Abstract
Cellulose-degrading bacteria were identified from distillery lees, and the strains were optimized for fermentation and enzyme production, providing effective strains for the resource utilization of distillery lees and developing cellulase. Based on the univariate test, the response surface test was used to optimize enzyme production conditions for fermentation. The screened strain JZ2 had a clear circle-to-colony diameter ratio of 2.0. The enzyme activities of exoglucanase, endoglucanase, and β-glucosidase were 4.341 ± 0.05 U/mL, 1.874 ± 0.04 U/mL, and 0.739 ± 0.02 U/mL, respectively. The bacterial colonies were large, and the bacterial cells’ morphology was rod-shaped. We identified Bacillus cereus (JZ2) from 16S rDNA sequence homology and phylogenetic tree analysis as belonging to the genus Bacillus in the thick-walled Bacillus phylum. The optimum production conditions of cellulase from strain JZ2 were a fermentation time of 2 d, a rotation rate of 180× g, and a temperature of 26 °C using the response surface method. The enzyme activity of JZ2 was 4.625 U/mL under optimal enzyme production conditions. In addition to good activity, the cellulase from the JZ2 strain may have the potential to convert distillery lees cellulose into useful compounds.
1. Introduction
Distillery lees are the main byproduct of the distillation industry [1,2] and play a key role in the circular economy [3]. China has a 2000-year history of brewing liquor [4] as a major producer and consumer. The annual production of distillery lees is as high as 58.66 million tons [5], ranking first in the world [6,7]. Distillery lees have high moisture content and acidity and are prone to corruption during storage. Traditional disposal methods, such as stacking and discarding, cause environmental pollution [4] and resource waste [8,9]. At present, distillery lees are mainly used in the production of protein feed, bio-organic fertilizers, chemical products, production fuel, etc. [10,11]. White distillery grains, however, can contain about 20% of raw fiber content, which livestock cannot digest and absorb properly [12,13]. Therefore, the resource utilization of distillery lees must be urgently solved.
The degradation of raw cellulose is important to the use of distillery lees. Physical and chemical methods for cellulose degradation have issues related to pollution, energy consumption, and high costs [14]. The biological method has attracted the attention of researchers for its advantages of environmental protection, strong targeting, low external cost, mild operating conditions, and low energy consumption [15,16,17]. In recent years, research at home and abroad has mainly focused on isolating high-yielding cellulose strains from the soil, conducting mutate treatment on existing strains in the laboratory, and optimizing enzyme-producing conditions to identify strains with strong cellulase production capacity [18,19,20,21]. Khosravi et al. used Congo red-CMC (carboxymethyl cellulose) plates to screen high-yielding cellulase strains [16]. Yu et al. used carboxymethylcellulose sodium medium to screen strains with cellulose degradation ability from distillery lees sludge, which provided a theoretical method for screening cellulose in distillery lees [22]. However, the need for new isolated cellulose-degrading microorganisms continues. There is relatively little research on cellulose in distillery lees. Compared to that produced by fungi, the cellulase produced by bacteria has more advantages, including acid alkali resistance, heat resistance, short production cycle, and easy cultivation [23,24]. Consequently, screening cellulose-degrading bacteria in distillery lees is significant.
In this study, cellulose-degrading bacteria were screened from distillery lees and identified via morphological observation, 16S rDNA sequence analysis, and physiological and biochemical identification. The enzyme production conditions of the selected dominant strains were optimized. Our aim for this study was to find a superior cellulase-producing bacterium strain that could increase the resource utilization of distillery lees.
2. Materials and Methods
2.1. Materials
The distillery lees were provided by Shanxi Baozi Liquor Industry Co., Ltd. Congo red staining solution, DNS (3,5-Dinitrosalicylic acid) reagent, acetic acid/sodium acetate buffer solution, sodium chloride solution, CMC–Na solution, microcrystalline cellulose solution, and salicylic solution reagent were purchased from Xi’an Dafeng Harvest Biotechnology Co., Ltd., Xi’an, China.
Enrichment medium (g/L): CMC–Na 5, Na2HPO4 1.6, MgSO4·7H2O 0.5, K2HPO4 0.9, KNO3 1, KCl 0.5, yeast powder 0.5, peptone 0.5, distilled water 1000, pH = 7, 121 °C, and sterilization for 20 min. Select medium (g/L): CMC–Na 10, peptone 10, yeast powder 5, KH2PO4 1, MgSO4·7H2O 0.2, NaCl 10, agar 20, distilled water 1000, pH = 7, 121 °C, and sterilization for 20 min. Fermentation medium (g/L): CMC–Na 10, KH2PO4 1, MgSO4·7H2O 0.2, NaCl 10, peptone 10, yeast powder 10, distilled water 1000, pH = 7, 121 °C, and sterilization for 20 min. Luria–Bertani medium (LB, g/L): BactoTM tryptone 10, sodium chloride 10, BactoTM yeast extract 5, agar 15, pH = 7, 121 °C, and sterilization for 20 min.
2.2. Test Method
2.2.1. Initial Screening of Cellulose-Degrading Bacteria
Initial screening of cellulose-degrading bacteria was performed according to the method of [25]. We added 1 g of distillery grains to 100 mL of sterile water and shook the mixture thoroughly for 30 min at 180× g (WH-25 constant temperature incubator, Beijing Guangming Medical Instrument Factory, Beijing, China). After layering, we added 10 mL of bacterial suspension to a 90 mL liquid enrichment medium and cultured them at 180× g, 30 °C for 24 h. Afterward, the supernatant was diluted to 10−7 using a 10-fold dilution method. An aliquot of 200 μL from each dilution was spread onto agar plates with CMC and incubated at 30 °C for 2 days. Then, we picked colonies with an inoculation needle and repeated their separation and purification in the selected medium. The purified single colony was inoculated into CMC-Na medium and incubated at 30 °C for 24–48 h. After incubation, all the plates were stained with Congo red staining solution for 25 min and washed with 1.0 M NaCl for 15 min. We determined the ratio of hydrolysis circle diameter (D) to colony diameter (d) and selected strains with a large ratio for rescreening.
2.2.2. Rescreening of Cellulose-Degrading Bacteria
Strains with large transparent circles were inoculated in a liquid enzyme-producing medium at 30 °C for 12 h and centrifuged at a rotation rate of 180× g. Afterward, the prepared seed solution was inoculated into 8 mL of an enzyme-producing medium with 5% inoculum and cultured for 24 h. The bacterial solution was then centrifuged at 4 °C for 5 min at a rotation rate of 100× g. We used the supernatant to determine cellulase activity and selected strains with high enzyme activity for bacterial identification.
The DNS method determined cellulase activities (endoglucanase, exoglucanase, and β-glucosidase) [26]. Endoglucanase activity was analyzed using CMC as a substrate in phosphate buffer (0.05 M, pH 5.5). Exoglucanase activity was estimated using microcrystalline cellulose as a substrate in phosphate buffer (0.05 M, pH 5.5). β-glucosidase activity was estimated using D-(-)-salicin as a substrate in phosphate buffer (0.05 M, pH 5.5). The selected strains were grown in LB broth medium for 24 h at 30 °C. After centrifuging for 5 min at 1000× g (TGL-18M high-speed centrifuge, Shanghai Lu Xiangyi Centrifuge Instrument Co., Ltd., Shanghai, China), 0.5 mL of enzyme solution (the supernatant) was mixed with 1 mL of 1% (m/v) CMC phosphate buffer, 1% (m/v) microcrystalline cellulose phosphate buffer, or 1% (m/v) D-(-)-salicin phosphate buffer in three different test tubes. Test tubes were incubated at 30 °C for 30 min at a rotation rate of 100× g. Then, we added 2.5 mL of DNS solution to a boiling water bath for an incubation time of 10 min. Afterward, we diluted the volume of the mixture to 25 mL and measured the absorbance at 540 nm (EV-2600 UV/VIS spectrophotometer, Shanghai Onla Instrument Co., Ltd., Shanghai, China). Enzyme activity is defined as the number of enzymes required to catalyze the production of 1 μ mol and reduce sugars from the substrate within 1 min of enzyme solution. Enzyme activity was calculated according to the following equation:
where C is the mass concentration of the solution (mg/mL) calculated from the absorbance value, V is the total volume of the reaction (mL), 180 is the molecular weight of glucose, v represents the volume of the test solution (mL), and t is the total reaction time (min).
2.2.3. Strains Identification
Physiological and Biochemical Identification
Selected isolates were incubated on LB solid medium at 30 °C for 24–48 h. Gram staining was performed for the morphological identification of bacteria. The cultured strains were pretreated and observed using a scanning electron microscope (Gemini SEM 360, Carl Zeiss Co., Ltd., Jena, Germany). We observed and recorded the colony morphology.
We physiochemically identified selected isolates using a series of tests such as nitrate broth, V-P, starch hydrolysis, mannitol, Simone’s citrate, gelatin liquefaction, lysozyme, and motive medium, according to Hussain’s method [26].
Molecular Identification
Molecular identification was performed for the taxonomic characterization of isolated strains [16]. Total DNA was extracted using the Ezup column bacterial genome DNA extraction kit following instructions from the manufacturer. Polymerase chain reaction (PCR) amplified the 16S rDNA gene from the strain’s genomic DNA using universal primers 27F (AGTTTGATCMTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT). AmplitronyxTM (Nyxtechnik Inc., San Diego, CA, USA), a gradient thermal cycler, performed PCR amplification for 30 cycles according to the following steps: 3 min at 94 °C for denaturation, 1 min at 55 °C for annealing, 2 min at 94 °C for extension, and 10 min at 94 °C for elongation. We maintained the reaction mixtures at 4 °C until their use. BLAST was used to analyze the obtained sequence results on the NCBI website, and MEGA 7.0 was used to construct a phylogenetic tree to determine the species of the strain.
2.2.4. Optimization of Enzyme Production Conditions for Strain Fermentation
Single Factor Test
A 250 mL Erlenmeyer flask was used for all microbial cultures. To obtain the optimum fermentation time, we selected different initial values (l. 2. 3, 4, or 5 d) to optimize the fermentation time for high-yielding cellulase production. We inoculated the stock solution with 5% inoculum in 100 mL of fermentation culture medium and shook the mixture in a rotary shaker (WH-25, Shanghai Longyue Instrument Equipment Co., Ltd., Shanghai, China) at 180× g and 25 °C. To determine the optimal rotation rate, we inoculated the stock solution with 5% inoculum in 100 mL of fermentation medium and incubated the mixture in a rotary shaker at 25 °C with different rotation rates (150, 160, 170, 180, or 190× g) for 3 d. To obtain the optimum temperature, we inoculated the stock solution with 5% inoculum in 100 mL of fermentation medium in a rotary shaker at 170× g for 3 d. The CMC enzyme activities of all samples were measured to determine the optimal singe factor conditions. We repeated each experiment three times and plotted their average values.
Response Surface Test
The single-factor test results, three factors of fermentation time, rotation rate, and culture temperature, were optimized to obtain high CMC enzyme activity. An analysis test for three-factor and three-level fermentation was designed to determine the optimal enzyme-producing conditions for JZ2 (Table 1).

Table 1.
Response surface factor level table.
2.3. Statistical Analysis
The experiment was replicated three times. The data are expressed as mean ± standard error. Excel 2016 was used to assess the data. Origin 2021 was used to create presentation graphics. SPSS 19.0 was used to complete statistical analysis, and p-values of <0.05 were considered statistically significant.
3. Results and Analysis
3.1. Isolation and Screening of Cellulose-Degrading Bacteria
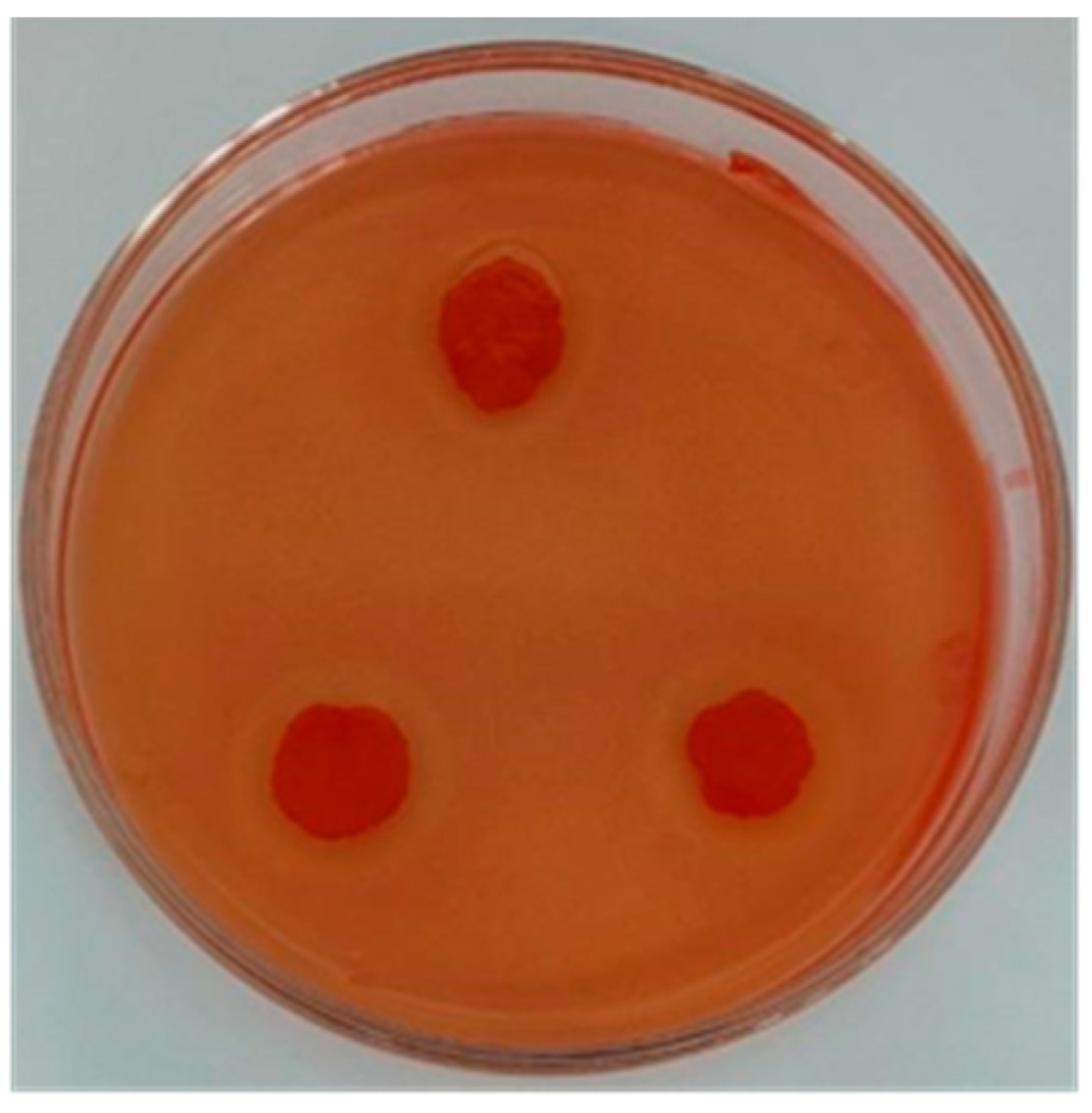
A total of five strains with large transparent circle diameters were isolated from the piled white distillery lees (Table 2). The D/d of strain JZ2 is 2, and the ratio was the largest. The transparent circle produced by strain JZ2 was large and clear (Figure 1). Cellulase activity can be measured by the ratio of the transparent circle size to the colony size to a certain extent. However, further determination of cellulase activity was needed to rescreen. Measuring enzyme activity can provide a better indication of a strain’s ability to produce cellulase.

Table 2.
Filter results of transparent circles.
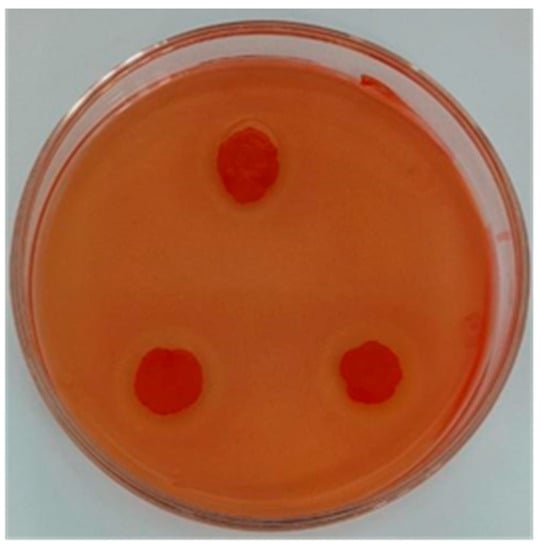
Figure 1.
Transparent circle produced by strain JZ2.
The strains selected via the initial screening were inoculated into a liquid enzyme-producing medium. We then determined endoglucanase activity, exoglucanase activity, and β-glucosidase activity in the fermentation broth. Table 3 shows the measurement results of enzyme activity. The enzyme activity of strain JZ2 was higher than in other strains. The values of exoglucanase activity, endoglucanase activity, and β-glucosidase activity were 4.341 ± 0.05 U/mL, 1.874 ± 0.04 U/mL, and 0.739 ± 0.02 U/mL, respectively. Zuo Yong et al. [27] isolated and purified a strain of Bacillus Vickers with cellulase production ability from distillery grains, producing exoglucanase, endoglucanase, and β-glucosidase activities of 2.66 U/mL, 0.60 U/mL, and 0.25 U/mL, respectively. The strains tested in this article had high enzyme activity, with the screened JZ2 strain having the highest activity. As a result, this has a huge advantage in the degradation of cellulose in distillery lees.

Table 3.
Determination results of enzyme activity.
3.2. Identification of Strains
3.2.1. Morphological Identification
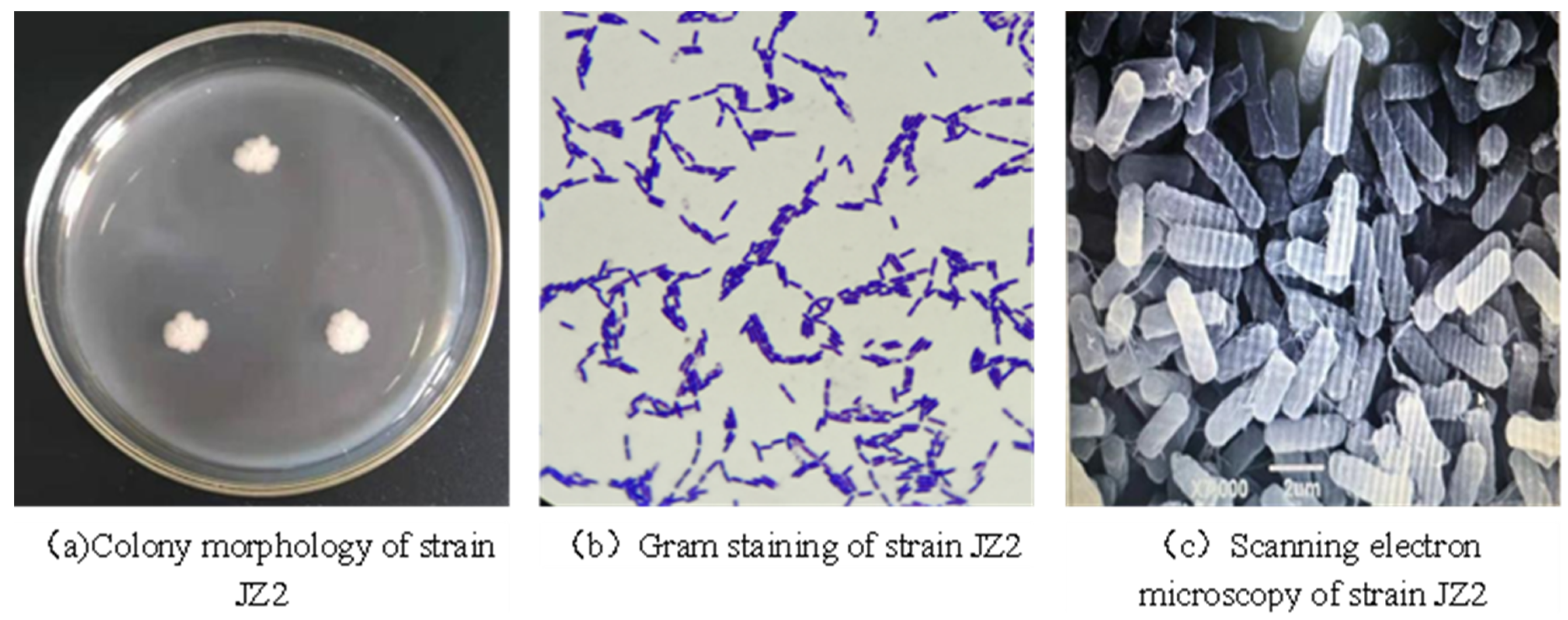
The isolated and screened strain JZ2 was inoculated into the medium for morphological observation. As shown in Figure 2, the colony generated on the culture medium was relatively large, approximately circular in shape, with neat and extended edges. The color was milky white with a slight luster. The texture was soft and sticky and appeared silky when lifted. The surface was rough, similar to frosted glass or melted wax, with a slight odor. After Gram staining, the strain was positive and produced spores (Figure 2b). The morphology of the JZ2 strain was observed using a scanning electron microscope [28]. The cells of the JZ2 strain were rod-shaped (as shown in Figure 2c).
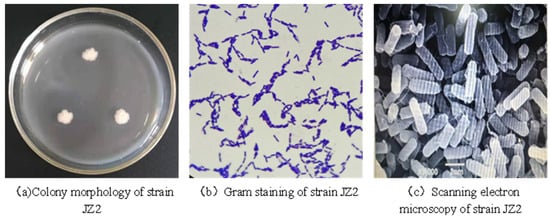
Figure 2.
Morphological characteristics of strain JZ2.
3.2.2. Physiological and Biochemical Identification
The strain JZ2 was inoculated in a biochemical identification tube. Changes in the identification tube were observed at 24, 48, and 72 h, respectively. As shown in Table 4, the appraisal results of the mannitol reaction test, Simon’s citrate utilization test, and the nitrate reduction test were negative, while the other tests were positive.

Table 4.
Physiological and biochemical characteristics of strain JZ2.
3.2.3. Molecular Identification
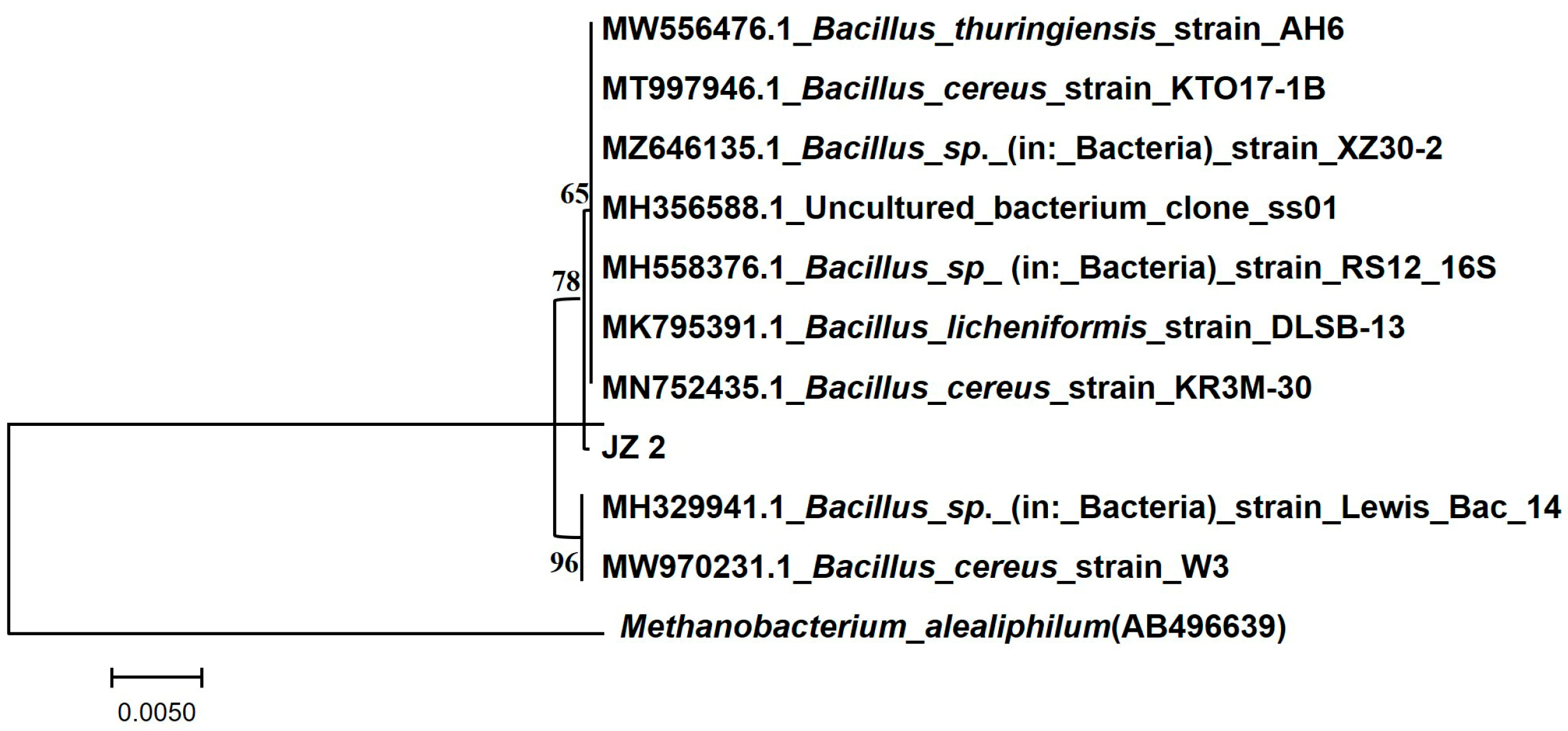
PCR sequencing of the JZ2 genomic DNA of cellulose-degrading bacteria showed that the strain sequence length was 1447 bp. We submitted the DNA sequences from strain JZ2 to NCBI for comparison. They found that the JZ2 strain had the highest homology, upwards of 99%, compared to Bacillus cereus 16S rDNA sequences. The strains associated with JZ2 were selected from NCBI comparison results. A phylogenetic tree was constructed using MEGA 7.0 (Figure 3). According to morphology, physiological and biochemical identification, 16S rDNA sequence homology analysis of the strain, and phylogenetic tree analysis, we determined that Bacillus cereus belongs to the genus Bacillus in the thick-walled Bacillus phylum. Abdelhamid et al. demonstrated that Bacillus cereus isolated from Egyptian soil is a potential producer of cellulolytic enzymes [29]. Hui et al. also reported Bacillus cereus as a bacterium that can produce degraded cellulose and polymer. They also found cellulase and xylanase genes in the B. cereus genome [30].
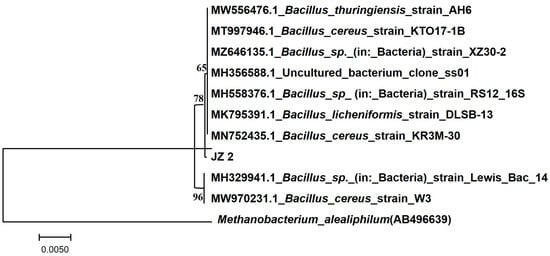
Figure 3.
Phylogenetic tree of strain JZ2.
3.3. Optimization of Enzyme Production Conditions for Strain Fermentation
3.3.1. Single Factor Results
Effect of Fermentation Time on Enzyme Activity
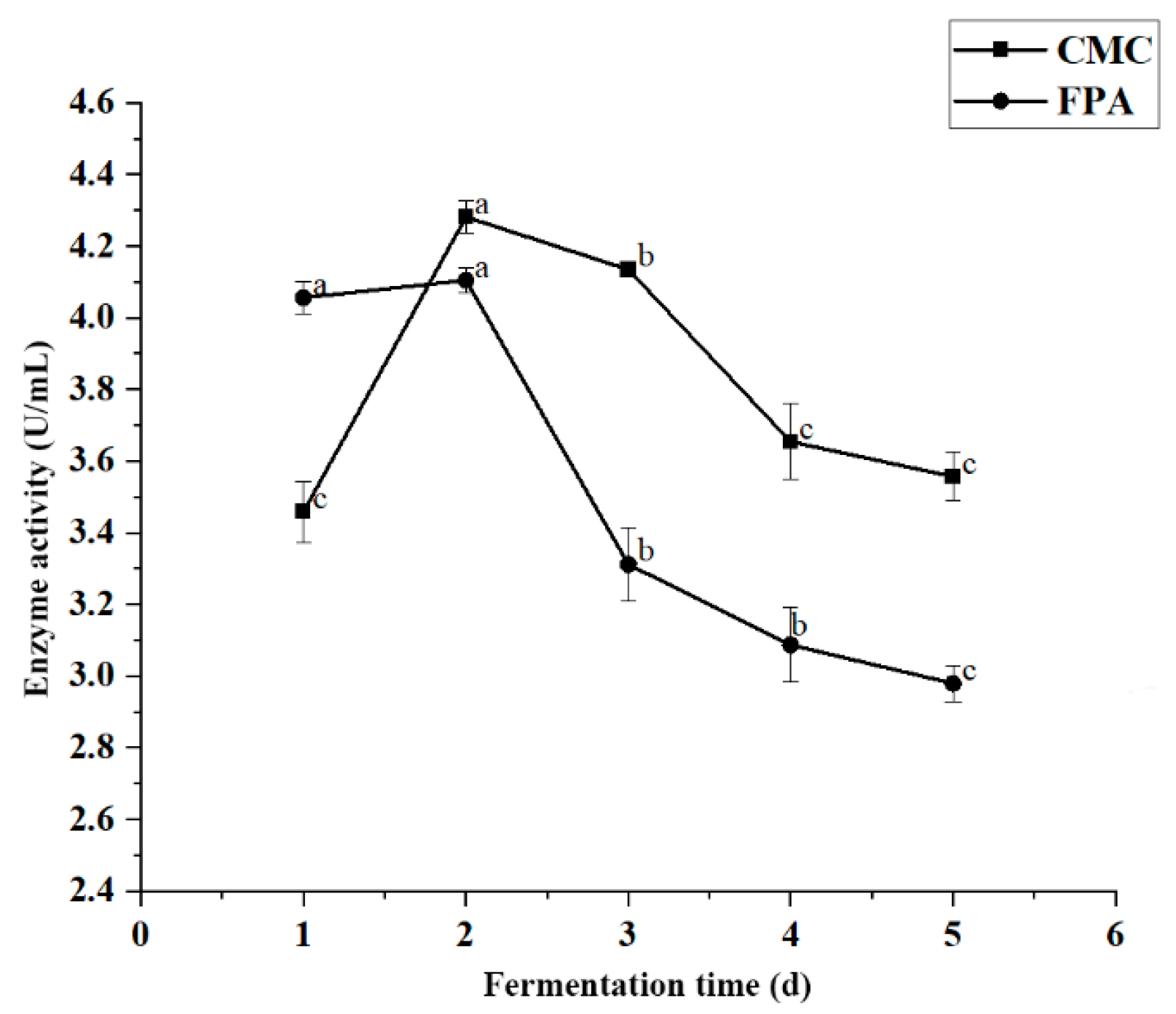
Fermentation time significantly affected enzyme activity (p < 0.05). The variation curve of cellulase activity from JZ2 at different fermentation times is shown in Figure 4. Our results showed that enzyme productivity first increased and then decreased, reaching its maximum productivity after 2 d of incubation. The maximum enzyme activity values of CMC and FPA for strain JZ2 were 4.282 U/mL and 4.105 U/mL, respectively. After 2 d, enzyme activity began to decrease. The increase in enzyme activity corresponds to the enzyme production stage of the strain. The maximum amount of cellulase was produced by continuously inducing cellulase with the substrate and the extension of fermentation time. The decrease in enzyme activity may be due to the production of other byproducts in the fermentation medium and the depletion of nutrients. These conditions cause bacterial stress, resulting in the inactivation of enzyme secretion [26,31].
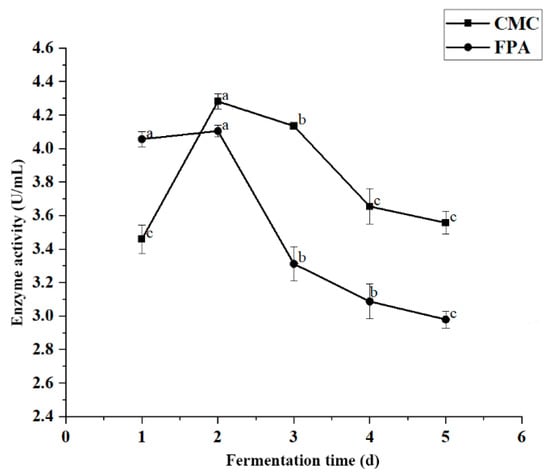
Figure 4.
Effect of fermentation time on enzyme production conditions for JZ2. Different letters (a–c) on same curve in the figure indicate significant differences (p < 0.05) for enzyme activity.
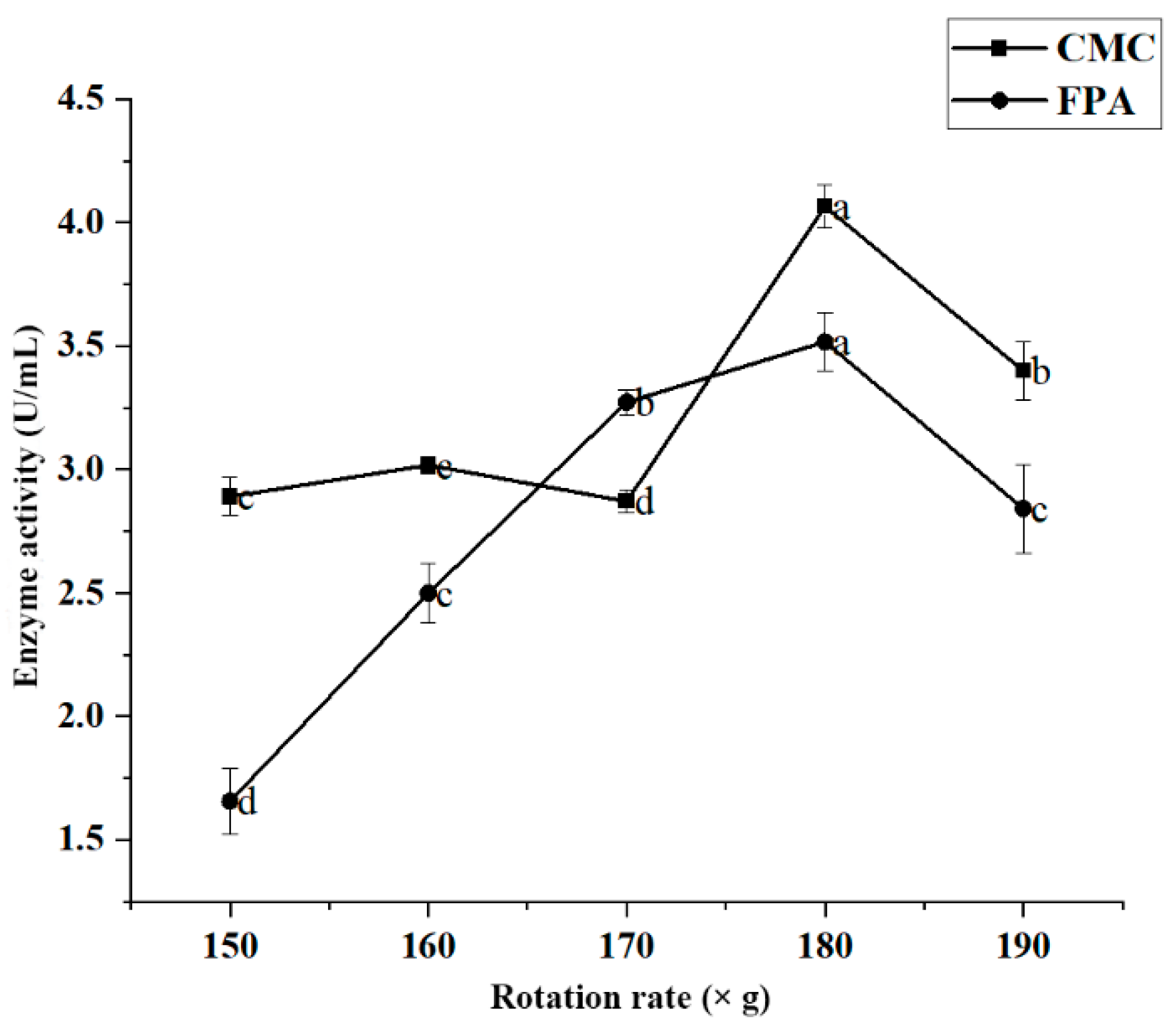
Effect of Rotation Rate on Enzyme Activity
Figure 5 shows the effect of rotation rate on enzyme activity in strain JZ2. The results showed that the enzyme activity of JZ2 increased with the increased rotation rate (150–180× g). When the rotation rate was 180× g, the CMC and FPA enzyme activity values reached 4.066 U/mL and 3.518 U/mL, respectively. The enzyme activity gradually decreased when the rotation rate was <180× g. According to previous studies, the rotation rate is important because the dissolved oxygen levels it creates in the culture broth affect the cell growth of cellulase-producing microorganisms [32]. However, higher rotation rates also inhibit enzyme activity [26,33]. This inhibition effect may be due to the impact of high shear forces on JZ2, leading to a decrease in enzyme production capacity.
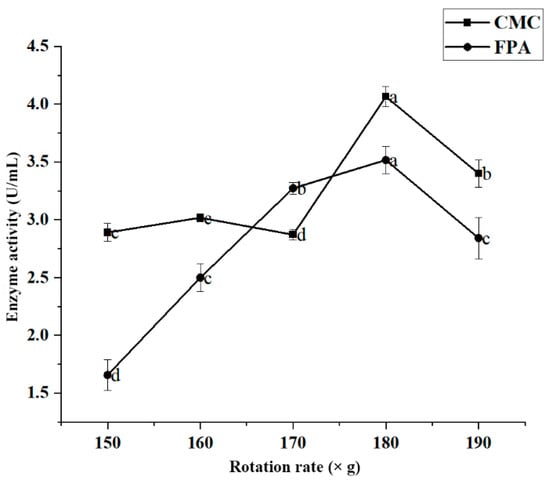
Figure 5.
Effect of rotation rate on the enzyme production conditions of JZ2. Different letters (a–d) on same curve in the figure indicate significant differences (p < 0.05) for enzyme activity.
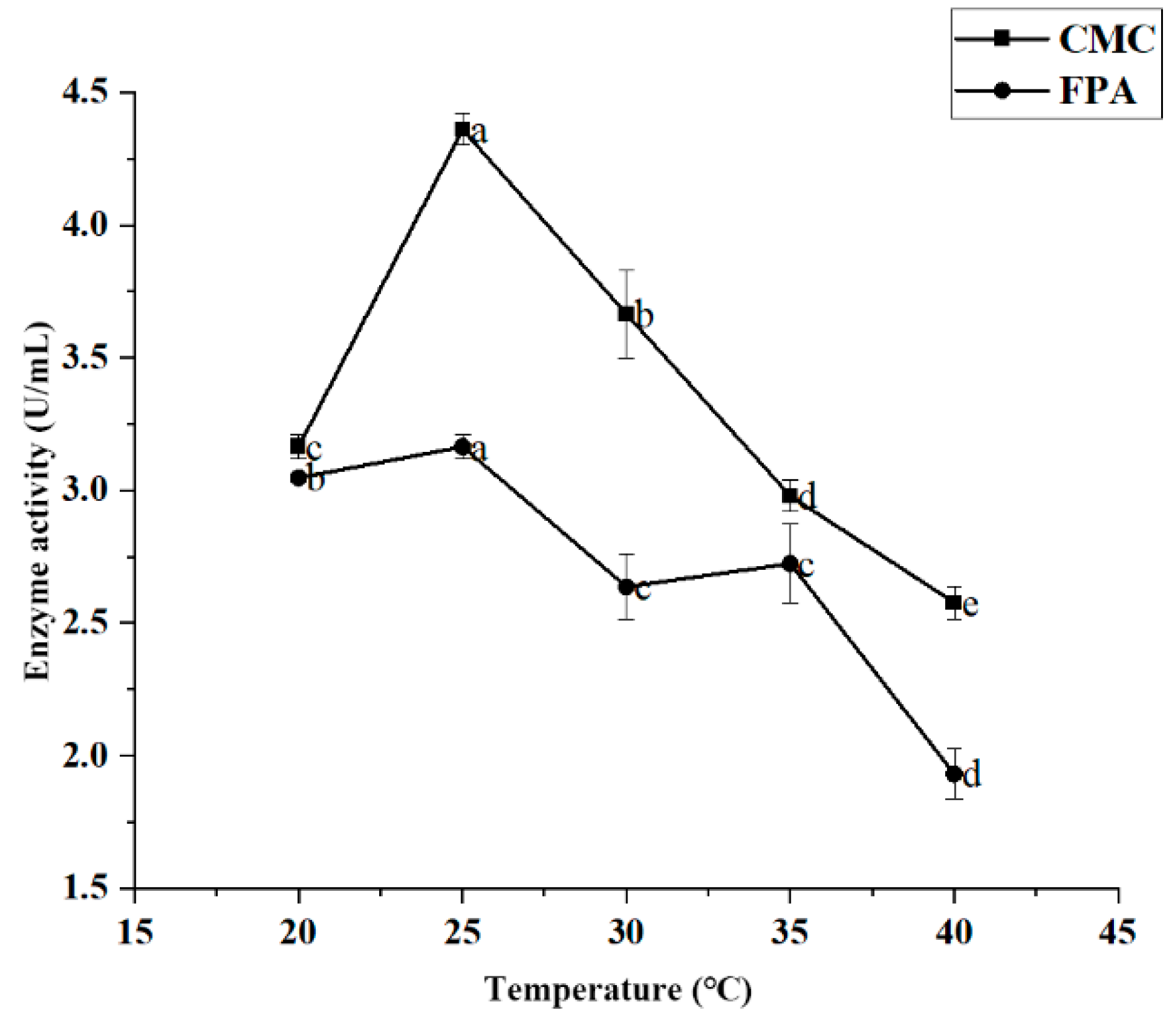
Effect of Incubation Temperature on Enzyme Activity
Figure 6 shows the variation curve of cellulase activity at different temperatures. For JZ2, maximum enzyme activity was achieved after 20 °C of incubation and decreased after 25 °C. The CMC and FPA enzyme activity values of strain JZ2 reached a maximum of 4.360 U/mL and 3.165 U/mL, respectively. High temperatures may affect the energy exchange between the strain and the external substance, potentially affecting enzyme production.
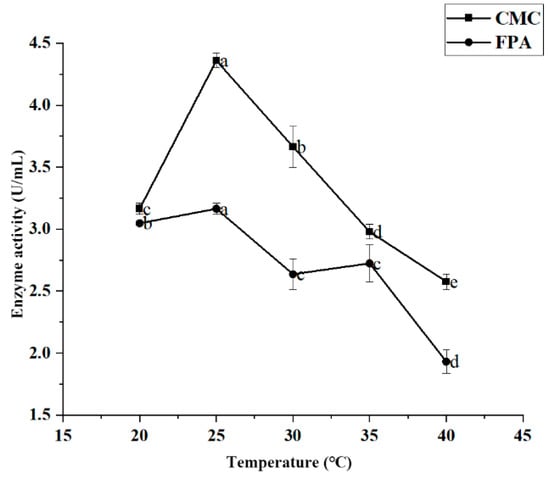
Figure 6.
Effect of temperature on JZ2 enzyme production conditions. Different letters (a–e) on same curve in the figure indicate significant differences (p < 0.05) for enzyme activity.
3.3.2. Response Surface Optimization Test Results
Design Expert 8.0 was used to analyze the experimental data. Table 5 shows the results of optimization. We obtained the quadratic regression equation for independent variables and CMC enzyme activity: Y = −265.9339 + 2.0392 A + 2.8952 B + 0.5815 C + 0.00025 AB − 0.03235 AC + 0.007345 BC − 0.3357 A2 − 0.008552 B2 − 0.03586 C2.

Table 5.
JZ2 response surface experimental design and results.
Table 6 shows the analysis of variance. The p-value of the regression model was 0.0013 < 0.01, indicating that the regression equation was statistically significant. The real relationship between the three factors and enzyme activity of the response value was indicated by the misfit term of the p-value, which was 0.1056 > 0.05. R2 = 0.9447 indicated a high fit between the equation and the experimental results [34]. The regression model was proven to be effective. The coefficient of variation (CV = 7.66%) indicated good experimental repeatability and the equation’s predictive ability for the experiment. Influencing factors on the enzyme activity of strain JZ2 were temperature (C) > fermentation time (A) > rotation rate (B). Temperature and rotation rate × temperature (BC) significantly affected CMC enzyme activity at the selected factor levels. Temperature affected bacteria biosynthesis by altering the physical properties of the fermentation broth, such as the rate of dissolved oxygen and degradation and the uptake of certain substrates [35,36]. The CMC cellulase activity results under the influence of temperature showed that the highest enzyme production occurred at 25.7 °C.

Table 6.
JZ2 response surface experiment variance analysis table.
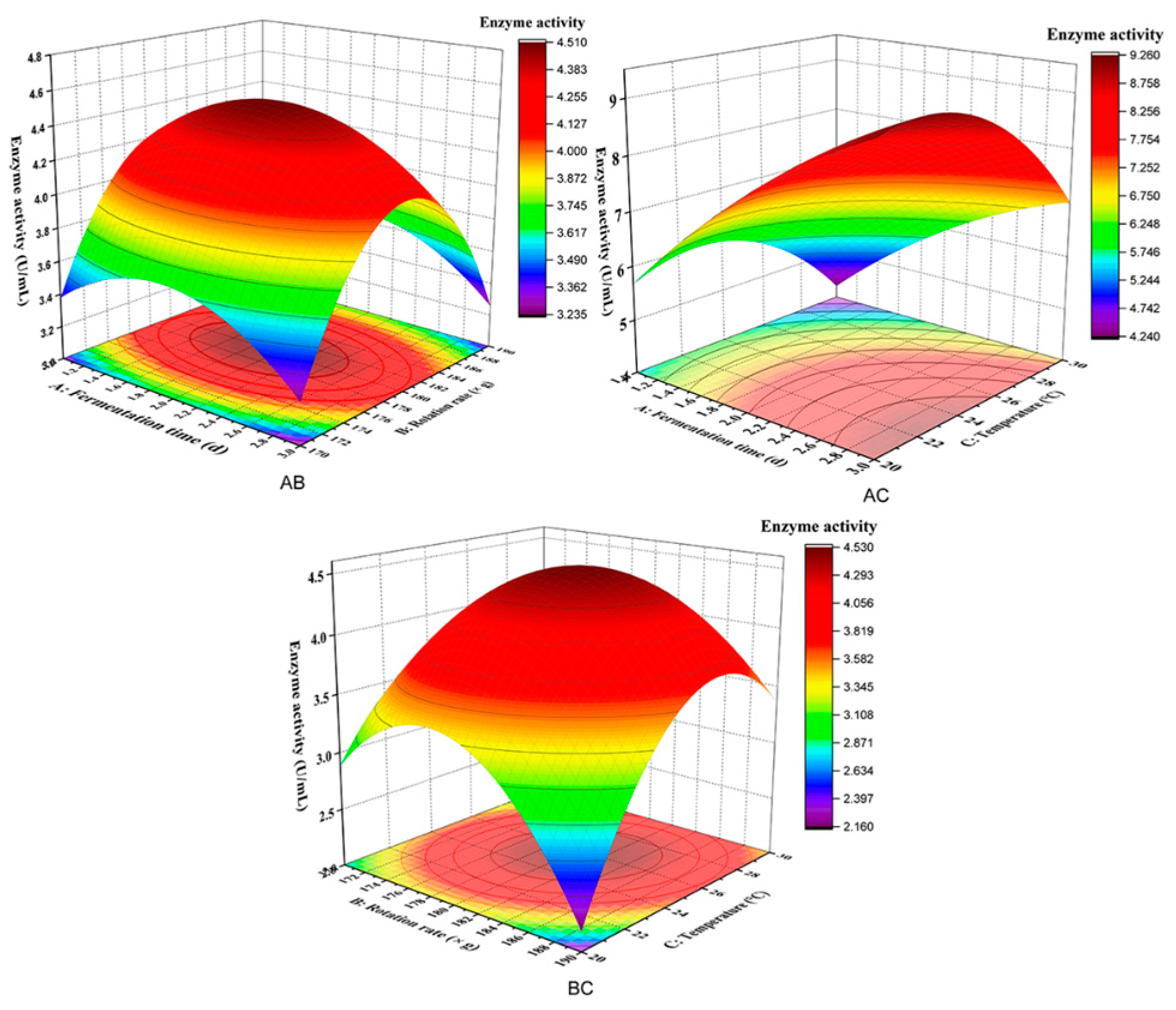
Figure 7 shows the influence of various factors on the interaction of response values. A moderate temperature of 25 °C led to an increase in CMC enzyme activity before decreasing with increasing fermentation time (1~3 days) or rotation rate (170~190× g). Compared with rotation rate, temperature significantly affected CMC enzyme activity. As shown in Figure 7AC, a moderate fermentation time of 2 days led to an increase in CMC enzyme activity before decreasing with the increase in rotation rate (170~190× g) or temperature (20~25 °C). The effect of temperature on CMC enzyme activity was more significant than fermentation time. As shown in Figure 7BC, when the rotation rate was maintained at a moderate level (180× g), the changes in CMC enzyme activity increased with increasing fermentation time (1~3 days) or temperature (20~25 °C). CMC enzyme activity was significantly affected by the interaction between rotation rate and temperature. The effect of temperature on CMC enzyme activity was more significant than that of the rotation rate.
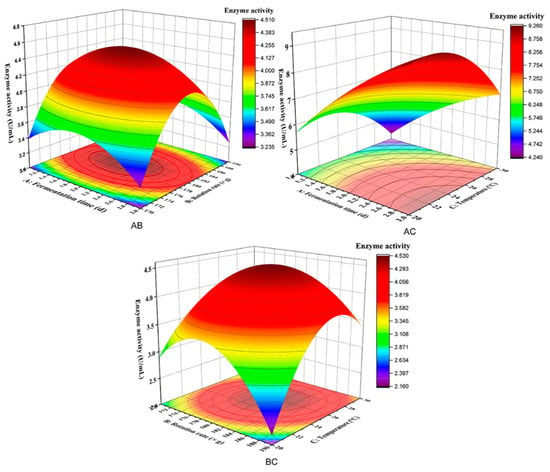
Figure 7.
Influence of various factors on the interaction of response values. AB indicates an interaction of fermentation time and rotation rate; AC indicates an interaction of fermentation time and temperature; BC indicates an interaction of rotation rate and temperature.
3.3.3. Validation Test
Optimizing the process conditions with the response surface methodology revealed the optimum production conditions. Our results showed that the fermentation time was 1.86 min, and the rotation rate and temperature were 180.36× g and 25.74 °C, respectively. We obtained the highest value of CMC enzyme activity at a value of 4.527 U/mL. To verify the model’s reliability, we changed the optimized fermentation enzyme production conditions to reflect the actual situation at a fermentation time of 2 d, a rotation rate of 180× g, and a temperature of 26 °C. Validation tests were conducted under these conditions. The enzyme activity of strain JZ2 was 4.625 U/mL, close to the model’s predicted value. Therefore, the model parameters optimized by the response surface analysis method were reliable and accurately optimized for the enzyme production conditions of the strain. According to the literature, Dong Dan et al. screened out the cellulase-producing bacillus from distillery lees, and its enzyme production activity was 1.2 U/mL [37]. The cellulosic strains screened in this paper had relatively high enzyme production.
4. Conclusions
In this study, a high-yielding cellulase strain, JZ2, was screened from white wine lees. We characterized and identified JZ2 by analyzing its colony characteristics through physiological and biochemical testing and phylogenetic analysis based on 16S rRNA gene sequencing. We found that JZ2 belongs to the genus Bacillus in the thick-walled Bacillus phylum. The colonies were large, and the bacterial cells were rod-shaped. JZ2 exhibited the ability to yield cellulase. The enzyme activities of exoglucanase, endoglucanase, and β-glucosidase were 4.341 ± 0.05 U/mL, 1.874 ± 0.04 U/mL, and 0.739 ± 0.02 U/mL, respectively. The optimal cellulase production conditions for JZ2 were a fermentation time of 2 d, a rotation rate of 180× g, and a temperature of 26 °C. The enzyme activity of JZ2 performed better than reporter strains at 4.625 U/mL in three validation tests under optimal enzyme production conditions. This strain is a cellulose-degrading bacterium with research and development potential, laying a foundation for further research on cellulose degradation in distillery lees.
5. Prospects
Future in-depth studies should be conducted in the following areas:
- (1)
- Further exploration of the fermentation conditions for strain JZ2 and the Saccharomyces cerevisiae complex to improve the strains’ synergistic effect;
- (2)
- Modification of strain JZ2 by mutagenesis or molecular means to improve its degradation ability and enzyme production conditions;
- (3)
- Develop a multi-strain complex, enhance the enzyme system through the synergistic effect of different strains, and improve the treatment of distillery lees.
Author Contributions
Conceptualization, A.-G.L.; methodology, Y.-Y.W.; validation, S.-S.X.; formal analysis, J.Z.; resources, J.-W.H.; data curation, S.-L.S.; writing—original draft preparation, A.-G.L.; writing—review and editing, A.-G.L. and B.-F.H.; funding acquisition, B.-F.H. All authors have read and agreed to the published version of the manuscript.
Funding
Research Project Supported by Shanxi Scholarship Council of China (2020-146); Shanxi Province Scholarship Program for Scientific and Technological Activities of Overseas Students (20210050); Shanxi Provincial Higher Education Solid State Brewing Engineering Technology Research Center (2022P022); Collaborative innovation center for high-value utilization of brewing by-product resources (No. jzxyxtcxzx202104).
Acknowledgments
This work was supported by the Merit-based Funding Project for the Scientific and Technological Activities of Overseas Students in Shanxi province (20210050). This research project was supported by the Shanxi Scholarship Council of China (2020-146).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, Y.F.; Ren, M.J.; Lei, H.X.; Li, H.L.; Zheng, X.G. Research progress on feed comprehensive utilization of distiller’s grains. Agric. Technol. Svcs. 2023, 40, 88–90. [Google Scholar]
- Ma, Y.C.; Hou, Y.X.; Huang, M.Q.; Ye, H.; Zheng, Y.; Wu, J.H.; Sun, B.G. Current status and prospects of research and utilization of Baijiu industry by-products as biomass resources. Food Ferment. Indus. 2022, 48, 292–306. [Google Scholar]
- Torres, M.A.; Valdez, A.L.; Angelicola, M.V.; Raimondo, E.E.; Pajot, H.F.; Nieto-Peñalver, C.G. Vinasse as a substrate for inoculant culture and soil fertigation: Advancing the circular and green economy. Sci. Total Environ. 2023, 887, 164014. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Ding, X.; Chen, X.; Yin, C.; Yang, E.; Sun, D.; Wang, W.; Guo, F. Multiple responses optimization of antioxidative components extracted from distiller’s grains using response surface methodology and identify their chemical compositions. J. Food Process Preserv. 2021, 45, e15885. [Google Scholar] [CrossRef]
- Nie, Y.Z. Reflection on the current situation and innovative application of comprehensive utilization of distiller’s grains. Lt. Indus. Tech. 2021, 37, 11–12. [Google Scholar]
- Chen, Y.; Xiong, K.N.; Chi, Y.K.; Xiao, H.; Chen, H. The development and utilization of Baijiu distiller’s grains as feed and its enlightenment to rocky desertification areas in Guizhou. China Feed. 2021, 31, 105–111. [Google Scholar]
- Fan, E.-D.; Jiang, M.-Y.; Feng, M.-X.; Chen, Y.-F.; Xiao, D.-G.; Guo, X.-W. Optimization of Fermentation Conditions for Production of Feed Containing Functional Components from Distiller’s Grains by Mixed Bacteria Fermentation. Biotechnol. Bull. 2021, 37, 91. [Google Scholar]
- Ao, T.; Ran, Y.; Chen, Y.; Li, R.; Luo, Y.; Liu, X.; Li, D. Effect of viscosity on process stability and microbial community composition during anaerobic mesophilic digestion of Maotai-flavored distiller’s grains. Bioresour. Technol. 2020, 297, 122460. [Google Scholar] [CrossRef]
- Abiodun, A.S.; Oladimeji, F.N.; Peter, O.M.; Raphael, B.B. Environmental management and uses of vinasse-review. Asian J. Curr. Res. 2016, 1, 46–60. [Google Scholar]
- Fan, E.D.; You, X.L.; Zhang, J.; Cheng, P.Y.; Hu, F.; Xu, S. Study on the Comprehensive Utilization of Baijiu Brewing Waste Distiller’s Grains. Brewing Tech. 2022, 42, 99–104. [Google Scholar]
- Taoka, Y.; Nagano, N.; Kai, H.; Hayashi, M. Degradation of distillery lees (Shochu kasu) by cellulase-producing thraustochytrids. J. Oleo Sci. 2017, 66, 31–40. [Google Scholar] [CrossRef][Green Version]
- Lan, X.; Chen, X.; Zhang, J.; Liang, Z.; Gu, Y. Investigation on degradation and reuse of cellulose in distiller’s grains. Anim. Husb. Feed. Sci. Inn. Mong. 2018, 39, 21–22. [Google Scholar]
- Deng, S.G. Screening of Fungi for Degradation of Crude Fiber from Baijiu Distiller’s Grains and Preliminary Study on Degradation Technology. Master’s Thesis, Jiangxi Agricultural University, Nanchang, China, 2018. [Google Scholar]
- Zhang, Y. Screening of Efficient Cellulose Degrading Composite Bacterial Strains and Their Degradation Functions. Master’s Thesis, Tianjin University, Tianjin, China, 2019. [Google Scholar]
- Asgher, M.; Wahab, A.; Bilal, M.; Iqbal, H.M. Delignification of lignocellulose biomasses by alginate–chitosan immobilized laccase produced from Trametes versicolor IBL-04. Waste Biomass Valorization 2018, 9, 2071–2079. [Google Scholar] [CrossRef]
- Khosravi, F.; Khaleghi, M.; Naghavi, H. Screening and identification of cellulose-degrading bacteria from soil and leaves at Kerman province, Iran. Arch. Microbiol. 2021, 204, 88. [Google Scholar] [CrossRef]
- Li, J.; Li, M.Y.; Wang, J.L.; Zhang, T.; Zhou, Q. Research progress in microbial degradation of cellulose. Food Indus. Tech. 2022, 43, 396–403. [Google Scholar]
- Lynd, L.R.; Weimer, P.J.; Van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [PubMed]
- Seidl, V.; Gamauf, C.; Druzhinina, I.S.; Seiboth, B.; Hartl, L.; Kubicek, C.P. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genom. 2008, 9, 327. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, S.; Yang, X.; Ju, M.T. Cellulose isolation from corn stalk treated by alkaline biochars in solvent systems. BioResources 2018, 13, 691–703. [Google Scholar] [CrossRef]
- Ning, L.J. Screening, Fermentation Conditions Optimization, and Application of Cellulose Degrading Bacteria from Distiller’s Grains Unpublished. Master’s Thesis, Hebei University, Baoding, China, 2022. [Google Scholar]
- Yu, M.X.; Zhu, Y.; Cheng, D.; Lu, J. Screening and identification of cellulose degrading bacteria in distiller’s grains sludge. J. Fuyang Norm. Univ. 2021, 38, 58–62. [Google Scholar]
- Behera, S.S.; Ray, R.C. Solid state fermentation for production of microbial cellulases: Recent advances and improvement strategies. Int. J. Biol. Macromol. 2016, 86, 656–669. [Google Scholar] [CrossRef]
- Song, L.L.; Wen, G.; Huo, S.H.; Hu, X.L.; Yang, X.; Zhang, Z.P. Isolation and screening of cellulase producing bacteria from Baijiu distiller’s grains and study on their enzymatic properties. Food Ferment. Indus. 2020, 46, 43–49. [Google Scholar]
- Yaoling, W.; Xiaoli, X.; Xianglian, Z.; Xibin, L.; Heyu, W.; Li, W. Isolation and Compost Application of High-Activity Cellulose-degrading Strains from Distiller’s Grains. J. Henan Agric. Sci. 2022, 51, 79. [Google Scholar]
- Hussain, A.A.; Abdel-Salam, M.S.; Abo-Ghalia, H.H.; Hegazy, W.K.; Hafez, S.S. Optimization and molecular identification of novel cellulose degrading bacteria isolated from Egyptian environment. J. Genet. Eng. Biotechnol. 2017, 15, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Qin, S.R.; He, S.J.; Xu, J.; Yang, J.F.; Huang, X.Q. Screening and identification of high cellulase producing strains from distiller’s grains. Feed Res. 2021, 44, 82–87. [Google Scholar]
- Zhou, L.; Chen, L.; Lu, H.M.; Mo, L.Q.; Xu, S. Isolation and identification of high-temperature bacteria from sauce flavored distiller’s grains in Maotai area. China Brewing. 2016, 35, 61–65. [Google Scholar]
- Abdelhamid, S.A.; El-Shatoury, E.H.; Asker, M.S.; Abd-El-Aal, S.K.; Mohamed, S.S. Hydrolysis of Cellulose Rich Agricultural Waste Using Two Potent Local Bacterial Isolates. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2023, 93, 225–234. [Google Scholar] [CrossRef]
- Li, H.; Zhou, S.; Johnson, T.; Vercruysse, K.; Lizhi, O.; Ranganathan, P.; Phambu, N.; Ropelewski, A.J.; Thannhauser, T.W. Genome Structure of Bacillus cereus tsu1 and Genes Involved in Cellulose Degradation and Poly-3-Hydroxybutyrate Synthesis. Int. J. Polym. Sci. 2017, 2017, 6192924. [Google Scholar] [CrossRef]
- Ariffin, H.; Abdullah, N.; Umi Kalsom, M.; Shirai, Y.; Hassan, M. Production and characterization of cellulase by Bacillus pumilus EB3. Int. J. Eng. Technol. 2006, 3, 47–53. [Google Scholar]
- Jo, K.-I.; Lee, Y.-J.; Kim, B.-K.; Lee, B.-H.; Chung, C.-H.; Nam, S.-W.; Kim, S.-K.; Lee, J.-W. Pilot-scale production of carboxymethylcellulase from rice hull by Bacillus amyloliquefaciens DL-3. Biotechnol. Bioprocess Eng. 2008, 13, 182–188. [Google Scholar] [CrossRef]
- Lejeune, R.; Baron, G. Effect of agitation on growth and enzyme production of Trichoderma reesei in batch fermentation. Appl. Microbiol. Biotechnol. 1995, 43, 249–258. [Google Scholar] [CrossRef]
- Li, H.; Dou, M.; Wang, X.; Guo, N.; Kou, P.; Jiao, J.; Fu, Y. Optimization of cellulase production by a novel endophytic fungus Penicillium oxalicum R4 isolated from Taxus cuspidata. Sustainability 2021, 13, 6006. [Google Scholar] [CrossRef]
- Dhakal, R.; Bajpai, V.K.; Baek, K.-H. Production of GABA (γ-aminobutyric acid) by microorganisms: A review. Braz. J. Microbiol. 2012, 43, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Nema, S.; Teagarden, D. Protein aggregation—Pathways and influencing factors. Int. J. Pharm. 2010, 390, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Che, Z.M.; Guan, T.W. Screening of cellulase producing strains from Baijiu distiller’s grains and detection of their enzyme activity. China Brewing. 2015, 34, 44–48. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).