3.1. Design of the Oligonucleotides
Spherical GNPs 13 ± 2 nm in diameter were used, as were model oligodeoxyribonucleotides that form the simplest duplex (i.e., the initial blunt-ended duplex designated as F–R) and duplexes carrying a 3′ single-stranded overhang of various nucleotide compositions and lengths (designated as FLn–R and FTn–R) (
Figure 1). FLn oligonucleotides (n = 1, 2, and 4) contained different numbers of an L motif (TTGTTG), which possesses enhanced affinity for the surface of GNPs [
23]. Oligonucleotides from the FTn series served as controls, in which, instead of the L motif, we used hexatimidylate; it has weaker affinity for the surface of GNPs [
23]. Single-stranded L motifs, as explained above, should play the role of DNA regions that predetermine the enhanced affinity of DNA molecules for GNPs, that is, they should act as “anchor” regions.
We studied in detail effects of the length of the L and T motifs on the affinity of the respective ssDNAs and dsDNAs for the surface of GNPs and on the capacity of GNPs for these DNAs (i.e., the highest possible number of bound DNA molecules per GNP). By the affinity of DNA for GNPs, we mean the energy of interaction of nucleotide units and of their motifs with the surface of GNPs. In this context, the multipoint nature of the interaction of DNA strands with GNPs should be taken into account. This phenomenon implies that the formation of several (many) contacts with a GNP by one DNA molecule reduces the area of the GNP available for adsorption of other identical molecules of DNA. Consequently, the capacity of GNPs for a DNA having stronger affinity for GNPs is less than that for a DNA having weaker affinity. It is the details addressed by this study (on the basis of principles of material balance) that allow us to elucidate the mechanism of (and problems with) the formation of noncovalent associates of DNA with GNPs (DNA/GNP). In our study, the 3′-terminal single-stranded motifs 6, 12, and 24 nucleotides long in the oligonucleotides of the L and T series underlie the comparative analysis of the formation of noncovalent associates of ssDNA or dsDNA with GNPs. It is important to emphasize again that (1) all formed duplexes bearing an overhang have a 3′-terminal single-stranded motif, and (2) for the assembly of all duplexes, the same oligonucleotide R was used, which is completely complementary to the F segment within oligonucleotides of any series (
Figure 1). At the same time, sequences F and R were chosen so as to minimize the likelihood of both intra- and intermolecular interactions [
38]. The last requirement was imposed because of evidence that the formation of secondary structures of DNA, that is, hairpins or duplexes, lowers the efficiency of the formation of DNA/GNP associates [
23,
39].
We deliberately employed 5′-
32P-labeled (radioactive) strands of ssDNA to exclude the effect of fluorescent labels on the interaction of DNAs with GNPs; this phenomenon has been documented by us earlier [
14]. For dsDNA, the
32P label was introduced into either one or the other strand (consecutively) to quantify the interaction of each strand with GNPs.
3.2. ssDNAs and GNPs
In the first stage of the study, we researched in detail the adsorption of a set of ssDNAs on GNPs (concentrations of GNPs: 0.5 nM) under standardized conditions (
Figure 2,
Table 2) and carried out a theoretical calculation (affinity-rating analysis) of relative affinity of all tested ssDNAs for GNPs, similarly to [
23]. The standardized conditions (24 h incubation at 25 °C) have previously been shown to be sufficient to achieve thermodynamic equilibrium in a heterophasic interaction of ssDNA with GNPs [
40]).
The base oligodeoxynucleotides (hereafter “initial oligodeoxynucleotides”) F and R have similar characteristics regarding their affinity for GNPs (
Figure 2). Langmuir constants (K
L) and surface capacity of GNPs are almost identical between these ssDNAs, which is important to consider below.
As expected, the introduction of one high-affinity hexameric L motif at the 3′ end of the F oligonucleotide led to a significant increase in the efficiency of adsorption of FL on GNPs. If we compare F with its extended version, FL, the surface capacity of GNPs increased from 36 to 56 molecules of DNA per nanoparticle; this increase is inversely proportional to a decrease in equilibrium interaction constant K
L (
Table 2), which characterizes the stability of ssDNA/GNP associates. Making the anchor L motif longer—from hexa- to dodeca- and tetracosenucleotides—caused a significant decrease in the capacity of GNPs for oligonucleotides FL2 and FL4 (capacity for FL2: 38 molecules per GNP, and for FL4: 17). The efficiency of binding of ssDNA to GNPs changed approximately by the same factor (2.2), judging by the K
L value. Therefore, one of possible determinants of this imbalance is an increase in electrostatic repulsion between elongated DNAs adsorbed on the surface of metal nanoparticles. We have reported a high probability of this type of phenomenon earlier, in a detailed analysis of an interaction of charged molecules with a metal nanoparticle [
41]. The second possible determinant is elevated likelihood of long oligonucleotides forming secondary structures (imperfect hairpins or intermolecular complexes, their stability was assessed by means of [
42]) that possess weaker affinity for GNPs [
39] (
Figures S1 and S2, Table S3). There is no doubt that, in this case, ssDNA somewhat loses its ability to bind to GNPs because it is the implementation of a network of direct ssDNA/GNP contacts that primarily determines the stability of the nanoassociates in question [
4,
23].
We could not resist the temptation to evaluate the influence of the nucleotide sequence of the “anchor” motif in ssDNA on the efficiency of its adsorption on the surface of GNPs. The presence of L motifs (TTGTTG sequence) within ssDNAs (FLn series) should significantly enhance the interaction of the oligonucleotide with GNPs [
23]. On the other hand, the use of a longer ssDNA can raise the probability of formation of its secondary structures that have lower affinity for GNPs and can increase electrostatic repulsion of DNA molecules between themselves and from the surface of citrate GNPs [
39,
43]. That is why we compared these data with the adsorption of the control set of ssDNAs: the FTn series. The homogenous composition of the single-stranded overhang in this series ensures that nitrogenous bases are less likely to take part in intra- or intermolecular NA–NA interactions while preserving the overall charge of the DNA molecules. One can see that, surprisingly, under these conditions, GNP affinity values of oligonucleotides of the same length are comparable between the two series (
Figure 2A,
Table 2). Only in the case of the dodecanucleotide extension (
n = 2, FL2) is the oligonucleotide capable of 35% stronger binding to the surface of GNPs as compared to its control counterpart (FT2).
Next, we assessed how the presence of the “anchor” hexanucleotide groups and their extension affect the outcome of loading of the surface of GNPs by the “main” segments (
Figure 1) of FLn (or FTn) in the noncovalent ssDNA/GNP associate (compare the first and second rows in
Table 2). Readers can see that the introduction of a single hexanucleotide L motif raises the capacity of GNPs for FL by more than 1.5-fold as compared to the initial oligonucleotide (F). Further extension proved to be fruitless: if we compare L1 with the L2 motif and then with the L4 motif under the standard conditions of assembly of nanoassociates (25 °C), there is no expected outcome. For instance, the introduction of the fourfold L motif (compare FL and FL4) causes a threefold decrease in the observed capacity of GNPs for ssDNA (from 56 ± 2 to 17 ± 2 molecules), while in terms of “anchor” nucleotides, there is an increase in capacity of only 1.2-fold. These data, overall, are consistent with experimental results [
44] concerning the relation between the extension of an oligonucleotide by means of high-affinity motifs (via introduction of homoadenylate strands) and surface capacity of GNPs for the oligonucleotide. At the same time, it should be noted that our use of the L motif as a gold-binding “anchor” yields greater affinity of these types of extended oligonucleotides for GNPs as compared to oligo(dA)-containing analogs [
23,
45,
46,
47]. In addition, the use of the L anchor is comparable—in terms of achieved efficiency of adsorption of DNA containing it on GNPs—with fluorescein-containing DNA. An increase in the efficiency of adsorption of fluorescently labeled DNAs on GNPs by severalfold as compared to native DNAs has been demonstrated by us elsewhere [
14].
3.3. Effectiveness of ssDNA Adsorption on GNPs Depends on Incubation Temperature
All the experiments described above were conducted under standardized conditions, that is, the adsorption of oligonucleotides on GNPs was studied at 25 °C and with a low-ionic-strength solution (4 mM sodium citrate, pH 5.8). The temperature at which the interaction is carried out is one of the decisive factors determining the effectiveness of DNA binding to GNPs. We have previously found that the noncovalent interaction of ssDNA with GNPs proceeds faster at elevated temperatures [
4]. Considering this and the fact that the extension of an ssDNA strand raises the probability of formation of its complicated structures, we next investigated in detail the influence of the incubation temperature of the reaction mixture (25, 37, or 45 °C) on the efficiency of formation of noncovalent ssDNA–GNP associates. One can see that the sequence of ssDNA significantly affects the nature of the dependence of its affinity for GNPs on temperature (
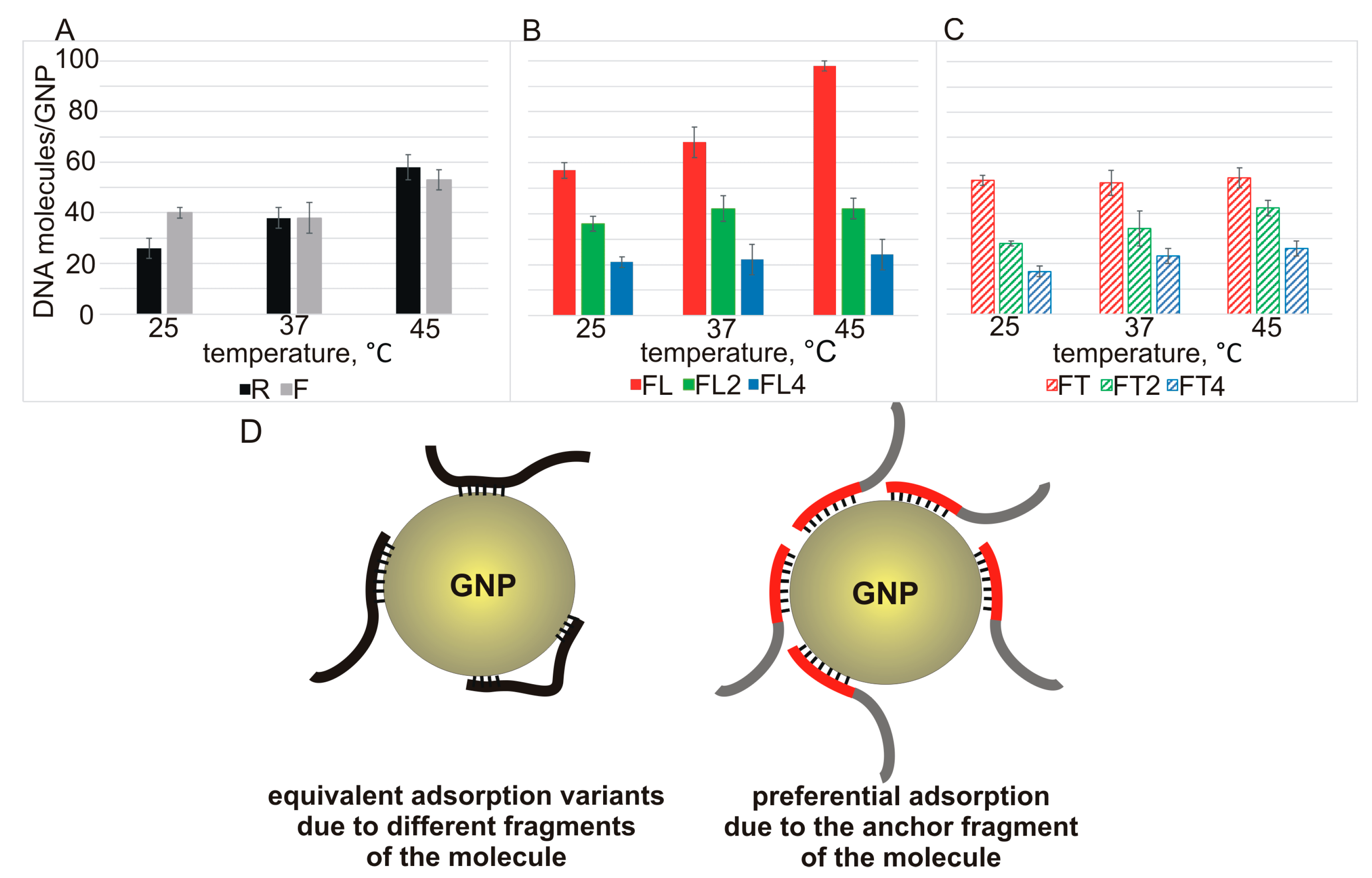
Figure 3A–C).
In the case of binding of 20-mer strands R and F with GNPs, there is a tendency for adsorption capacity to increase with temperature, as expected for ssDNA having a heterosequence that is not prone to the formation of secondary structures (
Figure 3A).
A significant temperature-dependent increase in the capacity of GNPs for ssDNA of the FLn series was observed only for FL (length: 26 nucleotides) (
Figure 3B, red solid bars). In the other cases (FLn at
n > 1), we did not notice an increase in the capacity with increasing temperature of the mixture incubation (
Figure 3B,C). This finding validates the notion of a balance of opposing factors: formation of intra- and/or intermolecular structures versus the enhanced anchoring ability of the L region.
Figure 3.
The dependence of the capacity of GNPs for ssDNAs R and F (A) or ssDNAs of the L series (B) or of the T series (C) on the temperature of incubation of the ssDNA with GNPs. Modes of adsorption (on GNPs) of the ssDNA oligonucleotides that do not contain (left) or do contain (right) an “anchor” motif (D).
Figure 3.
The dependence of the capacity of GNPs for ssDNAs R and F (A) or ssDNAs of the L series (B) or of the T series (C) on the temperature of incubation of the ssDNA with GNPs. Modes of adsorption (on GNPs) of the ssDNA oligonucleotides that do not contain (left) or do contain (right) an “anchor” motif (D).
Our data are suggestive of two possible modes of adsorption of ssDNA on GNPs. For instance, the initial oligonucleotides R and F, which have similar values of affinity for GNPs, and some FTn oligonucleotides (FT2 and FT4) at 25 °C probably form a network of equivalent contacts between their sequence regions and GNPs, without an obvious adsorption advantage for any of these regions. Raising the incubation temperature increases the degrees of freedom of ssDNA, thereby facilitating the interaction of ssDNA with the nanoparticles. This effect enables a greater number of ssDNA molecules to bind to the GNP surface (
Figure 3D, left). In the presence of the “anchor” L motif in FL, we observed a pronounced enlargement of the binding capacity with increasing temperature, and the maximum capacity of 98 molecules per GNP was reached at 45 °C (
Figure 3D, right).
Oligonucleotides FL2 and FL4 (they have an extended “anchor”) each occupy a greater contact area on the surface of a GNP and at the same time have an elevated propensity for intra-and/or intermolecular structures. These circumstances (taking into account greater electrostatic repulsion with the extension of ssDNA strands) do not allow for a significant increase in the efficiency of this interaction with increasing temperature. Nevertheless, the very presence of gold-binding sequences in FLn (at
n > 1) during the formation of the respective noncovalent associates with GNPs enables the mode of predominantly “anchor” adsorption (
Figure 3D, right), thereby minimizing electrostatic repulsion during the adsorption of such a large number of molecules. For FTn oligonucleotides, there is probably no clear advantage of one mode, as in the case of F and R, as explained below in the affinity-rating analysis section.
3.4. Affinity-Rating Analysis
The methodology of rating (affinity-rating analysis) the gold affinity in different DNA hexamers, allowing us to theoretically calculate the relative affinity of ssDNA for GNPs, has been proposed by us earlier [
23]. By testing this method on the affinity of the present series of ssDNAs for GNPs, we found that the statistically determined (theoretical) affinity of the studied FLn oligonucleotides for GNPs is well consistent with actually observed adsorption efficiency (
Table 3).
Readers can see that the rating of theoretical affinity for GNPs increases in the series F < FL < FL2 < FL4. At the same time, only FL has elevated capacity for GNPs (
Figure 2) relative to the initial oligonucleotide, F, and the elongation of the L motif reduces the efficiency of binding of ssDNA to GNPs.
The rating method makes it possible to explain the role of Ln and Tn “anchors” in the adsorption of ssDNA on GNPs.
FL consists of two regions: F has a rating of 5.0 × 10−5, and the L motif (6 nucleotides long) 12.0 × 10−5, i.e., the affinity of these two segments of FL, when normalized to the number of nucleotides, differs by a factor of 2.4. As a consequence, the interaction of FL with GNPs is most likely mediated by the short 6-nt L motif. This interaction provides much greater occupancy of the surface of GNPs by FL molecules as compared to F. After the extension of the L motif to 12 and 24 nt, its theoretically expected affinity strengthens to 62.0 × 10−5 in FL2 and to 86.0 × 10−5 in FL4. Thus, with the increase in the length of the “anchor,” the affinity difference between the “anchor” and the “main” region of the molecule increases even more, by 12- and 17-fold, respectively. Now, the longer “anchor” is even more likely to be adsorbed on GNPs while occupying a larger proportion of the nanoparticle surface, thus leading to the observed decrease in capacity for FL2 and FL4. Moreover, the probability of formation of secondary structures in the elongated oligonucleotides is higher, which also results in lower capacity of GNPs for FL2 and FL4 as compared to FL.
The hexathymidylate motif tends to lower the rating of the FTn containing it; that is, this motif in ssDNA no longer plays the role of an “anchor” that has paramount importance for the adsorption of ssDNA on GNPs. Its introduction into DNA (and extension) does not result in preferential adsorption of the “anchor” part of DNA on GNPs. In other words, we see different levels of capacity for FT2 and FT4 with increasing temperature owing to the equivalent adsorption of different segments of their molecules, in contrast to FL2 and FL4 (
Table 2). It can be said that the greater capacity of GNPs for FT compared to F is explained by (i) higher probability of a larger number of different binding modes with the extension of the molecule (
Figure 3D), (ii) a slight increase in electrostatic repulsion after minimal elongation, and (iii) the absence of secondary structures. The electrostatic repulsion and its negative contribution to the capacity strengthen with further lengthening of the anchor (in FT2 and FT4).
Therefore, during the interaction of the tested ssDNAs with GNPs, different binding modes are realized:
- (1)
The presence of one short high-affinity sequence of the L motif significantly promotes binding precisely due to the direct contact of this motif with the surface of the metal nanoparticle;
- (2)
The presence of the Tn motif, which does not have pronounced affinity for GNPs, causes predominant or equivalent interaction (with GNPs) of the other (“main”) part of the oligonucleotide: in our case, F;
- (3)
The presence of any long additional motif diminishes the capacity of GNPs for the oligonucleotides, and this phenomenon is most likely explained by (i) the preferential interaction of GNPs with DNA in the single-stranded state (elevated probability of secondary structures) and (ii) a greater role of electrostatic repulsion between negatively charged oligonucleotides adsorbed on the GNP surface.
Striking the right balance between the above factors mainly determines the effectiveness of the ssDNA interaction with GNPs. Apparently, such a balance makes the FL oligonucleotide significantly better than all the other tested ssDNAs (FLn and FTn series and initial oligonucleotide F).
3.6. Adsorption of dsDNA on GNPs
Next, we compared the adsorption (on GNPs) of dsDNAs carrying single-stranded overhangs (L series and T series 6, 12, and 24 nucleotides long) and of the initial blunt-ended duplex (
Figure 1). It should be stressed that we assessed adsorption parameters of dsDNA by means of the balance of all the interacting ssDNAs binding to the nanoparticle (
Figure 4,
Table 4). For this purpose, a radioactive 5′
32P label was introduced into one or the other DNA strand. One can see that even at high concentrations of dsDNA (all duplexes were stable), we did not observe well-pronounced and equivalent efficiency levels of adsorption of both complementary DNA strands for any of the tested duplexes (
Figure 4). A question arises: are such complementary ssDNAs adsorbed on the surface of GNPs in the form of a duplex?
The saturation curves of dsDNAs F*–R and FLn*–R, which contain radiolabeled F strands, have a local maximum in the range of low concentrations of ssDNAs (20–40 nM,
Figure 4A–D), when the presence of a substantial amount of the ssDNA form in the reaction mixture should still be expected. Obviously, an increase in the concentration of the interacting strands to 100 nM should considerably reduce the probability of “complementarity-mismatched” interaction of individual DNA strands with GNPs. In this context, it is important to take into consideration that the strands released from dsDNA, which have a propensity for intra- and/or intermolecular interactions, can also lose the ability to independently bind to GNPs [
15]. In the extreme case, an increase in the concentration of dsDNA and the consequent increase in Tm values (
Table S2) determine the convergence of capacity values when we examine saturation curves of complementary ssDNAs (with the exception of FL–R) (
Figure 4A,C,D). It is for this reason that values of saturation capacity of the GNP surface for components F and FL2 relative to component R become significantly closer (
Figure 4A,D), that is, under the conditions that are most conducive to the formation of dsDNA.
Let us examine the same duplexes when the R strand is radiolabeled (
Figure 4A–D). One can see that radiolabeled 20-mer R* in a stoichiometric mixture with any DNA of the L series adsorbs worse at most concentrations. Only ~14 molecules of R* per GNP when mixed with F, FL, or FL2, and only six molecules of R* when mixed with long FL4, bind to a GNP, whereas 20 to almost 40 molecules of the L series of oligonucleotides were found cover the GNP. The use of the long L4 overhang (having high Au affinity, occupying greater specific area on the GNP surface, and creating the strongest electrostatic repulsion between adsorbed DNA strands within the associates with GNPs) further complicates the adsorption of R. As already mentioned, an important experimental finding here is the difference in saturation capacity of GNPs between FLn*–R and FLn–R* (at
n = 1, 2, and 4).
The capacity of GNPs for the labeled R* strand in mixtures with each FTn oligonucleotide was ~13 molecules per GNP. This confirms that adsorption efficiency of even the longest T-containing 44-mer FT4 does not give it an advantage in affinity for GNPs when FT4 competes with R. This is an important result, especially in contrast to FL4, which carries the long, gold-binding L4 motif (
Table 4) [
4,
23,
32,
37].
In the comparison of DNA duplexes radiolabeled on different strands (FLn*, FTn*, or R*), it becomes obvious that, overall, T motifs are inferior to L motifs in the efficiency of formation of DNA/GNP nanoassociates. This observation vividly reflects the difference in features (affinity) of individual parts of the oligonucleotides and is consistent with the presented data on ssDNAs in
Figure 3B,C. Thus, the observed differences between complementary ssDNAs of a duplex in the adsorption correlate with the affinities of individual ssDNAs for GNPs. Probably, gold affinity levels of complementary oligonucleotides must be very similar to ensure their adsorption on GNPs in the form of a DNA duplex. This is evident in the F–R duplex, where the capacity on the F* side is 20 ± 2 molecules and, on the R* side, it is 16 ± 1 molecules (
Table 4).
3.7. The Dependence of Adsorption Efficiency of a Mixture of ssDNAs to GNPs on Incubation Temperature
Next, we investigated the dependence of the capacity of GNPs for dsDNA on the incubation temperature of the reaction mixture (25, 37, and 45 °C). Such data should reveal how the adsorption of a stoichiometric mixture of complementary ssDNAs occurs and is realized on GNPs: individually or as part of the duplex. To distinguish between processes of adsorption of dsDNA and the competition of individual ssDNA strands with each other, we conducted experiments on the adsorption of each ssDNA (FLn or FTn) paired with a noncomplementary oligonucleotide, T20, under the same conditions. Oligothymidylate T20 by itself has GNP affinity similar to that of R (
Figure S3), i.e., T20 should manifest exceptionally competitive behavior when coadsorbed with any of the ssDNAs on GNPs.
Let us analyze the adsorption (on GNPs) of DNAs of the FLn series in a mixture with the R oligonucleotide, which has almost the same affinity for GNPs as the F oligonucleotide does. For duplexes F*–R and FL*–R, two significantly different binding modes are seen at 25 and 45 °C (gray and red solid bars in
Figure 5A). Duplexes FL2*–R and FL4*–R manifested the same binding mode at all temperatures (green and blue solid bars in
Figure 5A). Consequently, the adsorption of dsDNA via a long high-affinity albeit structured “anchor” is more favorable at all temperatures as compared to all other adsorption modes. R* paired with any DNA of the L series showed a tendency for increased capacity with increasing temperature (
Figure 5C). By contrast, the capacity for T20* in the presence of DNA of the L series either diminished or remained at the same level with increasing temperature (
Figure 5E). Namely, under competitive conditions, lower-affinity T20 is inferior to higher-affinity DNAs of the L series. Because T20 is a homosequence, preferential adsorption of any of its regions can be ruled out, and we observed the same magnitude of adsorption at all temperatures. Capacity values of mixtures with T20* did not exceed those for R* under the corresponding conditions (
Figure 5C,E). This finding can be explained by partial formation of a duplex between R and DNA of the L series, with this process bringing a greater number of R molecules to the surface of GNPs.
For oligonucleotide pairs (FTn* and R), similar capacity values were observed regardless of incubation temperature (
Figure 5B, e.g., 21 molecules of FT2*–R per GNP at 25, 37, and 45 °C: green striped bars), in agreement with previously published data [
4]. Capacity values of R* in mixtures with DNA of the FT series increased with increasing temperature (
Figure 5D), i.e., partial duplex formation takes place, as in the case of FLn–R* (
Figure 5E). Note that FT4–R* is adsorbed almost two times more (
Figure 5D) as compared to mixture FL4–R* (
Figure 5C) at both 25 and 45 °C. This result reflects more favorable and/or faster adsorption of the shorter and higher-affinity R compared to FT4 (in contrast to the ratio of gold affinity levels in the FL4–R pair) [
43]. The substitution of FLn with FTn resulted in a significant increase in the adsorption of T20*, thereby reflecting a difference in the affinity between the L and T motifs, which allows more T20 molecules to be adsorbed during competition with lower-affinity FTn (
Figure 5E,F).
At the same time, we should highlight the absence of a temperature dependence in the adsorption of T20* in pairs with FTn (
Figure 5F) or with FLn (
Figure 5F), in contrast to the behavior of R*, whose capacity increases with temperature, thus confirming that the adsorption of complementary DNAs is not exclusively competitive (
Figure 5C,D).
Therefore, it can be concluded that when interacting with GNPs, ssDNAs complementary to each other compete with each other, while some of them form a duplex, which is also adsorbed on GNPs.
Figure 5.
The dependence of the capacity of GNPs for a mixture of DNAs on the temperature of incubation of the DNAs with GNPs. (A) DNAs of the L series in a mixture with complementary R, (B) DNAs of the T series in a mixture with complementary R, (C) complementary R mixed with DNA of the L series, (D) complementary R mixed with DNA of the T series, (E) competitive T20 mixed with DNA of the L series, and (F) competitive T20 mixed with DNA of the T series. * denotes radioactively labeled DNA.
Figure 5.
The dependence of the capacity of GNPs for a mixture of DNAs on the temperature of incubation of the DNAs with GNPs. (A) DNAs of the L series in a mixture with complementary R, (B) DNAs of the T series in a mixture with complementary R, (C) complementary R mixed with DNA of the L series, (D) complementary R mixed with DNA of the T series, (E) competitive T20 mixed with DNA of the L series, and (F) competitive T20 mixed with DNA of the T series. * denotes radioactively labeled DNA.
3.8. Evaluation of the Mobility of DNA–GNP Associates by Agarose Gel Electrophoresis
Determination of surface capacity of GNPs for DNA characterizes the average occupancy of the nanoparticles by oligonucleotides in a given sample. The gel electrophoresis method enables researchers (i) to separate DNA/GNP associates by their charge and size and (ii) to estimate the proportion of associates having different levels of oligonucleotide occupancy in one sample. It has been previously reported that the mobility of associates of GNPs with unstructured ssDNAs is determined only by the charge of these associates [
4]. In general, the higher the capacity of a GNP for ssDNA, the greater the negative charge on the surface of GNPs (owing to negatively charged internucleotide phosphate groups in DNA) and the higher the electrophoretic mobility of ssDNA/GNP associates and the proportion of maximally mobile GNPs in the sample [
4,
49].
Analyzing scans and autoradiographs of the same agarose gel containing GNPs with radioactively labeled ssDNA, we found that the mobility of the associates indeed directly depends on the number of adsorbed molecules of ssDNA (
Figure 6). On the contrary, images of associates consisting of GNPs and a pair of complementary ssDNAs did not correlate with the surface capacity of GNPs for dsDNA (
Figure 7).
We hypothesized that for associates of GNPs and a pair of complementary ssDNAs, the relation between the capacity and mobility in the gel differs from this relation for associates of GNPs with one ssDNA. Therefore, to analyze the mobility of DNA–/GNP associates, in each lane of the gel image, we identified two parts having the same geometric area (marked with a blue dotted line in
Figure 6: the upper part contained “slow” associates, and the lower one contained “fast” ones). We calculated the proportion “slow” and “fast” associates relative to the total staining intensity of the entire lane for each associate formation temperature (25, 37, and 45 °C).
Let us first examine the electrophoresis of ssDNA/GNP associates using FL/GNP associates as an example (
Figure 7A). At 25 °C and at a concentration of ssDNA from 30 to 50 nM, the capacity in FL–GNP associates increases from 47 to 54 molecules of FL per GNP (
Figure 7A, rows [DNA] and FL*/GNP). These seven FL molecules correspond to an addition of 175 internucleotide phosphate groups (
Figure 7A, row “charge”). In the meantime, the increase in the lower fraction (“fast” associates) in the gel was 14% (
Figure 7A, row “% bottom”). For the 50 nM concentration of FL, while changing the temperature from 25 to 37 and to 45 °C, we observed an increase in FL/GNP capacity by 21 and 23 FL molecules, that is, by 525 and 575 phosphate groups. The proportion of “fast” associates grew to 86% and 93%, respectively. Furthermore, it is visible that the increase in the FL concentration and in incubation temperature (with the increase in capacity and charge up to 2450 phosphate groups) did not raise the highest mobility of the associates under electrophoresis conditions. This outcome is due to the highest occupancy of the binding sites on the surface of GNPs by ssDNA molecules [
4,
14]. Thus, for high-affinity unstructured ssDNA FL, the expected picture was observed: with increasing capacity, the charge increased, and the proportion of “fast” associates grew. FL/GNP associates are compact, their movement is not hindered by the size of the gel pores, and their velocity is determined only by the electric field parameters used. For the second ssDNA, R, which is complementary to FL, a similar pattern was registered (
Figure S4).
The associates obtained by incubation of GNPs with a mixture of FL and R at 25 °C in the concentration range of 30–50 nM ssDNA (
Figure 7) had the same capacity for ssDNA FL (40 molecules on average, row FL*–R/GNP in the figure) and for ssDNA R (11 molecules on average, row FL–R*/GNP in the figure), corresponding to 1134–1303 internucleotide phosphate groups (
Figure 7B, row “charge”). Nonetheless, 72–81% of FL–R/GNP associates were “slow” rather than “fast,” as might be expected from their high negative charge (
Figure 7B). According to the FL–R melting point (
Table S2), the ssDNA should be in a duplex at 25 °C [
48]. Although occupancy levels of GNPs by FL and R molecules are not equal, it is expected that some of the adsorbed DNAs formed a duplex because we see a decrease in the mobility of the FL–R/GNP associate, probably owing to an increase in the size of the associate itself.
Raising the temperature of incubation of GNPs with mixture FL–R up to 37 °C enlarged the capacity of GNPs (on average) by 16 molecules of FL and by 10 molecules of R (an increase of 690 phosphate groups in row “charge” in
Figure 7). At this temperature, the DNA in the incubation solution can be of single- and double-stranded form (
Tables S1 and S2). Consequently, some oligonucleotides could be adsorbed in a single-stranded state, thereby forming compact “fast” associates. Moreover, with increasing temperature, the greater increase in capacity was due to FL. This effect widened the imbalance between the amounts of adsorbed FL and R even more and increased the proportion of associates of GNPs with ssDNA. Overall, for FL–R/GNP associates, these changes elevated the proportion of “fast” associates (from 28% at 30 nM to 56% at 50 nM,
Figure 7B, for 37 °C, row “% bottom”).
At 45 °C, the DNA should be in a single-stranded state (
Tables S1 and S2). Accordingly, we predicted the formation of mostly “fast” associates at all DNA concentrations. Nevertheless, the proportion of “fast” associates was only 36% at a DNA concentration of 30 nM and a large total charge of associates: 1952 phosphate groups (
Figure 7B, for 45 °C, row “Charge”). Considering the high capacity of GNPs for oligonucleotides, especially FL, at 45 °C, FL/GNP associates are most likely formed via DNA adsorption through the “anchor” and a vertical arrangement of the “main” part (F) of the FL oligonucleotide with respect to the GNP surface. These circumstances should facilitate the complementary interaction of adsorbed FL molecules with R molecules adsorbed on another GNP. As a result, highly charged but large and “slow” associates (FL/GNP + R/GNP) can come into being. With a further increase in the concentration of FL and R at 45 °C, the same trends were observed as those at 37 °C: the proportion of “fast” associates increased from 36% to 74% (
Figure 7B, for 45 °C, row “% bottom”).
The presence of large and “slow” associates (i.e., a redistribution of some oligonucleotides from filled GNPs to naked GNPs) was confirmed in experiments involving (i) separation of mixture (FL + R) /GNPs (
Figure 7B) and of a mixture of associates obtained beforehand (FL–GNP + R–GNP) (
Figure S5A) and (ii) mixing of additional citrated GNPs with ssDNA/GNP associates, followed by incubation for 0, 2, and 24 h (
Figure S5A,B).
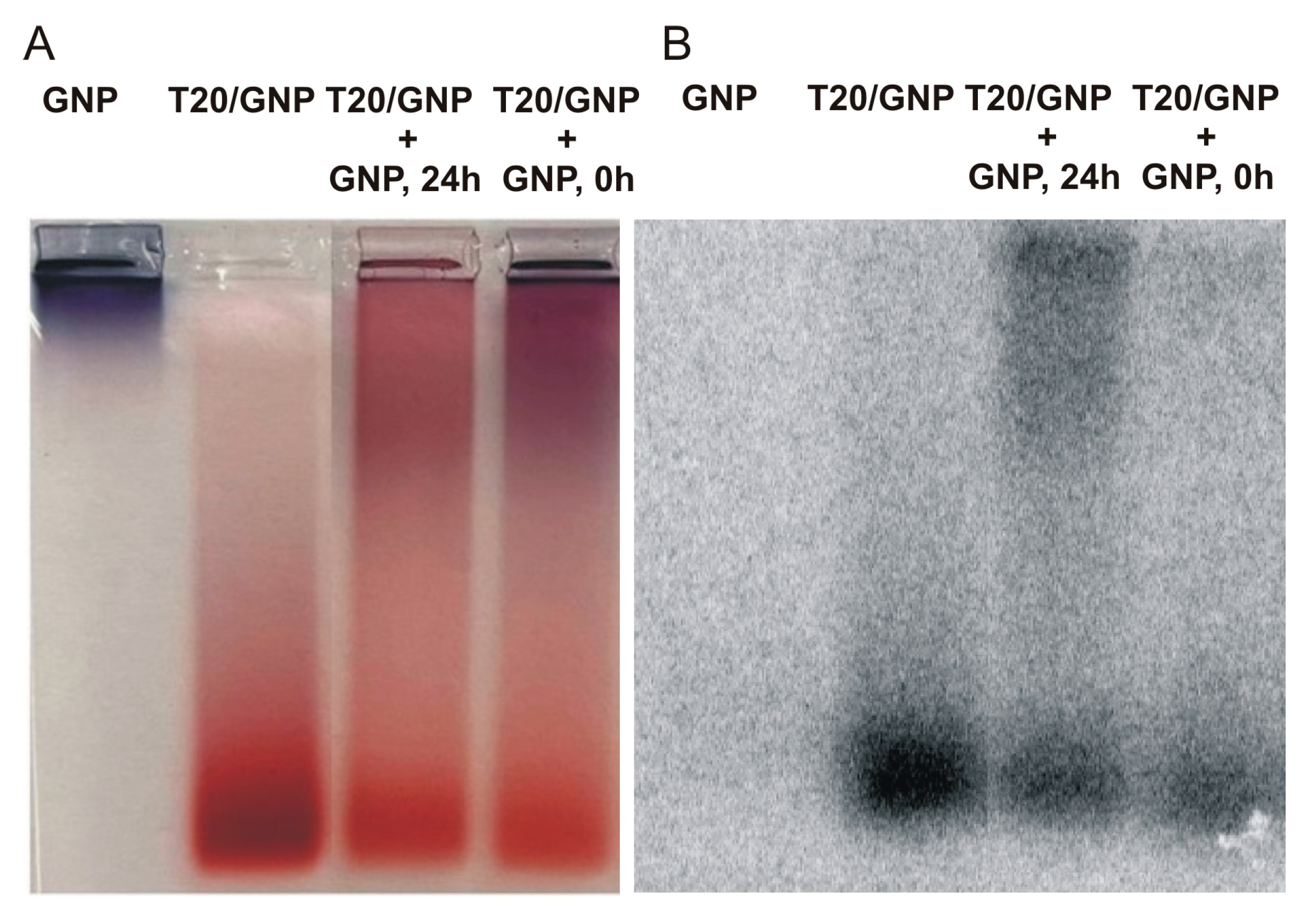
Analysis by means of radiolabeled T20 clearly confirmed the redistribution of some oligonucleotides from loaded GNPs to naked GNPs (
Figure 8).
The initial ratio of the amount of T20 adsorbed on the particles of the upper and lower fractions (according to electrophoretic mobility) was 21% and 79% (
Figure 8A,B, lane T20–GNP). Immediately after the addition of the naked GNPs to T20*/GNPs, this ratio shifted to 29% and 71%, respectively (
Figure 8A,B, lane T20/GNP+GNP, 0 h). After 24 h, the amounts of DNA in the upper and lower particle fractions equalized and amounted to 50% each (
Figure 8A,B, lane T20/GNP+GNP, 24 h). Moreover, immediately after mixing, we noticed desorption of some T20 molecules (22% or 7 out of 32 molecules), and after 24 h, almost no desorption was detectable (6% or 2 out of 32 molecules). In other words, upon addition of naked GNPs, there was probably desorption of some DNA molecules from the surface of the initial associates into the solution. These molecules were then adsorbed onto the surface of the added GNPs. As a consequence, after 24 h, DNA desorption into the solution was not detectable.
Therefore, after the mixing of two types of associates (FL/GNP and R/GNP), desorption of the appreciable proportion of DNA into the solution at 1 day after the mixing indicates the formation of a duplex by these DNAs in solution (
Figure S6).
Thus, the ratios of the capacity and of electrophoretic mobility—of GNP associates with ssDNA or with a mixture of complementary ssDNAs—indirectly confirm possible formation of DNA duplexes upon adsorption on GNPs at all tested concentrations and temperatures. It is known that the method of DNA binding to the surface of GNPs affects the size of the associate and its mobility in an agarose gel [
50]. Indirectly, the existence of such a relation has been demonstrated for associates of GNPs with thiol-containing and native oligonucleotides. The associates of GNPs with thiol-containing ssDNAs have a larger radius as compared to the associates of GNPs with thiol-dsDNA or with thiol-free ssDNA [
49]. As a consequence, associates of GNPs with thiol-bound dsDNAs migrate more slowly in an agarose gel than do associates of GNPs with noncovalently bound dsDNAs [
50].