Dealing with Unreliable Annotations: A Noise-Robust Network for Semantic Segmentation through A Transformer-Improved Encoder and Convolution Decoder
Abstract
:1. Introduction
- 1.
- Inspired by the previous success of CNN and ViT, a ViT-based modified encoder and CNN-based decoder UNet-style segmentation network is proposed.
- 2.
- To simulate real clinical scenarios, noise is manually added to the ground truth to create noisy labels, constructing a CT spine segmentation dataset with noisy labels for evaluation purposes.
- 3.
- A simple and efficient adaptive denoising learning (ADL) is proposed for segmentation network to achieve a noise-robust segmentation framework.
- 4.
- The proposed segmentation network with ADL attains competitive performance against other baseline methods across various evaluation metrics under the same data conditions with both accurate and noisy label sets.
2. Methods
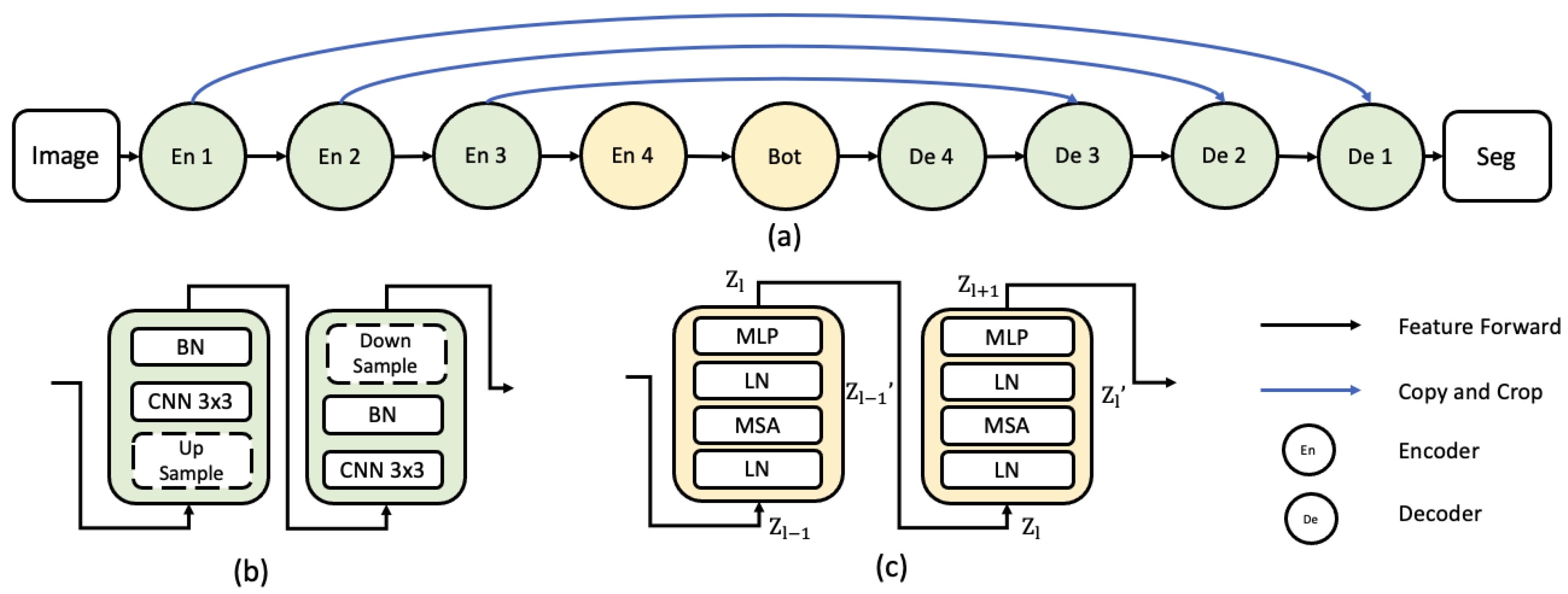
2.1. Vision Transformer Layer
- (i)
- The process of tokenization converts the input image x of dimensions into a sequence of flattened 2D patches denoted . Each patch measures , and accounts for the total number of image patches, thereby defining the input sequence length.
- (ii)
- These patches are subsequently mapped onto vectors in a latent D-dimensional embedding space using a trainable linear projection. To capture the spatial information inherent in the patches, learnable position embeddings are added to the patch embeddings, as follows:In this expression, represents the patch embedding projection, is the position embedding, and provides the feature map input to the first ViT layer.
- (iii)
- Comprising L layers, each incorporating an MSA and a MLP, the transformer encoder’s output from the layer is illustrated aswhere stands for the layer normalization operator, and stands for the encoded image.
- (iv)
- The MLP is a fully connected feedforward neural network that consists of multiple layers of nodes. In the proposed NR-UNet, the MLP refines the features extracted by the MSA mechanism. The MLP involves two linear layers interspersed with a GELU activation function [35], defined asIn this equation, and denote the first and second linear layers. The subsequent MSA is composed of multiple self-attention heads that operate in parallel to capture different aspects of the input tokens. Each self-attention head calculates attention scores employing Query (Q), Key (K), and Value (V) matrices, which are derived from the input tokens via linear transformations:The matrices Query (Q), Key (K), and Value (V) are derived from the input tokens through linear transformations:where , , and are learnable weight matrices [21].
- (v)
- The attention scores are computed by taking the dot product of the Q and K matrices, subsequently scaling and normalizing through softmax:where denotes the dimensionality of the Key vectors.
- (vi)
- The individual outputs of the self-attention heads are concatenated and linearly transformed to generate the final output of the MSA:where is the output of the i-th self-attention head, H is the total number of heads, and is another learnable weight matrix.
2.2. ViT Encoder to CNN Decoder
2.3. Noisy Labels
- a
- Starting with a dataset containing perfect annotations, we randomly select a subset of annotations to be altered, and the ratio of noisy labels to the whole dataset is denoted as .
- b
- For the selected annotations, we apply image-processing operations such as erosion, dilation, and elastic transformation to generate noisy labels. These operations mimic the types of noise that could be present in real-world clinical scenarios, where annotations might be imperfect due to various factors. Erosion and dilation are fundamental morphological operations. Let A be the binary annotation mask and B be a structuring element. The erosion (⊖) and dilation (⊕) operations can be defined aswhere represents the pixel coordinates in the image.Elastic transformation is a nonlinear deformation technique that can simulate the local warping of shapes. Given an image and two displacement fields and , which are generated by Gaussian smoothing of random fields, an elastic transformation can be defined aswhere is the deformation scale factor.
- c
- The altered annotations are then used to replace their corresponding original annotations in the dataset, creating a new dataset with a mix of perfect and noisy labels. By simulating noisy labels in this manner, we create a dataset that can challenge experiments, enabling us to assess the noise-robustness of our proposed method and compare its performance against existing high-performing techniques.
2.4. Adaptive Denoising Learning Strategy
3. Experiments and Results
3.1. Dataset
3.2. Experimental Setup
3.3. Metrics
3.4. Experiments in a Noise-Free Setup
3.5. Experiments in a Noisy Setup
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Wang, Z.; Voiculescu, I. Quadruple augmented pyramid network for multi-class COVID-19 segmentation via CT. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 2956–2959. [Google Scholar]
- Gao, Y.; Guo, J.; Fu, C.; Wang, Y.; Cai, S. VLSM-Net: A Fusion Architecture for CT Image Segmentation. Appl. Sci. 2023, 13, 4384. [Google Scholar] [CrossRef]
- Noble, J.A.; Boukerroui, D. Ultrasound image segmentation: A survey. IEEE Trans. Med. Imaging 2006, 25, 987–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Voiculescu, I. Triple-view feature learning for medical image segmentation. In Proceedings of the Resource-Efficient Medical Image Analysis: First MICCAI Workshop, REMIA 2022, Singapore, 22 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 42–54. [Google Scholar]
- Benjdira, B.; Ouni, K.; Al Rahhal, M.M.; Albakr, A.; Al-Habib, A.; Mahrous, E. Spinal cord segmentation in ultrasound medical imagery. Appl. Sci. 2020, 10, 1370. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z. Deep learning in medical ultrasound image segmentation: A review. arXiv 2020, arXiv:2002.07703. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 fourth international conference on 3D vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar]
- Kolařík, M.; Burget, R.; Uher, V.; Říha, K.; Dutta, M.K. Optimized high resolution 3d dense-u-net network for brain and spine segmentation. Appl. Sci. 2019, 9, 404. [Google Scholar] [CrossRef] [Green Version]
- Guan, S.; Khan, A.A.; Sikdar, S.; Chitnis, P.V. Fully Dense UNet for 2-D Sparse Photoacoustic Tomography Artifact Removal. IEEE J. Biomed. Health Inform. 2019, 24, 568–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. Resunet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote. Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Tang, Y.M.; Wang, Z.; Yu, K.M.; To, S. Atrous residual interconnected encoder to attention decoder framework for vertebrae segmentation via 3D volumetric CT images. Eng. Appl. Artif. Intell. 2022, 114, 105102. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Z.; Voiculescu, I. RAR-U-Net: A residual encoder to attention decoder by residual connections framework for spine segmentation under noisy labels. In Proceedings of the 2021 IEEE International Conference on Image Processing (ICIP), Anchorage, AK, USA, 19–22 September 2021; pp. 21–25. [Google Scholar]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. Unet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the Computer Vision–ECCV 2022 Workshops, Tel Aviv, Israel, 23–27 October 2022; Part III. Springer: Berlin/Heidelberg, Germany, 2023; pp. 205–218. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Wang, Z.; Zheng, J.Q.; Voiculescu, I. An uncertainty-aware transformer for MRI cardiac semantic segmentation via mean teachers. In Proceedings of the Medical Image Understanding and Analysis: 26th Annual Conference, MIUA 2022, Cambridge, UK, 27–29 July 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 494–507. [Google Scholar]
- Wang, J.; Zhang, H.; Yi, Z. CCTrans: Improving Medical Image Segmentation with Contoured Convolutional Transformer Network. Mathematics 2023, 11, 2082. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 20–25 June 2021; pp. 10012–10022. [Google Scholar]
- Bortsova, G.; Dubost, F.; Hogeweg, L.; Katramados, I.; De Bruijne, M. Semi-supervised medical image segmentation via learning consistency under transformations. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part VI 22. Springer: Berlin/Heidelberg, Germany, 2019; pp. 810–818. [Google Scholar]
- Li, S.; Zhang, C.; He, X. Shape-aware semi-supervised 3D semantic segmentation for medical images. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part I 23. Springer: Berlin/Heidelberg, Germany, 2020; pp. 552–561. [Google Scholar]
- Liu, X.; Yuan, Q.; Gao, Y.; He, K.; Wang, S.; Tang, X.; Tang, J.; Shen, D. Weakly supervised segmentation of COVID19 infection with scribble annotation on CT images. Pattern Recognit. 2022, 122, 108341. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jeong, W.K. Scribble2label: Scribble-supervised cell segmentation via self-generating pseudo-labels with consistency. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part I 23. Springer: Berlin/Heidelberg, Germany, 2020; pp. 14–23. [Google Scholar]
- Kervadec, H.; Dolz, J.; Wang, S.; Granger, E.; Ayed, I.B. Bounding boxes for weakly supervised segmentation: Global constraints get close to full supervision. In Proceedings of the Medical Imaging with Deep Learning, PMLR, Montreal, QC, Canada, 6–8 July 2020; pp. 365–381. [Google Scholar]
- Lu, Z.; Fu, Z.; Xiang, T.; Han, P.; Wang, L.; Gao, X. Learning from weak and noisy labels for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 486–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Liu, X.; Li, C.; Xu, Z.; Ruan, J.; Zhu, H.; Meng, T.; Li, K.; Huang, N.; Zhang, S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging 2020, 39, 2653–2663. [Google Scholar] [CrossRef] [PubMed]
- Ba, J.L.; Kiros, J.R.; Hinton, G.E. Layer normalization. arXiv 2016, arXiv:1607.06450. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian error linear units (gelus). arXiv 2016, arXiv:1606.08415. [Google Scholar]
- Huang, J.; Qu, L.; Jia, R.; Zhao, B. O2u-net: A simple noisy label detection approach for deep neural networks. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 3326–3334. [Google Scholar]
- Yao, J.; Burns, J.E.; Munoz, H.; Summers, R.M. Detection of vertebral body fractures based on cortical shell unwrapping. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nice, France, 1–5 October 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 509–516. [Google Scholar]
- Chaurasia, A.; Culurciello, E. Linknet: Exploiting encoder representations for efficient semantic segmentation. In Proceedings of the 2017 IEEE Visual Communications and Image Processing (VCIP), St. Petersburg, FL, USA, 10–13 December 2017; pp. 1–4. [Google Scholar]
- Kim, S.W.; Kook, H.K.; Sun, J.Y.; Kang, M.C.; Ko, S.J. Parallel feature pyramid network for object detection. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 234–250. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Pmlr, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]



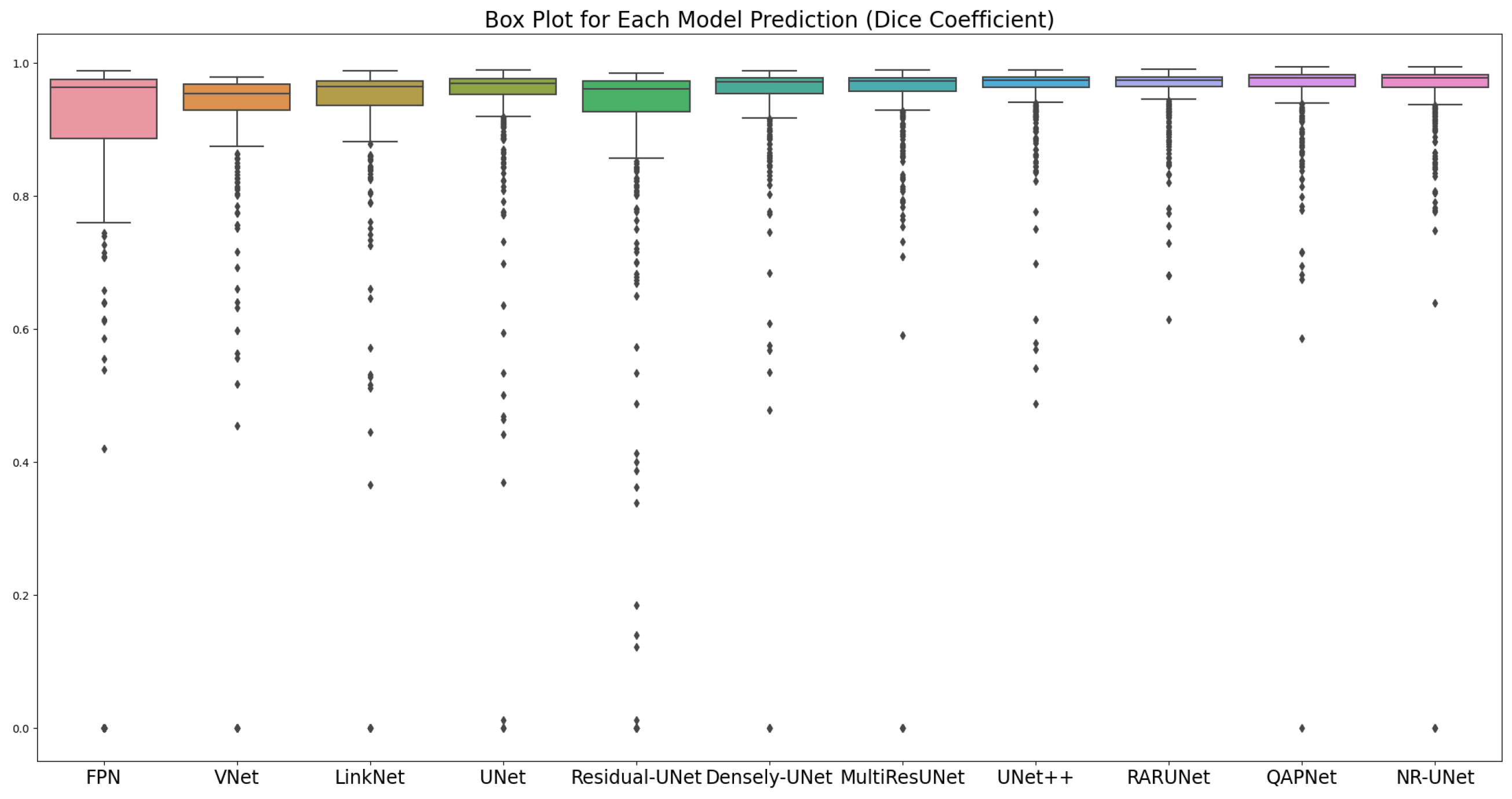


| Network | Dice | IoU | Acc | Pre | Rec | Spe | Par |
|---|---|---|---|---|---|---|---|
| FPN [39] | 0.9373 | 0.8821 | 0.9944 | 0.9191 | 0.9563 | 0.9961 | 17.59 |
| Residual UNet [13] | 0.9416 | 0.8897 | 0.9949 | 0.9481 | 0.9353 | 0.9976 | 9.90 |
| VNet [10] | 0.9446 | 0.8950 | 0.9950 | 0.9202 | 0.9703 | 0.9961 | 14.74 |
| LinkNet [38] | 0.9524 | 0.9091 | 0.9959 | 0.9662 | 0.9390 | 0.9985 | 20.32 |
| UNet [1] | 0.9580 | 0.9193 | 0.9963 | 0.9619 | 0.9541 | 0.9983 | 8.64 |
| Dense UNet [13] | 0.9612 | 0.9252 | 0.9966 | 0.9600 | 0.9624 | 0.9982 | 15.47 |
| MultiRes UNet [19] | 0.9644 | 0.9312 | 0.9969 | 0.9633 | 0.9655 | 0.9983 | 7.76 |
| UNet++ [2] | 0.9659 | 0.9340 | 0.9970 | 0.9676 | 0.9642 | 0.9985 | 8.86 |
| RARUNet [17] | 0.9674 | 0.9369 | 0.9972 | 0.9721 | 0.9629 | 0.9987 | 11.79 |
| QAPNet [3] | 0.9690 | 0.9399 | 0.9973 | 0.9715 | 0.9666 | 0.9987 | 15.14 |
| NR-UNet | 0.9703 | 0.9424 | 0.9974 | 0.9740 | 0.9667 | 0.9988 | 182.90 |
| Proportion () | Network | ADL | Dice | IoU |
|---|---|---|---|---|
| 75% | Residual UNet | ✗ | 0.7962 | 0.6614 |
| ✓ | 0.8210 | 0.6964 | ||
| 75% | UNet | ✗ | 0.8004 | 0.6672 |
| ✓ | 0.8337 | 0.7148 | ||
| 75% | NR-UNet | ✗ | 0.8196 | 0.6943 |
| ✓ | 0.8466 | 0.7340 | ||
| 50% | Residual UNet | ✗ | 0.8179 | 0.6919 |
| ✓ | 0.8453 | 0.7321 | ||
| 50% | UNet | ✗ | 0.8188 | 0.6932 |
| ✓ | 0.8564 | 0.7489 | ||
| 50% | NR-UNet | ✗ | 0.8362 | 0.7185 |
| ✓ | 0.8832 | 0.7908 | ||
| 25% | Residual UNet | ✗ | 0.9002 | 0.8185 |
| ✓ | 0.9213 | 0.8541 | ||
| 25% | UNet | ✗ | 0.9084 | 0.8322 |
| ✓ | 0.9303 | 0.8697 | ||
| 25% | Dense UNet | ✗ | 0.9096 | 0.8342 |
| ✓ | 0.9284 | 0.8664 | ||
| 25% | NR-UNet | ✗ | 0.9101 | 0.8350 |
| ✓ | 0.9532 | 0.9106 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Voiculescu, I. Dealing with Unreliable Annotations: A Noise-Robust Network for Semantic Segmentation through A Transformer-Improved Encoder and Convolution Decoder. Appl. Sci. 2023, 13, 7966. https://doi.org/10.3390/app13137966
Wang Z, Voiculescu I. Dealing with Unreliable Annotations: A Noise-Robust Network for Semantic Segmentation through A Transformer-Improved Encoder and Convolution Decoder. Applied Sciences. 2023; 13(13):7966. https://doi.org/10.3390/app13137966
Chicago/Turabian StyleWang, Ziyang, and Irina Voiculescu. 2023. "Dealing with Unreliable Annotations: A Noise-Robust Network for Semantic Segmentation through A Transformer-Improved Encoder and Convolution Decoder" Applied Sciences 13, no. 13: 7966. https://doi.org/10.3390/app13137966
APA StyleWang, Z., & Voiculescu, I. (2023). Dealing with Unreliable Annotations: A Noise-Robust Network for Semantic Segmentation through A Transformer-Improved Encoder and Convolution Decoder. Applied Sciences, 13(13), 7966. https://doi.org/10.3390/app13137966







