A Coordinate-Regression-Based Deep Learning Model for Catheter Detection during Structural Heart Interventions
Abstract
Featured Application
Abstract
1. Introduction
- Tracking the tip of a catheter for future use in a mixed-reality navigation system.
- Addressing the limited accuracy, low availability, and high cost of EM sensor tracking systems.
- Proposing a catheter tip coordinate regression detection methodology leveraging deep convolutional neural networks to reduce the time-consuming task of generating ground truth masks.
2. Materials and Methods
2.1. Dataset
2.2. Architectures
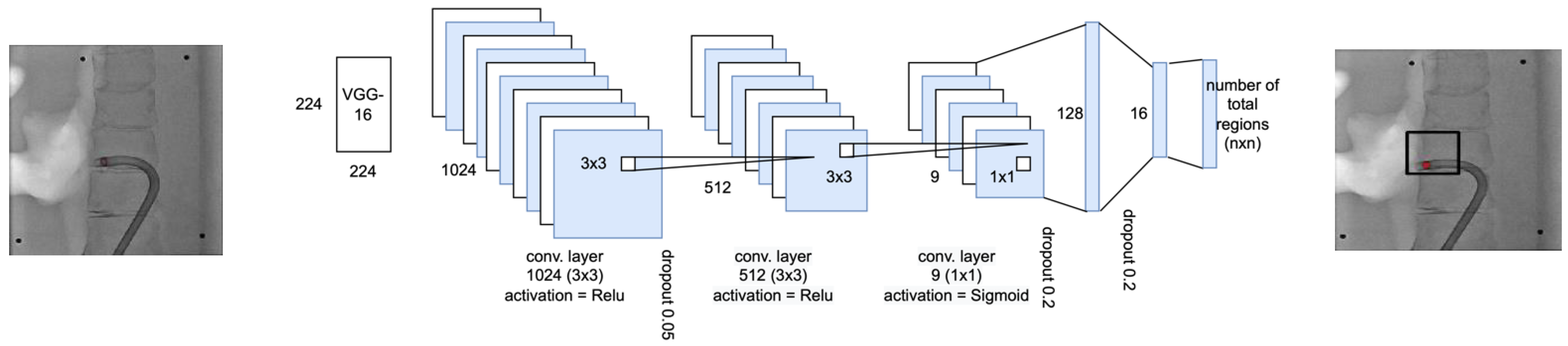
2.2.1. Region Selection Network
2.2.2. Localizer Network
2.3. Dual Network Inference
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Schmitto, J.D.; Mokashi, S.A.; Cohn, L.H. Minimally-invasive valve surgery. J. Am. Coll. Cardiol. 2010, 56, 455–462. [Google Scholar] [CrossRef]
- Endo, Y.; Nakamura, Y.; Kuroda, M.; Ito, Y.; Hori, T. The Utility of a 3D Endoscope and Robot-Assisted System for MIDCAB. Ann. Thorac. Cardiovasc. Surg. 2019, 25, 200–204. [Google Scholar] [CrossRef]
- Dieberg, G.; Smart, N.A.; King, N. Minimally invasive cardiac surgery: A systematic review and meta-analysis. Int. J. Cardiol. 2016, 223, 554–560. [Google Scholar] [CrossRef]
- Jiang, Z.; Qu, H.; Zhang, Y.; Zhang, F.; Xiao, W.; Shi, D.; Gao, Z.; Chen, K. Efficacy and Safety of Xinyue Capsule for Coronary Artery Disease after Percutaneous Coronary Intervention: A Systematic Review and Meta-Analysis of Randomized Clinical Trials. Evid. Based. Complement. Altern. Med. 2021, 2021, 6695868. [Google Scholar] [CrossRef]
- Bansilal, S.; Castellano, J.M.; Fuster, V. Global burden of CVD: Focus on secondary prevention of cardiovascular disease. Int. J. Cardiol. 2015, 201 (Suppl. S1), S1–S7. [Google Scholar] [CrossRef]
- Little, S.H. Structural Heart Interventions. Methodist Debakey Cardiovasc. J. 2017, 13, 96–97. [Google Scholar] [CrossRef]
- Wasmer, K.; Zellerhoff, S.; Kobe, J.; Monnig, G.; Pott, C.; Dechering, D.G.; Lange, P.S.; Frommeyer, G.; Eckardt, L. Incidence and management of inadvertent puncture and sheath placement in the aorta during attempted transseptal puncture. Europace 2017, 19, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Faletra, F.F.; Pedrazzini, G.; Pasotti, E.; Moccetti, T. Side-by-side comparison of fluoroscopy, 2D and 3D TEE during percutaneous edge-to-edge mitral valve repair. JACC Cardiovasc. Imaging 2012, 5, 656–661. [Google Scholar] [PubMed]
- Arujuna, A.V.; Housden, R.J.; Ma, Y.; Rajani, R.; Gao, G.; Nijhof, N.; Cathier, P.; Bullens, R.; Gijsbers, G.; Parish, V. Novel system for real-time integration of 3-D echocardiography and fluoroscopy for image-guided cardiac interventions: Preclinical validation and clinical feasibility evaluation. IEEE J. Transl. Eng. Health Med. 2014, 2, 1–10. [Google Scholar]
- Sra, J.; Krum, D.; Choudhuri, I.; Belanger, B.; Palma, M.; Brodnick, D.; Rowe, D.B. Identifying the third dimension in 2D fluoroscopy to create 3D cardiac maps. JCI Insight 2016, 1, e90453. [Google Scholar] [CrossRef] [PubMed]
- Celi, S.; Martini, N.; Emilio Pastormerlo, L.; Positano, V.; Berti, S. Multimodality imaging for interventional cardiology. Curr. Pharm. Design 2017, 23, 3285–3300. [Google Scholar]
- Biaggi, P.; Fernandez-Golfín, C.; Hahn, R.; Corti, R. Hybrid imaging during transcatheter structural heart interventions. Curr. Cardiovasc. Imaging Rep. 2015, 8, 1–14. [Google Scholar]
- Falk, V.; Mourgues, F.; Adhami, L.; Jacobs, S.; Thiele, H.; Nitzsche, S.; Mohr, F.W.; Coste-Manière, È. Cardio navigation: Planning, simulation, and augmented reality in robotic assisted endoscopic bypass grafting. Ann. Thorac. Surg. 2005, 79, 2040–2047. [Google Scholar] [PubMed]
- Muraru, D.; Badano, L.P. Physical and technical aspects and overview of 3D-echocardiography. In Manual of 3D Echocardiography; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–44. [Google Scholar]
- Jang, S.-J.; Torabinia, M.; Dhrif, H.; Caprio, A.; Liu, J.; Wong, S.C.; Mosadegh, B. Development of a Hybrid Training Simulator for Structural Heart Disease Interventions. Adv. Intell. Syst. 2020, 2, 2000109. [Google Scholar] [CrossRef]
- Liu, J.; Al’Aref, S.J.; Singh, G.; Caprio, A.; Moghadam, A.A.A.; Jang, S.-J.; Wong, S.C.; Min, J.K.; Dunham, S.; Mosadegh, B. An augmented reality system for image guidance of transcatheter procedures for structural heart disease. PLoS ONE 2019, 14, e0219174. [Google Scholar] [CrossRef]
- Torabinia, M.; Caprio, A.; Jang, S.-J.; Ma, T.; Tran, H.; Mekki, L.; Chen, I.; Sabuncu, M.; Wong, S.C.; Mosadegh, B. Deep learning-driven catheter tracking from bi-plane X-ray fluoroscopy of 3D printed heart phantoms. Mini-Invasive Surg. 2021, 5, 32. [Google Scholar] [CrossRef]
- Southworth, M.K.; Silva, J.R.; Silva, J.N.A. Use of extended realities in cardiology. Trends Cardiovas. Med. 2020, 30, 143–148. [Google Scholar] [CrossRef]
- Jung, C.; Wolff, G.; Wernly, B.; Bruno, R.R.; Franz, M.; Schulze, P.C.; Silva, J.N.A.; Silva, J.R.; Bhatt, D.L.; Kelm, M. Virtual and Augmented Reality in Cardiovascular Care: State-of-the-Art and Future Perspectives. JACC Cardiovasc. Imaging 2022, 15, 519–532. [Google Scholar] [CrossRef]
- Kasprzak, J.D.; Pawlowski, J.; Peruga, J.Z.; Kaminski, J.; Lipiec, P. First-in-man experience with real-time holographic mixed reality display of three-dimensional echocardiography during structural intervention: Balloon mitral commissurotomy. Eur. Heart J. 2020, 41, 801. [Google Scholar] [CrossRef]
- Arjomandi Rad, A.; Vardanyan, R.; Thavarajasingam, S.G.; Zubarevich, A.; Van den Eynde, J.; Sa, M.; Zhigalov, K.; Sardiari Nia, P.; Ruhparwar, A.; Weymann, A. Extended, virtual and augmented reality in thoracic surgery: A systematic review. Interact. Cardiovasc. Thorac. Surg. 2022, 34, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Jia, Y.; Shelhamer, E.; Donahue, J.; Karayev, S.; Long, J.; Girshick, R.; Guadarrama, S.; Darrell, T. (Eds.) Caffe: Convolutional architecture for fast feature embedding. In Proceedings of the 22nd ACM International Conference on Multimedia, Orlando, FL, USA, 3–7 November 2014. [Google Scholar]
- Sermanet, P.; Eigen, D.; Zhang, X.; Mathieu, M.; Fergus, R.; LeCun, Y. Overfeat: Integrated recognition, localization and detection using convolutional networks. arXiv 2013, arXiv:13126229. [Google Scholar]
- Chandan, G.; Jain, A.; Jain, H. Real time object detection and tracking using Deep Learning and OpenCV. In Proceedings of the 2018 International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 11–12 July 2018. [Google Scholar]
- Rajchl, M.; Lee, M.C.; Oktay, O.; Kamnitsas, K.; Passerat-Palmbach, J.; Bai, W.; Damodaram, M.; Rutherford, M.A.; Hajnal, J.V.; Kainz, B. Deepcut: Object segmentation from bounding box annotations using convolutional neural networks. IEEE Trans. Med. Imaging 2016, 36, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.-W.; Heng, P.-A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Zhang, J.; Jin, Y.; Xu, J.; Xu, X.; Zhang, Y. Mdu-net: Multi-scale densely connected u-net for biomedical image segmentation. arXiv 2018, arXiv:181200352. [Google Scholar] [CrossRef]
- Jin, Q.; Meng, Z.; Pham, T.D.; Chen, Q.; Wei, L.; Su, R. DUNet: A deformable network for retinal vessel segmentation. Knowl.-Based Syst. 2019, 178, 149–162. [Google Scholar]
- Jin, Q.; Meng, Z.; Sun, C.; Cui, H.; Su, R. RA-UNet: A hybrid deep attention-aware network to extract liver and tumor in CT scans. Front. Bioeng. Biotechnol. 2020, 8, 1471. [Google Scholar] [CrossRef]
- Dolz, J.; Ben Ayed, I.; Desrosiers, C. Dense multi-path U-Net for ischemic stroke lesion segmentation in multiple image modalities. In International MICCAI Brainlesion Workshop; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Guo, J.; Deng, J.; Xue, N.; Zafeiriou, S. Stacked dense u-nets with dual transformers for robust face alignment. arXiv 2018, arXiv:181201936. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv 2018, arXiv:180910486. [Google Scholar]
- Clèrigues, A.; Valverde, S.; Bernal, J.; Freixenet, J.; Oliver, A.; Lladó, X. Acute and sub-acute stroke lesion segmentation from multimodal MRI. Comput. Meth. Prog. Biol. 2020, 194, 105521. [Google Scholar] [CrossRef]
- Dolz, J.; Desrosiers, C.; Ben Ayed, I. IVD-Net: Intervertebral disc localization and segmentation in MRI with a multi-modal UNet. In International Workshop and Challenge on Computational Methods and Clinical Applications for Spine Imaging; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Zhuang, J. LadderNet: Multi-path networks based on U-Net for medical image segmentation. arXiv 2018, arXiv:181007810. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:180403999. [Google Scholar]
- Urschler, M.; Ebner, T.; Štern, D. Integrating geometric configuration and appearance information into a unified framework for anatomical landmark localization. Med. Image Anal. 2018, 43, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Artico, J.; Fontana, M.; Moon, J.C.; Davies, R.H.; Kellman, P. Landmark Detection in Cardiac MRI by Using a Convolutional Neural Network. Radiol. Artif. Intell. 2021, 3, e200197. [Google Scholar] [CrossRef] [PubMed]
- Dabbah, M.A.; Murphy, S.; Pello, H.; Courbon, R.; Beveridge, E.; Wiseman, S.; Wyeth, D.; Poole, I. Detection and location of 127 anatomical landmarks in diverse CT datasets. Med. Imaging Image Process. 2014, 9034, 284–294. [Google Scholar] [CrossRef]
- Ibragimov, B.; Likar, B.; Pernus, F.; Vrtovec, T. Computerized Cephalometry by Game Theory with Shape-and Appearance-Based Landmark Refinement. In Proceedings of the International Symposium on Biomedical imaging (ISBI), Prague, Czech Republic, 13–16 April 2016. [Google Scholar]
- Zheng, Y.; John, M.; Liao, R.; Nottling, A.; Boese, J.; Kempfert, J.; Walther, T.; Brockmann, G.; Comaniciu, D. Automatic aorta segmentation and valve landmark detection in C-arm CT for transcatheter aortic valve implantation. IEEE Trans. Med. Imaging 2012, 31, 2307–2321. [Google Scholar] [CrossRef]
- Oktay, O.; Bai, W.; Guerrero, R.; Rajchl, M.; de Marvao, A.; O’Regan, D.P.; Cook, S.A.; Heinrich, M.P.; Glocker, B.; Rueckert, D. Stratified Decision Forests for Accurate Anatomical Landmark Localization in Cardiac Images. IEEE Trans. Med. Imaging 2017, 36, 332–342. [Google Scholar] [CrossRef]
- Rohr, K. Landmark-Based Image Analysis using Geometric and Intensity Models; Springer: Berlin/Heidelberg, Germany, 2001. [Google Scholar]
- Zheng, Y.F.; Liu, D.; Georgescu, B.; Nguyen, H.; Comaniciu, D. 3D Deep Learning for Efficient and Robust Landmark Detection in Volumetric Data. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Volume 9349, pp. 565–572. [Google Scholar] [CrossRef]
- Xu, Z.; Huang, Q.; Park, J.; Chen, M.; Xu, D.; Yang, D.; Liu, D.; Zhou, S.K. Supervised Action Classifier: Approaching Landmark Detection as Image Partitioning. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017. [Google Scholar]
- Yang, D.; Zhang, S.T.; Yan, Z.N.; Tan, C.W.; Li, K.; Metaxas, D. Automated Anatomical Landmark Detection on Distal Femur Surface Using Convolutional Neural Network. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (Isbi), Brooklyn, NY, USA, 16–19 April 2015; pp. 17–21. [Google Scholar]
- Nibali, A.; He, Z.; Morgan, S.; Prendergast, L. Numerical coordinate regression with convolutional neural networks. arXiv 2018, arXiv:180107372. [Google Scholar]
- Toshev, A.; Szegedy, C. DeepPose: Human Pose Estimation via Deep Neural Networks. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014. [Google Scholar]
- Chu, X.; Yang, W.; Ouyang, W.L.; Ma, C.; Yuille, A.L.; Wang, X.G. Multi-Context Attention for Human Pose Estimation. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition (Cvpr 2017), Honolulu, HI, USA, 21–26 July 2017; pp. 5669–5678. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Mao, J.; Li, G.; Ma, R. (Eds.) End-to-End Coordinate Regression Model with Attention-Guided Mechanism for Landmark Localization in 3D Medical Images. International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Dünnwald, M.; Betts, M.J.; Düzel, E.; Oeltze-Jafra, S. Localization of the Locus Coeruleus in MRI via Coordinate Regression. In Bildverarbeitung für die Medizin 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 10–15. [Google Scholar]
- Jin, H.; Liao, S.; Shao, L. Pixel-in-pixel net: Towards efficient facial landmark detection in the wild. Int. J. Comput. Vis. 2021, 129, 3174–3194. [Google Scholar] [CrossRef]
- Ramadani, A.; Bui, M.; Wendler, T.; Schunkert, H.; Ewert, P.; Navab, N. A survey of catheter tracking concepts and methodologies. Med. Image Anal. 2022, 82, 102584. [Google Scholar] [CrossRef]
- Lessard, S.; Lau, C.; Chav, R.; Soulez, G.; Roy, D.; de Guise, J.A. Guidewire tracking during endovascular neurosurgery. Med. Eng. Phys. 2010, 32, 813–821. [Google Scholar] [CrossRef]
- Vandini, A.; Glocker, B.; Hamady, M.; Yang, G.-Z. Robust guidewire tracking under large deformations combining segment-like features (SEGlets). Med. Image Anal. 2017, 38, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, T.; Zhu, Y.; Zhang, W.; Zhou, S.K.; Comaniciu, D. Robust guidewire tracking in fluoroscopy. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Zweng, M.; Fallavollita, P.; Demirci, S.; Kowarschik, M.; Navab, N.; Mateus, D. Automatic guide-wire detection for neurointerventions using low-rank sparse matrix decomposition and denoising. In Proceedings of the Workshop on Augmented Environments for Computer-Assisted Interventions, Munich, Germany, 6 October 2015. [Google Scholar]
- Ambrosini, P.; Ruijters, D.; Niessen, W.J.; Moelker, A.; Walsum, T.v. Fully automatic and real-time catheter segmentation in X-ray fluoroscopy. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017. [Google Scholar]
- Nguyen, A.; Kundrat, D.; Dagnino, G.; Chi, W.; Abdelaziz, M.E.; Guo, Y.; Ma, Y.; Kwok, T.M.; Riga, C.; Yang, G.-Z. End-to-end real-time catheter segmentation with optical flow-guided warping during endovascular intervention. In Proceedings of the 2020 IEEE International Conference on Robotics and Automation (ICRA), Paris, France, 1 June 2020. [Google Scholar]
- Subramanian, V.; Wang, H.; Wu, J.T.; Wong, K.C.; Sharma, A.; Syeda-Mahmood, T. Automated detection and type classification of central venous catheters in chest X-rays. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Zhou, Y.-J.; Xie, X.-L.; Zhou, X.-H.; Liu, S.-Q.; Bian, G.-B.; Hou, Z.-G. A real-time multifunctional framework for guidewire morphological and positional analysis in interventional X-ray fluoroscopy. IEEE Trans. Cognit. Dev. Syst. 2020, 13, 657–667. [Google Scholar] [CrossRef]
- Li, R.-Q.; Bian, G.; Zhou, X.; Xie, X.; Ni, Z.; Hou, Z. A two-stage framework for real-time guidewire endpoint localization. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Vlontzos, A.; Mikolajczyk, K. Deep segmentation and registration in X-ray angiography video. arXiv 2018, arXiv:180506406. [Google Scholar]
- Vernikouskaya, I.; Bertsche, D.; Rottbauer, W.; Rasche, V. Deep learning-based framework for motion-compensated image fusion in catheterization procedures. Comput. Med. Imaging Graph. 2022, 98, 102069. [Google Scholar] [CrossRef]
- Liu, D.; Tupor, S.; Singh, J.; Chernoff, T.; Leong, N.; Sadikov, E.; Amjad, A.; Zilles, S. The challenges facing deep learning–based catheter localization for ultrasound guided high-dose-rate prostate brachytherapy. Med. Phys. 2022, 49, 2442–2451. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR2015), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition (CVPR2009), Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:170404861. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]







| Model | Region Number | Mean Error | Standard Deviation of Error | Training Time | Inference Time | Output Type |
|---|---|---|---|---|---|---|
| Pixels from 512 × 512 Image | Average for Each Image | |||||
| Mobile Net [73] | 5 × 5 | 19.29 | 9.77 | 3.8 s | 3.6 ms | Landmark |
| ResNet [74] | 5 × 5 | 3.35 | 6.23 | 4.9 s | 4.4 ms | Landmark |
| Dense Net [75] | 5 × 5 | 3.86 | 7.29 | 21.5 s | 11.2 ms | Landmark |
| U-Net [17] | 1 | 1.00 | 6.13 | 0.2 s | 10 ms | Mask |
| VGG [71] | 1 | 7.36 | 4.90 | 2.1 s | 1.7 ms | Landmark |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aghasizade, M.; Kiyoumarsioskouei, A.; Hashemi, S.; Torabinia, M.; Caprio, A.; Rashid, M.; Xiang, Y.; Rangwala, H.; Ma, T.; Lee, B.; et al. A Coordinate-Regression-Based Deep Learning Model for Catheter Detection during Structural Heart Interventions. Appl. Sci. 2023, 13, 7778. https://doi.org/10.3390/app13137778
Aghasizade M, Kiyoumarsioskouei A, Hashemi S, Torabinia M, Caprio A, Rashid M, Xiang Y, Rangwala H, Ma T, Lee B, et al. A Coordinate-Regression-Based Deep Learning Model for Catheter Detection during Structural Heart Interventions. Applied Sciences. 2023; 13(13):7778. https://doi.org/10.3390/app13137778
Chicago/Turabian StyleAghasizade, Mahdie, Amir Kiyoumarsioskouei, Sara Hashemi, Matin Torabinia, Alexandre Caprio, Muaz Rashid, Yi Xiang, Huzefa Rangwala, Tianyu Ma, Benjamin Lee, and et al. 2023. "A Coordinate-Regression-Based Deep Learning Model for Catheter Detection during Structural Heart Interventions" Applied Sciences 13, no. 13: 7778. https://doi.org/10.3390/app13137778
APA StyleAghasizade, M., Kiyoumarsioskouei, A., Hashemi, S., Torabinia, M., Caprio, A., Rashid, M., Xiang, Y., Rangwala, H., Ma, T., Lee, B., Wang, A., Sabuncu, M., Wong, S. C., & Mosadegh, B. (2023). A Coordinate-Regression-Based Deep Learning Model for Catheter Detection during Structural Heart Interventions. Applied Sciences, 13(13), 7778. https://doi.org/10.3390/app13137778






