4.1. Model Parameter Experiment
The obtained results were grouped according to the optimizer used, and the average, standard deviation, and coefficients of variation in the results were calculated (see
Table 4).
The SGD optimizer shows the lowest accuracy, AUC, precision, recall, and F1 values compared to the other two optimizers. Additionally, the variation coefficients are quite large, indicating a greater distribution of results. The Adam optimizer shows the best results in all measurement indicators: highest accuracy, AUC, precision, recall, and F1. Additionally, the variation coefficients for this optimizer are smaller, indicating a smaller distribution of results, which is desirable. The RMSprop optimizer shows better results than SGD but worse than Adam. Its accuracy, AUC, precision, recall, and F1 indicators are good, but the variation coefficients are higher than those for the Adam optimizer but lower than those for SGD.
Based on this analysis, the Adam optimizer is the best choice for further analysis because it has the highest accuracy, AUC, precision, recall, and F1, and its variation coefficients are smaller, indicating a smaller distribution of results. RMSprop is in second place, and the SGD optimizer is unsuitable in this situation due to its lower performance and higher result distribution. Therefore, the results obtained using the Adam and RMSprop optimizers will be analyzed further.
The effect of several model parameter combinations on AUC, accuracy, and F1 was compared for different optimizers.
Figure 1,
Figure 2 and
Figure 3 compare the impact of dropout rate and learning rate.
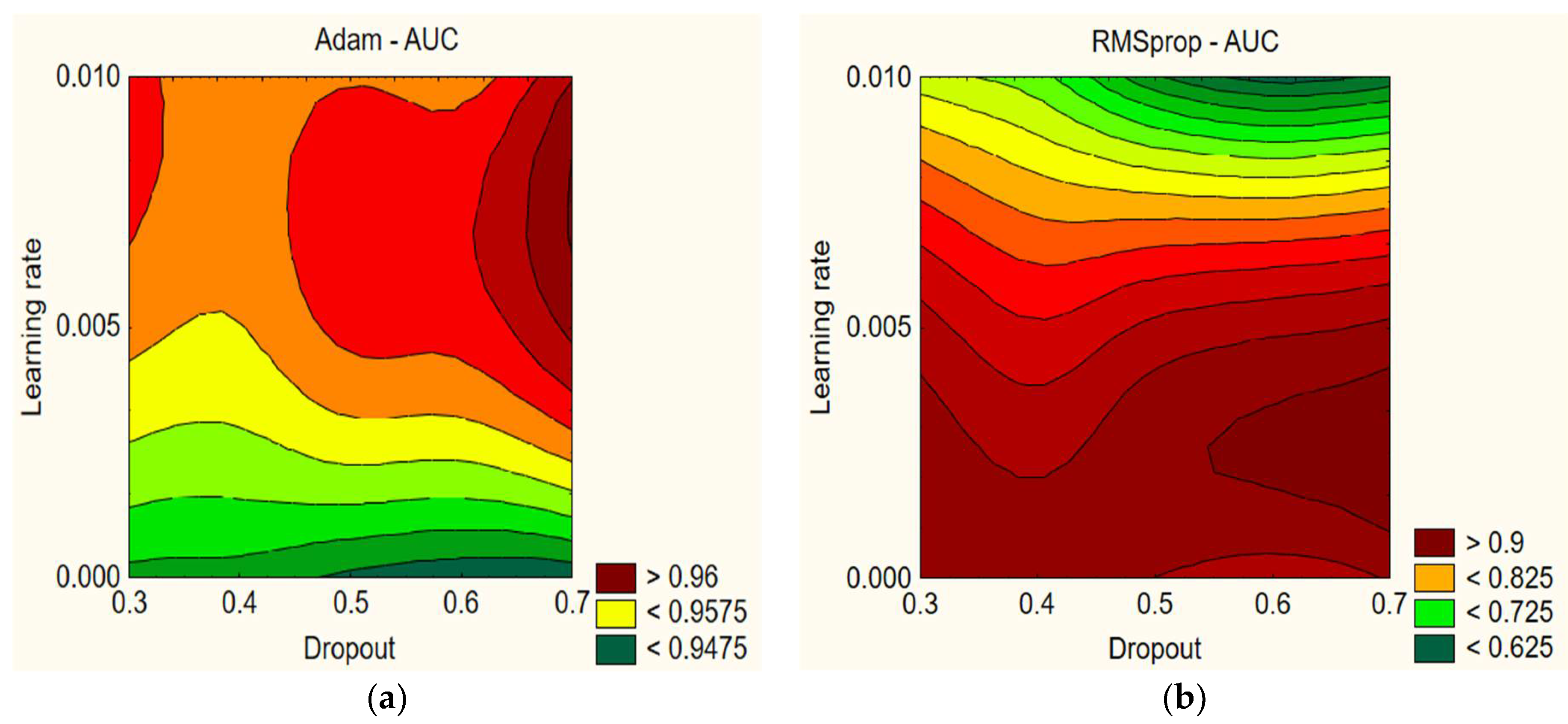
In
Figure 1, we can see that the AUC values are relatively high and range between 92.79% and 97.45% with the Adam optimizer. The AUC values slightly vary depending on the learning rate and dropout. Furthermore, it can be concluded that a model that uses a higher learning rate and dropout (for example, a learning rate of 0.005 and dropout of 0.7) can achieve a better AUC value. With the RMSprop optimizer, lower dropout percentages (0.3–0.4) and lower learning rates (0.001) tend to result in higher AUC values. On the other hand, combinations of higher dropout percentages (0.6–0.7) and higher learning rates (0.01) tend to result in lower AUC values, which may indicate that these combinations can cause too much fluctuation during training and hinder the model’s learning ability. Based on these findings, it can be concluded that selecting the appropriate learning rate and dropout percentage combination has a significant impact on the AUC variations. Properly selecting these parameters can improve the model’s performance and increase AUC values (see
Table 5).
Using the Adam optimizer, the highest AUC is achieved with a learning rate of 0.005 and dropout of 0.7, resulting in an AUC of 97.45%, whereas the lowest AUC is achieved with a learning rate of 0.01 and dropout of 0.6, resulting in an AUC of 92.79%. With the RMSprop optimizer, the highest AUC is achieved with a learning rate of 0.005 and dropout of 0.4, resulting in an AUC of 96.82%, whereas the lowest AUC is achieved with a learning rate of 0.01 and dropout of 0.4, resulting in an AUC of 50%. Overall, the Adam optimizer shows better results on average.
In
Figure 2, using the Adam optimizer, when the learning rate is 0.005 and dropout rate is 0.7, the model achieves the highest accuracy result (94.6%), whereas the lowest accuracy (87.73%) is obtained with a learning rate of 0.001 and dropout rate of 0.3. With RMSprop, the highest accuracy (93.8%) was achieved with a learning rate of 0.005 and dropout rate of 0.6, whereas the lowest accuracy (21%) was obtained with a learning rate of 0.01 and dropout rate of 0.7. Overall, all Adam optimizer results provide good results, whereas the RMSprop results are highly variable.
With the Adam optimizer, lower dropout rates generally result in better accuracy, whereas higher dropout rates (0.7) and lower learning rates (0.001) can decrease accuracy. Both learning rate and dropout rate have an impact on the model’s accuracy indicators. When analyzing the presented data with RMSprop, we can also see that different learning rates and dropout rates have varying effects on the model’s accuracy. For example, a network with a learning rate of 0.01 generally has poorer accuracy results (even up to 21%), whereas lower dropout rates tend to produce better accuracy (see
Table 6).
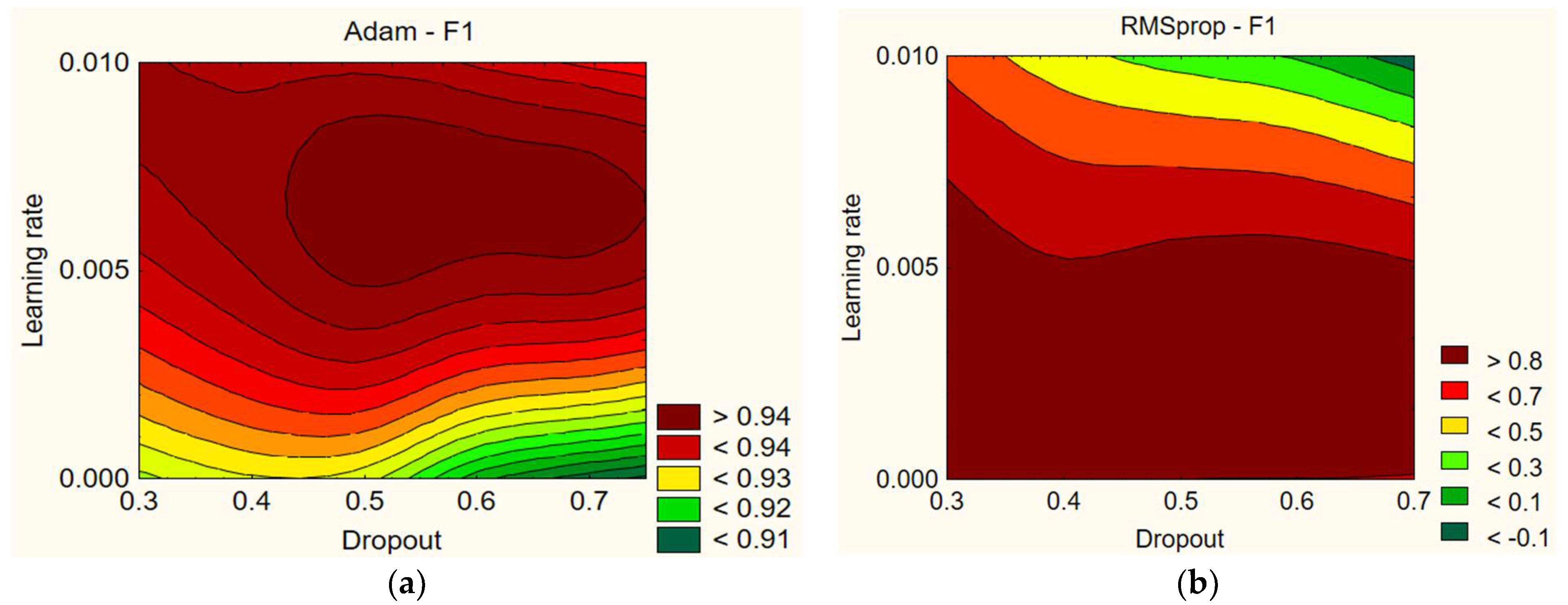
In
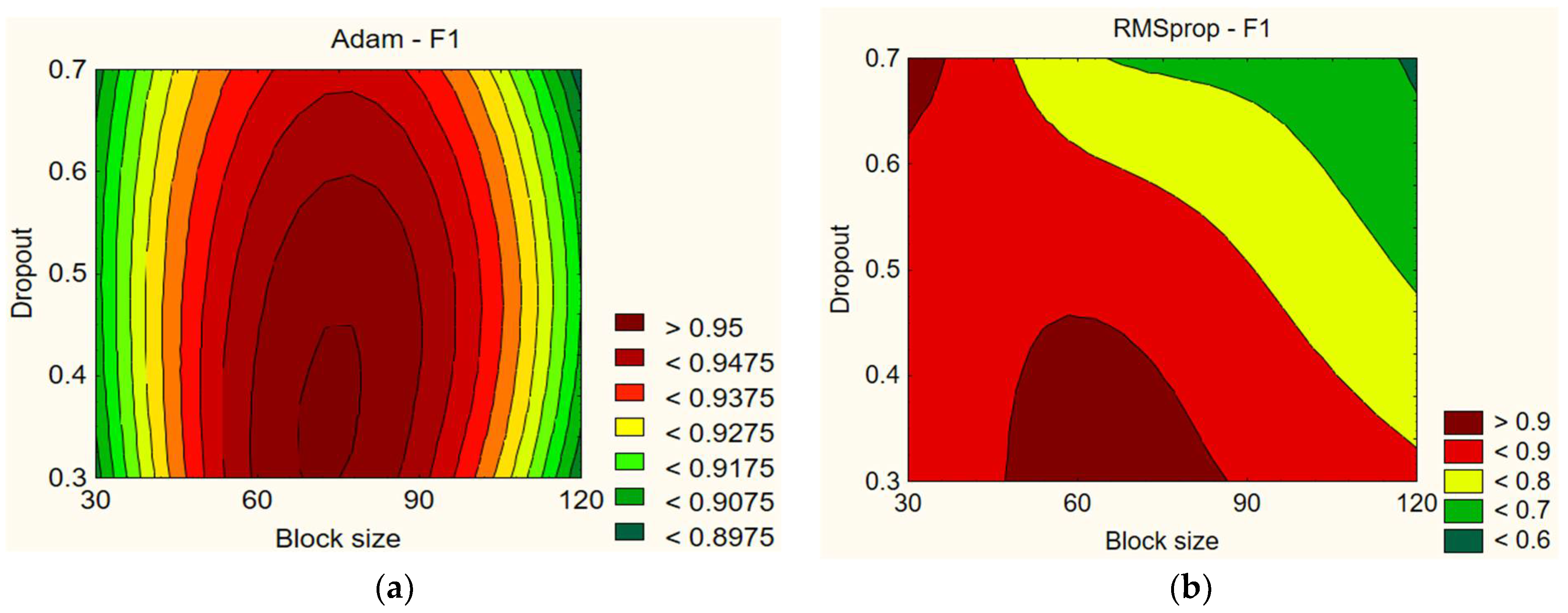
Figure 3, the data show that different values of learning rate and dropout have a small effect on the F1 score. For example, with the Adam optimizer, using a dropout of 0.7 and a learning rate of 0.005 gives the highest F1 score of 96.45%. Meanwhile, with a dropout of 0.6 and a learning rate of 0.001, the F1 score decreases to 90.74%. Overall, the F1 scores for the Adam optimizer are sufficiently high, but the data show that it is difficult to say whether the learning rate and dropout have a certain influence on the F1 score. However, it can be noted that the F1 score is lower with lower learning rate values (0.001). The results with the RMSprop optimizer are highly variable. Using a dropout of 0.3 and a learning rate of 0.005 gives the highest F1 score of 96.14%. Meanwhile, with a dropout of 0.7 and a learning rate of 0.01, the F1 score drops to 0%. Overall, the F1 scores for the RMSprop optimizer are widely dispersed, but the data show that the learning rate and dropout have a certain influence on the F1 score. Lower dropout rates and lower learning rates lead to better results. Comparing Adam and RMSprop, higher learning rate values in both optimizers resulted in worse F1 scores (see
Table 7).
Figure 4,
Figure 5 and
Figure 6 compare the impact of block size and learning rate. In
Figure 4, we can see from the Adam data that it can be observed that AUC values vary depending on the learning rate and the block size. It can also be concluded that a model that uses a smaller block size and a higher learning rate (for example, input length of 30–60 and learning rate of 0.005–0.01) can achieve a better AUC value. For instance, when the block size is 30 and the learning rate is 0.01, the AUC is 96.81%, whereas it decreases to 92.79% when the block size is 120, which is almost 4% lower. With RMSprop, it can be observed that combinations of shorter block sizes (30) and lower learning rates (0.001) tend to result in higher AUC values. Conversely, combinations of longer input lengths (120) and higher learning rates (0.01) result in lower AUC values (see
Table 8).
For instance, with a block size of 30 and a learning rate of 0.001, the AUC is 96.69%. However, it decreases to 93.67% when the block size increases to 120. From these observations, we can conclude that an appropriate combination of learning rate and block size significantly influences the variation in the AUC.
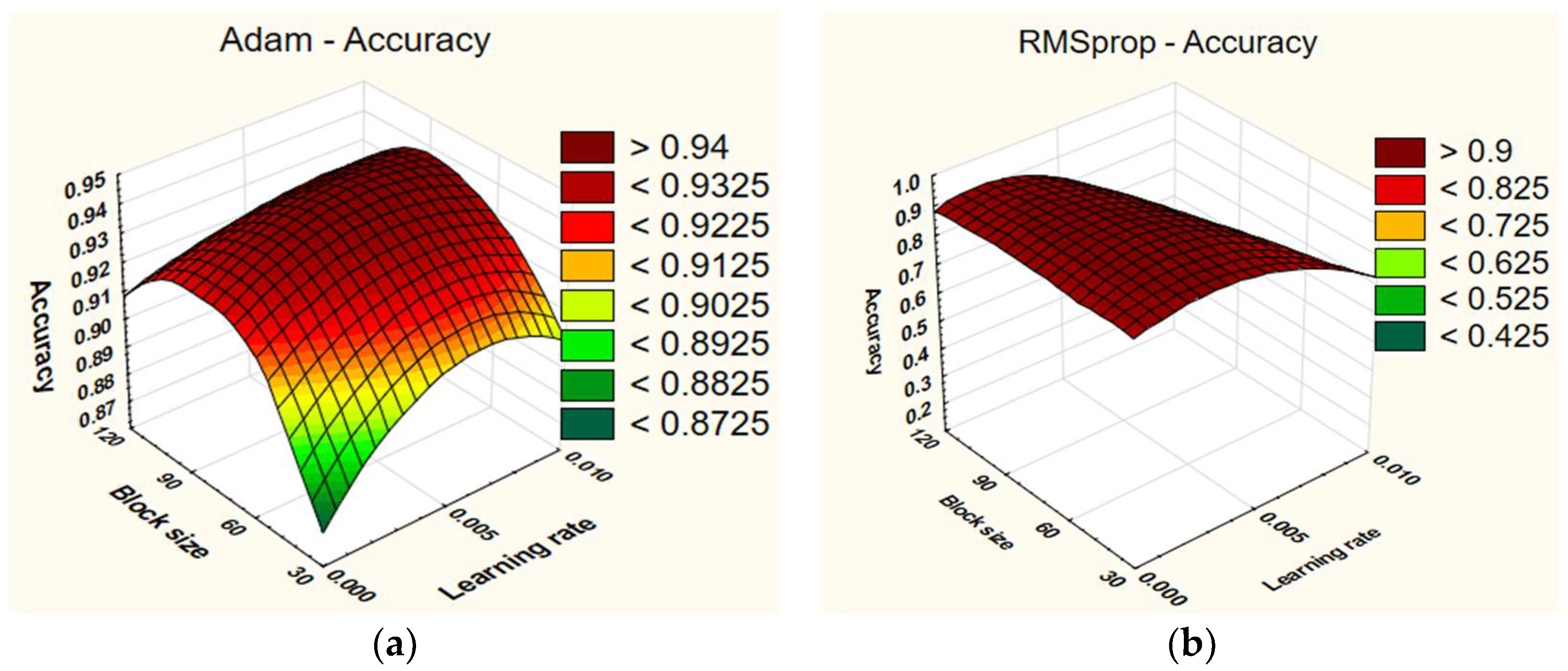
From
Figure 5, it is evident that the highest accuracy (93.8%) was achieved with a learning rate of 0.005 and a block size of 60when using RMSprop. Conversely, the lowest accuracy (21%) was observed when using a learning rate of 0.01 across all block sizes (30, 60, and 120). When employing Adam, the highest accuracy result (94.6%) was obtained with a learning rate of 0.005 and a block size of 60. On the other hand, the lowest accuracy (87.33%) was achieved with a learning rate of 0.001 and a block size of 30.
The block size does not have a significant impact on the results when using the RMSprop optimizer. Generally, all block size options yield similar performance, but slightly worse results are observed with a block size of 120. For instance, a network with a learning rate of 0.001 and a block size of 120 achieves an accuracy of 90.4%. With a block size of 30, the accuracy is 91%, and the accuracy is 92% with a block size of 60. On the other hand, the learning rate does have an impact, with poorer results observed when using a learning rate of 0.01. With the Adam optimizer, it can be observed that the learning rate has a lesser impact on the results. The results across different learning rates are relatively similar. However, the block size has a more pronounced effect. The best results are obtained with a block size of 60, whereas the worst results are observed with a block size of 30. The results for a block size of 120 fall approximately in the middle. For example, a network with a block size of 60 achieves an accuracy of 92.4%, whereas the accuracy is 89.46% with a block size of 30. With a block size of 120, the accuracy is 91.86% (see
Table 9).
Observations with the Adam optimizer (refer to
Figure 6) indicate that the choice of block size and learning rate significantly influences the F1 score. Generally, increasing the block size from 30 to 60 results in an enhancement in the F1 score. However, further increases up to 120 do not consistently yield positive effects on the F1 score. This behavior might be attributed to the larger block size, facilitating the model’s understanding of longer contexts, but excessively large block sizes could adversely affect training due to the increased complexity and memory requirements. Regarding the learning rate, the F1 scores’ distribution is quite diverse, suggesting that the learning rate does not have a significant influence. In contrast, with RMSprop, we observe that the learning rate has a more pronounced impact compared to the block size. A learning rate of 0.01 yields the poorest results, whereas both 0.005 and 0.001 provide better outcomes. In terms of recording length, optimal results are typically achieved with a block size of 60. However, satisfactory results can also be achieved with block sizes of 30 and 120, although the latter yields comparatively poorer average results. The impact of learning rate and block size on the F1 score is detailed in
Table 10.
Figure 7,
Figure 8 and
Figure 9 compare the impact of block size and learning rate.
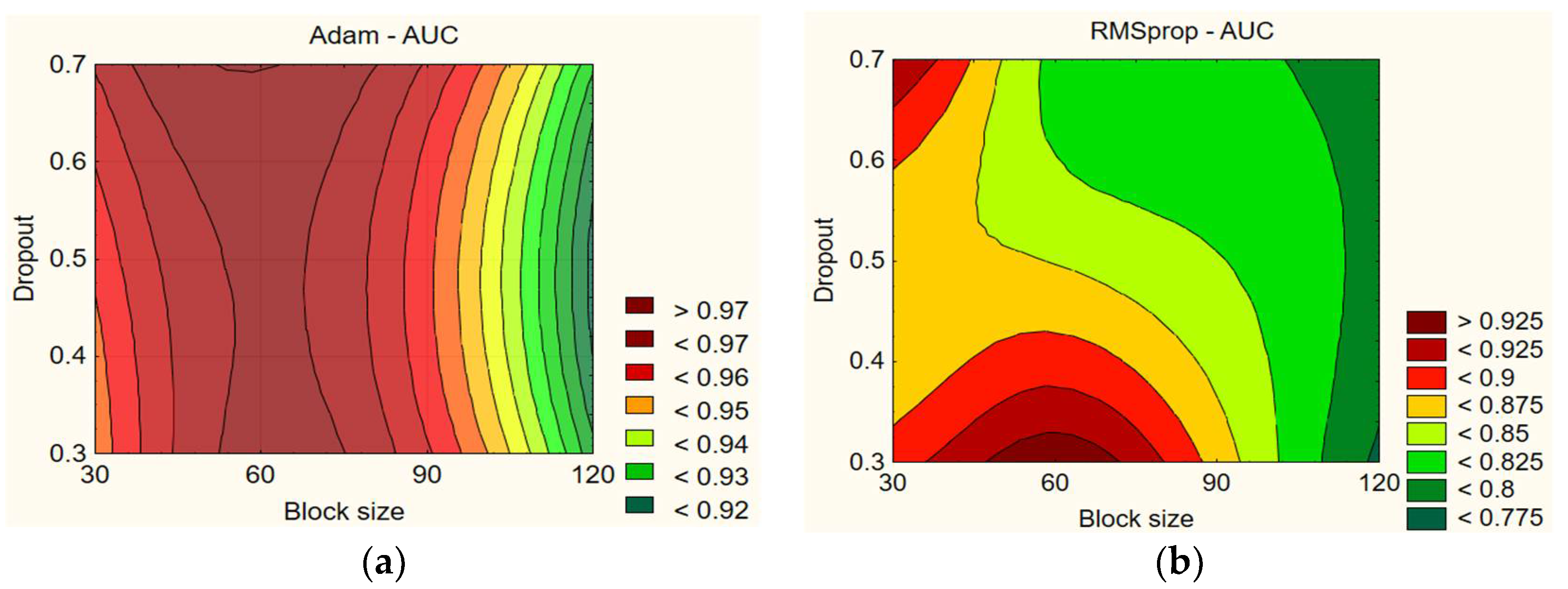
In
Figure 7, when examining the data for Adam, it can be noticed that higher dropout (0.6–0.7) with a block size of 60 generally leads to higher AUC values. For example, with a dropout of 0.7 and a block size of 60, the AUC reaches 97.45%, and it is 97.29% with a dropout of 0.6. However, worse AUC values are obtained with longer block sizes. For example, with a dropout of 0.7 and a block size of 120, the AUC is 95.51%, whereas it is 93.91% with a dropout of 0.6. When the dropout is lower (0.3–0.4), worse results are obtained. For example, with a block size of 30 and a dropout of 0.7, the AUC is 97.11%, whereas it is 95.93% with a dropout of 0.3. With RMSprop, lower AUC values are obtained with a block size of 120, whereas it depends on the dropout with other block size combinations. For example, with a dropout of 0.3 and a block size of 60, the AUC reaches 96.38%, whereas it is 95.39% with a dropout of 0.7 and a block size of 30. The highest values are obtained with a block size of 60 and a low dropout or with a smaller block size of 30 and a higher dropout (0.6–0.7). The results on dropout and block size impact on AUC are presented in
Table 11.
In
Figure 8, the data for the Adam optimizer reveals that the dropout rate has a lesser impact compared to the block size. Generally, higher accuracy values are achieved with a block size of 60. For instance, with a dropout rate of 0.7 and a block size of 60, the accuracy reaches 94.6%. Conversely, with different block sizes, lower accuracy values are observed. For example, with a dropout rate of 0.7 and a block size of 120, the accuracy reaches 92.2%, whereas it is 89.13% with a dropout rate of 0.3 and a block size of 30. With RMSprop, the dropout rate also has a lesser impact compared to the block size. However, better accuracy is obtained with smaller block sizes, specifically in the range of 30–60. For instance, with a dropout rate of 0.3 and a block size of 60, the accuracy reaches 93.73%, whereas it is 86.46% with a block size of 120. However, with a dropout rate of 0.7 and a block size of 120, the accuracy improves to 90.6%. Results are presented in
Table 12.
In
Figure 9, the data demonstrate that different values of block sizes and dropout have an impact on the F1 score. When using the Adam optimizer, lower dropout values (e.g., 0.3 and 0.4) result in higher F1 scores compared to higher dropout values (e.g., 0.6 and 0.7), and both 30 and 120 block sizes lead to worse results. For instance, the F1 score is 92.14% using a dropout of 0.7 and a block size of 30, and it is 90.81% for a block size of 120. However, with a dropout of 0.3 and a block size of 60, the F1 score improves to 95.51%. Overall, the results from the Adam optimizer indicate that dropout and block size have an impact on the F1 score (see
Table 13).
With the RMSprop optimizer, the results show slight differences. Significantly worse results are observed only with a block size of 120, whereas similar results are obtained with other parameter combinations. The best result is achieved with a dropout of 0.3 and a block size of 60, yielding an F1 score of 96.14%. Using a dropout of 0.7 and a block size of 30, the F1 score is 95.44%. However, when using a dropout of 0.7 and a block size of 120, the F1 score drops to 90.08%. Overall, the data from RMSprop demonstrate that dropout and block size impact the F1 score. When comparing Adam and RMSprop, the F1 scores are very similar, but the scores obtained with the Adam optimizer are slightly higher.
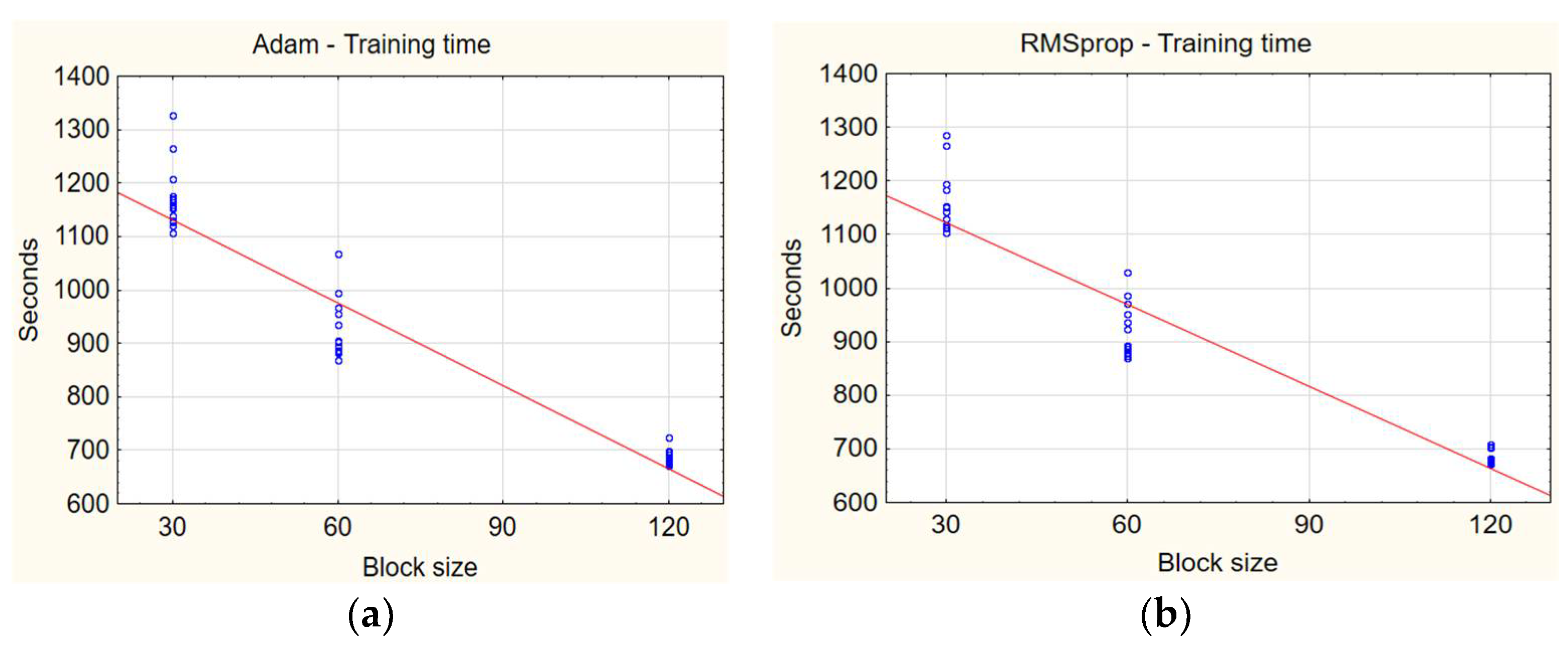
Figure 10 compares the training times based on different block sizes.
Firstly, a larger block size of 120 generally results in shorter training times compared to smaller block sizes of 30 and 60. This can be explained by the fact that a larger block size provides more efficient data utilization, reducing the number of updates per epoch. To better understand the relationship between time and block size, it is worth reviewing some average times by block size:
Block size 30: about 1100 s;
Block size 60: about 900 s;
Block size 120: about 700 s.
From these results, we see that increasing the record length from 30 to 120 reduces the average training time by about 37%. However, it is important to note that although a larger block size can help reduce training time, it affects the accuracy of the model.
Summarized results of the experiment are provided in
Table 14.
The initial reproduced data exhibit quite high results, especially the accuracy of g1—63.58%, the highest of all other models, and other results: g2 accuracy—87.18%; overall accuracy—92.93%; AUC—96.80%; Precision—96.76%; Recall—93.33%; and F1—95.02%. The article provided limited information on the c23 data level, but the AUC is very high—97.3%. The research results with Adam achieve the highest results among all models, in almost all criteria, except for g1—51.87% and Recall—95.27%. Furthermore, g2 accuracy—88.66%; overall accuracy—97.45%; AUC—97.45%; Precision—97.66%; Recall—95.27%; and F1—96.45%. RMSprop with the best parameters also exhibits high indicators, but overall has inferior results compared to the research results of Adam: g1 accuracy—57.31%; g2 accuracy—86.62%; overall accuracy—92.86%; AUC—96.82%; Precision—94.76%; Recall—96.20%; and F1—95.47%.
The main parameters are overall accuracy, AUC, and F1, and, during the research, the best results with the Adam optimizer surpass the AUC results indicated in the article by 0.1%. Compared to the reproduced model results, AUC is improved by 0.6%. Additionally, compared to the reproduced model results, g2 is improved by 1.5%, overall accuracy by 1.7%, and F1 by 1.4%. However, when comparing the reproduced model results with the best RMSprop research results, RMSprop results are slightly inferior or almost identical, except for the F1 result, which is better by 0.4%.
Based on this analysis, the best Adam research results indicate that there are parameters for this model that produce better results, and these parameters are selected for further analysis because they exhibit the highest results in almost all measurement indicators. These best parameters are:
Learning rate—0.005;
Dropout rate—0.7;
Sequence length—60;
Optimizer—Adam.
Therefore, by choosing these new parameters, the model is trained using previous training data, but with 5-fold cross-validation to further verify that the training is consistently better than the results reported in the article. The testing data are left the same as specified in the article. The cross-validation training results of the model are shown in
Table 15.
In this table, the reproduced results of the article, the results reported in the article, and the average results obtained by performing a 5-fold cross-validation with the best parameters found during the experiment are presented. The best parameters’ “raw” results show a g1 accuracy of 21.71%, a g2 accuracy of 90.56%, an overall accuracy of 95.31%, an AUC of 97.43%, a precision of 96.79%, a recall of 97.93%, and an F1 score of 97.36%. The results show that none of the parameters are improved compared to the reproduced or reported results with these parameters and “raw” dataset. Compared to the reported results, the accuracy is about 5% lower, and the AUC is about 2% lower, with g1 being completely poor and practically not working correctly. With the “c23” dataset, the results show a g1 accuracy of 21.71%, a g2 accuracy of 91.54%, an overall accuracy of 96.43%, an AUC of 98.02%, a precision of 97.42%, a recall of 98.14%, and an F1 score of 97.78%. Compared to the AUC result reported in the article, it was improved by 0.7%, and the other results were not reported in the article for this dataset level. Compared to the reproduced model results, all results are improved except for g1. The accuracy is improved by 3.5%, the AUC by 1.2%, and the F1 score by 2.7%. Finally, with the “c40” dataset, the results show a g1 accuracy of 32.64%, a g2 accuracy of 82.91%, an overall accuracy of 89.66%, an AUC of 90.68%, a precision of 90.88%, a recall of 97.25%, and an F1 score of 93.94%. Compared to the “c40” results reported in the article, all results are slightly lower by a few percentage points, e.g., the accuracy is 1.7% lower, and the AUC is 5% lower. Compared to the reproduced model results, the accuracy and AUC results are still lower, but the g1 accuracy is 0.5% better, and the recall is 1.7% better. In summary, this new model results in slightly improved “c23” AUC results.
To verify that the results obtained with the new parameters are consistent, the standard deviations and variation coefficients of the cross-validation results are presented in
Table 16.
When comparing the standard deviations of different data sets, the “raw” data set has larger standard deviation sizes for g2 accuracy, overall accuracy, and AUC compared to the “c23” data set. The “c40” data set has larger standard deviation sizes for all parameters compared to the “raw” and “c23” data sets, except for g1 accuracy, whose standard deviation is equal to 0. However, overall, the standard deviations are low for all data sets. Comparing the variation coefficient of different data sets, the “raw” data set has larger coefficient of variation sizes for g2 accuracy, overall accuracy, AUC, precision, and recall, compared to the “c23” data set. The “c40” data set has a significantly larger variation coefficient for g1 accuracy compared to other models, as well as larger variation coefficient sizes for all other parameters compared to the “raw” and “c23” models. The AUC standard deviation of the “c23” data set results (0.0050) is smaller than the standard deviations of the “raw” (0.0064) and “c40” (0.0254) models. A smaller standard deviation indicates that the “c23” AUC results are more concentrated around the mean, and, therefore, the model is more stable and has less result dispersion. In addition, the “c23” data set AUC variation coefficient (0.5237) is smaller than the variation coefficients for the “raw” (0.6819) and “c40” (2.8382) data sets. A smaller variation coefficient indicates that the “c23” AUC results are less dispersed compared to the results of other data sets.
Based on these comparisons, we can conclude that the model is more efficient and more stable when it comes to AUC results with the “c23” data set, and these results effectively improve the AUC results reported in the article.
4.2. Improved Model
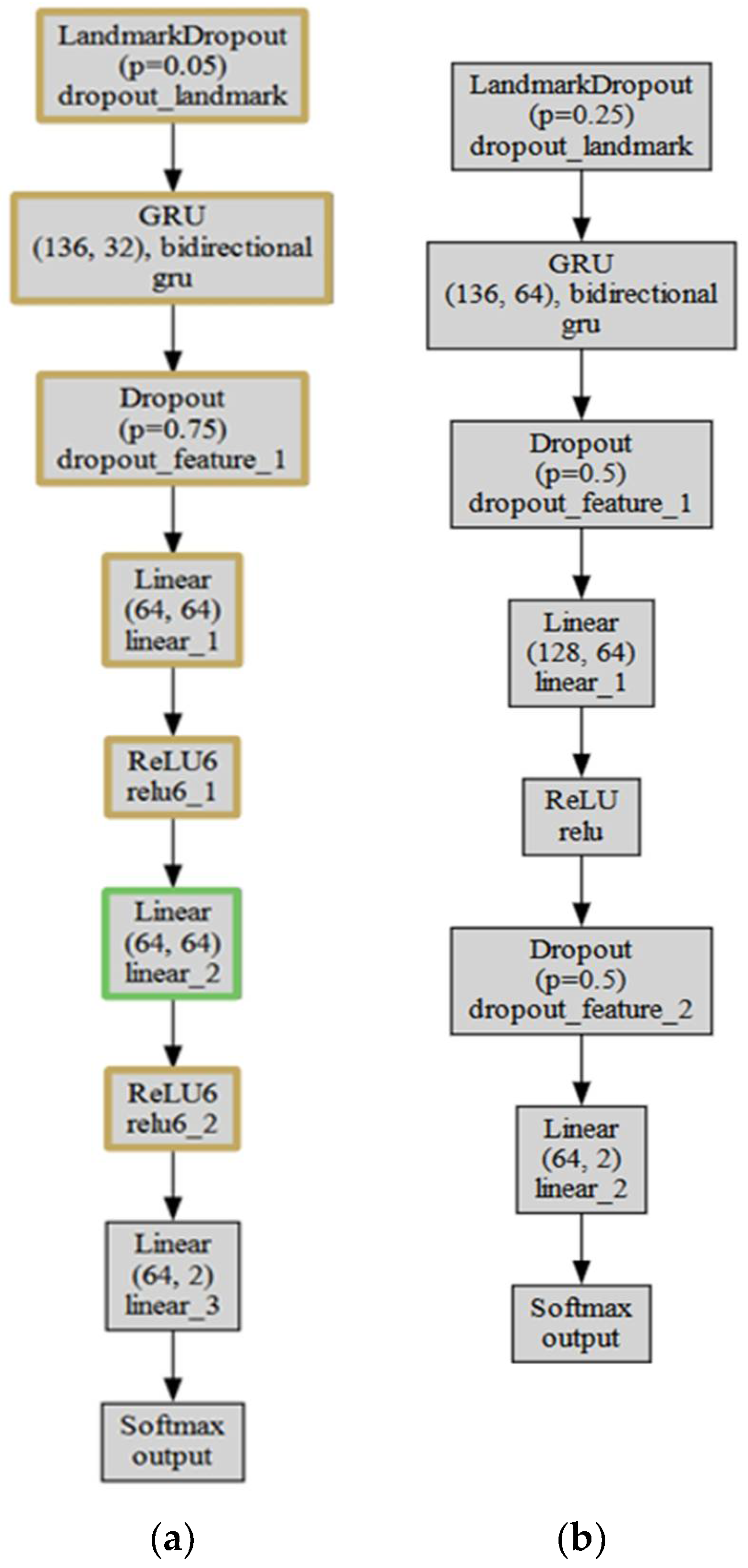
Based on the results obtained in the previous section, a new improved model version is presented in
Figure 11. When creating this model, the goal was to improve the AUC results and accuracy for the “c23” data level, compared to the results reported in the article or during the experiment. The “c23” data level is also important because these data are subject to data compression, which is often encountered on the internet; thus, this data could be more important considering the relevance of the models’ usage. AUC and accuracy are the most important indicators of model effectiveness in distinguishing true images from fake ones.
The main differences between the two models are the first dropout rate, the number of GRU hidden neurons, the number of dropout layers, and the configuration of linear layers and activation functions. The improved model has a smaller number of GRU hidden neurons and additional linear and ReLU6 layers, whereas LRNet has a higher first dropout rate and one additional dropout layer.
When comparing the two models, the improved model and LRNet, several key differences in their structure are observed. First, the improved model uses a smaller first dropout rate—0.05—compared to LRNet’s of 0.25. Second, the GRU hidden neuron count is smaller in the improved model—32—compared to LRNet’s 64. In addition, the improved model has one dropout layer with a 0.75 dropout rate, whereas LRNet has two dropout layers with a 0.5 dropout rate. The configuration of linear layers and activation functions is also different. The improved model has three linear layers with ReLU6 activation functions, whereas LRNet has two linear layers with ReLU activation functions. Additionally, the number of neurons in the linear layers of the improved model is smaller since the output of the GRU layer is smaller. The linear layers of the improved model are: 64 × 64, 64 × 64, 64 × 2, whereas LRNet has 128 × 64, 64 × 2. Finally, both models have a Softmax output layer with a single output. The improved model is more complex, with additional linear layers and ReLU6 activation functions, whereas LRNet has a higher first dropout layer coefficient and one additional dropout layer.
The model was trained using previous training data but with 5-fold cross-validation to further verify training errors and ensure that the model consistently achieved better results. However, the testing data remained the same as reported in the article. The cross-validation results for the improved model are shown in
Table 17.
This table presents the reproduced results, reported results, the average of the results obtained from the cross-validation with the best parameters found during the experiment, and the average of the results obtained from the cross-validation with the improved model. The improved model’s “raw” variant achieved very similar results to the reproduced model’s “raw” and the best parameter model’s “raw” results. This indicated that the improved model with the “raw” dataset had slightly lower performance indicators, such as g1 accuracy (79.78%), g2 accuracy (90.70%), overall accuracy (94.54%), AUC (96.92%), precision (95.96%), recall (98.11%), and F1 (97.02%). Compared to the reproduced model, AUC was 1% lower, with the reported model AUC 3% lower, and with the best parameter model AUC 0.5% lower. Secondly, the improved model’s “c23” variant achieved better results than the reproduced model in almost all criteria. g1 accuracy increased from 63.58% to 77.77%, g2 accuracy increased from 87.18% to 92.40%, overall accuracy increased from 92.93% to 96.17%, AUC increased from 96.80% to 98.39%, precision did not change, recall increased from 93.33% to 98.71%, and F1 increased from 95.01% to 97.72%. Compared with the data reported in the article, AUC improved by 1.08%, compared to the best parameter model by 0.36%, and compared to the reproduced model by 1.58%. Finally, the improved model’s “c40” variant showed similar results to the reproduced model. g1 accuracy decreased from 81.53% to 76.71%, but g2 accuracy increased from 82.44% to 83.46%, overall accuracy decreased from 91.28% to 88.54%, AUC decreased from 93.71% to 89.19%, precision decreased from 93.53% to 89.14%, recall increased from 95.53% to 98.29%, and F1 decreased from 94.52% to 93.48%. Compared to the best parameter model, almost all results were about 1% lower. In summary, the improved model showed the best AUC results with the “c23” dataset among all models and the best results for other parameters but showed slightly lower results with other datasets.
To ensure or verify that the results obtained with the improved model were consistent,
Table 18 presents the standard deviations and variation coefficients of the cross-validation results.
When comparing the standard deviations of different datasets, the “raw” dataset exhibits smaller standard deviation sizes for g2 accuracy, overall accuracy, AUC, and F1, in comparison to the “c40” dataset. It also demonstrates smaller standard deviations for g2 and overall accuracy when compared to the “c23” dataset, albeit with a negligible difference. Conversely, the “c40” dataset displays larger standard deviation sizes for all parameters when compared to the “raw” and “c23” datasets. Overall, the standard deviations are minimal for all datasets, but the noteworthy finding is that the AUC and F1 standard deviations are the smallest with the “c23” dataset.
Regarding the coefficient of variation in different datasets, it was observed that the “c40” dataset exhibits larger coefficients of variation for g1 and g2 accuracy, overall accuracy, AUC, precision, recall, and F1 when compared to the “c23” and “raw” datasets. The “raw” dataset demonstrates the smallest coefficients of variation, except for AUC, precision, and F1 results. The “c23” dataset showcases the best AUC, precision, and F1 results.
The AUC standard deviation (0.0049) of the “c23” dataset is smaller than the standard deviations of the “raw” (0.0102) and “c40” (0.0316) datasets. A smaller standard deviation indicates that the AUC results of the “c23” dataset are more concentrated around the mean, suggesting a higher level of stability and reduced result variability for the model. Furthermore, the AUC variation coefficient (0.5056) of the “c23” dataset is smaller than the variation coefficients of the “raw” (1.0610) and “c40” (3.5487) datasets. A smaller variation coefficient indicates that the AUC results of the “c23” dataset are less dispersed in comparison to the results of the other datasets.
From these comparisons, it can be concluded that the model is more effective and operates more stable than with other datasets with the “c23” dataset, especially when it comes to AUC. This indicates that the enhanced model consistently improves upon the performance of the original model, enabling the better identification of manipulated images from real ones. Moreover, the overall accuracy of the improved model was enhanced compared to the results of the replicated model. Unfortunately, with other datasets and model parameters, the model did not achieve better results.
Overall, with the improved model and the best parameters found in our experiment, the analyses of the “c23”, “raw”, and “c40” datasets indicate that the model performs most optimally and stably with the “c23” dataset, particularly regarding the AUC measure. With this dataset, the model’s effectiveness in discerning real images from forged ones and its overall accuracy is improved. However, the model’s performance does not show similar improvements with other datasets and parameters. Comparing standard deviations and coefficients of variation, the “c23” dataset consistently exhibits less dispersion and variation in results, underlining its stability and efficiency. In contrast, the “raw” and “c40” datasets showed larger standard deviations and coefficients of variation for most parameters, highlighting their relatively less stable and consistent performance. The improvements we observed in the model’s accuracy for the “c23” dataset are a direct result of the specific optimizations we made, which were tailored based on the characteristics and patterns identified during our experimentation phase. However, it’s important to note that these optimizations were not necessarily designed to enhance performance on other datasets, such as the raw and “c40” datasets. This explains why the accuracy values for these datasets did not show similar improvements. The optimizations that improved performance for the “c23” dataset may not be effective for other datasets due to their unique characteristics and data composition. Consequently, the improvements significantly contributed to enhancing the model’s performance and stability, particularly in relation to AUC results.