, H.; Winograd, N.; Benkovic, S.J. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science 2020, 368, 283–290. [Google Scholar] [CrossRef]
- Fowlkes, E.B.; Mallows, C.L. A method for comparing two hierarchical clustering. J. Amer. Statist. Assoc. 1983, 78, 553–569. [Google Scholar] [CrossRef]
- Davide, H.; Giuseppe, J. A statistical comparison between Matthews correlation coefficient (MCC), prevalence threshold, and Fowlkes–Mallows index. J. Biomed. Inform. 2023, 144, 104426. [Google Scholar]
- Kaufman, L.; Rousseeuw, P. Clustering by Means of Medoids; North-Holland: Amsterdam, The Netherlands, 1987. [Google Scholar]
- Karypis, G.; Aggarwal, R.; Kumar, V.; Shekhar, S. Multilevel hypergraph partitioning: Applications in VLSI domain. IEEE Trans. VLSI Sys. 1999, 7, 69–79. [Google Scholar] [CrossRef]
- Cong, Q.; Anishchenko, I.; Ovchinnikov, S.; Baker, D. Protein interaction networks revealed by proteome coevolution. Science 2019, 365, 185–189. [Google Scholar] [CrossRef] [PubMed]


| Predicted Class | |||
|---|---|---|---|
| + | − | ||
| Actual class | + | TP 1 | FN 2 |
| − | FP 3 | TN 4 | |
| Dataset | Number of Proteins | Number of Interactions | Attribute Values |
|---|---|---|---|
| FAA4 | 789 | 636 | 1140 |
| Natoc_0297 | 991 | 1031 | 1227 |
| 16 items | 1106 | 2611 | 2115 |
| HOXA10 | 2151 | 4367 | 2446 |
| NNMT | 2612 | 5258 | 2676 |
| MBP | 3121 | 7423 | 3235 |
| Dataset | Avg. Clustering Coefficient | Avg. Number of Neighbors | Network Density | Network Heterogeneity | Network Centralization |
|---|---|---|---|---|---|
| FAA4 | 0.651 | 13.978 | 0.155 | 0.562 | 0.341 |
| Natoc_0297 | 0.701 | 22.659 | 0.252 | 0.636 | 0.470 |
| 16 items | 0.695 | 11.638 | 0.112 | 0.740 | 0.180 |
| HOXA10 | 0.630 | 44.596 | 0.297 | 0.584 | 0.489 |
| NNMT | 0.652 | 27.648 | 0.307 | 0.566 | 0.493 |
| MBP | 0.631 | 23.521 | 0.196 | 0.677 | 0.445 |
| Method | Dataset | |||||
|---|---|---|---|---|---|---|
| FAA4 | Natoc_0297 | 16 Items | HOXA10 | NNMT | MBP | |
| kh | 70.97 | 66.19 | 55.18 | 60.77 | 50.15 | 62.95 |
| kD | 77.80 | 74.16 | 50.29 | 51.40 | 46.64 | 61.01 |
| Ah | 44.14 | 47.51 | 24.34 | 38.51 | 19.33 | 49.20 |
| AD | 56.47 | 40.58 | 21.90 | 25.11 | 26.72 | 33.17 |
| HDHPA | 80.46 | 73.39 | 70.13 | 65.60 | 49.55 | 73.18 |
| Method | Dataset | |||||
|---|---|---|---|---|---|---|
| FAA4 | Natoc_0297 | 16 Items | HOXA10 | NNMT | MBP | |
| kh | 77.62 | 74.50 | 34.98 | 56.97 | 46.95 | 67.06 |
| kD | 82.83 | 78.15 | 47.60 | 70.47 | 49.92 | 75.40 |
| Ah | 34.34 | 33.56 | 13.47 | 18.52 | 14.23 | 29.60 |
| AD | 58.67 | 50.91 | 38.12 | 46.33 | 58.95 | 48.71 |
| HDHPA | 81.13 | 80.60 | 49.29 | 67.81 | 57.56 | 79.88 |
| Method | MNIST | |
|---|---|---|
| FM-Index | ARI | |
| kh | 45.96 | 43.06 |
| kD | 46.52 | 48.40 |
| Ah | 63.97 | 57.24 |
| AD | 39.51 | 34.22 |
| HDHPA | 70.60 | 83.37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, R.; Hu, F.; Wang, F.; Bai, L. Hypergraph-Clustering Method Based on an Improved Apriori Algorithm. Appl. Sci. 2023, 13, 10577. https://doi.org/10.3390/app131910577
Chen R, Hu F, Wang F, Bai L. Hypergraph-Clustering Method Based on an Improved Apriori Algorithm. Applied Sciences. 2023; 13(19):10577. https://doi.org/10.3390/app131910577
Chicago/Turabian StyleChen, Rumeng, Feng Hu, Feng Wang, and Libing Bai. 2023. "Hypergraph-Clustering Method Based on an Improved Apriori Algorithm" Applied Sciences 13, no. 19: 10577. https://doi.org/10.3390/app131910577
APA StyleChen, R., Hu, F., Wang, F., & Bai, L. (2023). Hypergraph-Clustering Method Based on an Improved Apriori Algorithm. Applied Sciences, 13(19), 10577. https://doi.org/10.3390/app131910577







