End-to-End 3D Liver CT Image Synthesis from Vasculature Using a Multi-Task Conditional Generative Adversarial Network
Abstract
1. Introduction
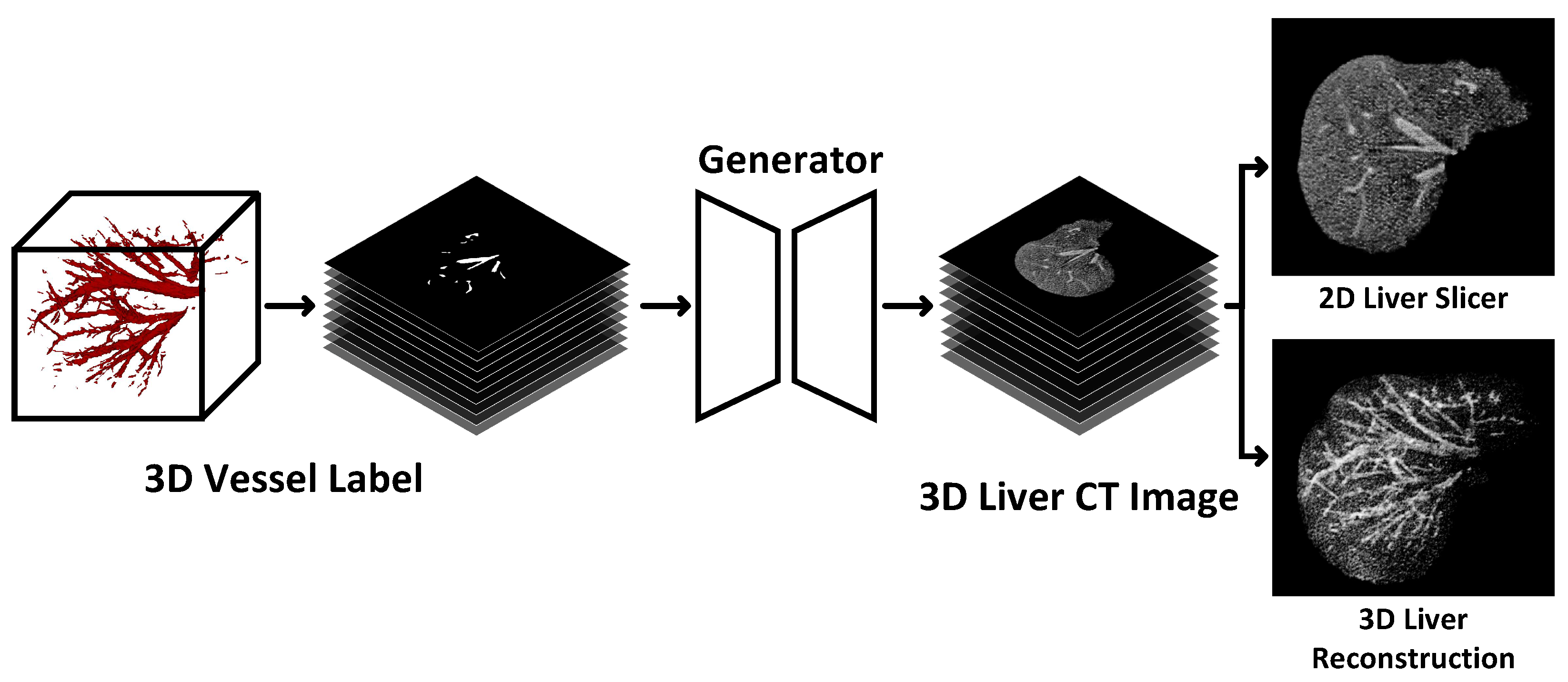
- We propose a multi-task conditional generation adversarial model that synthesizes 3D liver CT images by inputting liver vessel labels to provide a priori anatomical structure information and using real liver segmentation labels for guidance. To the best of our knowledge, this is the first application for 3D liver CT image generation.
- We introduce a robust multi-gradient method to optimize multiple tasks, specifically by balancing the weights of individual tasks in a multi-task framework.
- Extensive experiments were conducted on the collected real data, and the experimental results show that the performance of our method outperforms existing state-of-the-art methods. In addition, this study demonstrates the possibility of using vascular images to synthesize images of the liver.
2. Related Work
2.1. GAN-Based Model
2.2. Medical Image Generation
2.3. Multi-Task Learning
3. Materials and Methods
3.1. Dataset Description and Processing
3.2. 3DMT-GAN
3.2.1. Overview
3.2.2. Generation Task Based on Segmentation Task Guide
3.2.3. Multi-Task Generator Optimized by MGDA-UB
3.2.4. Patch Discriminator
4. Experimental Results
4.1. Implementation Settings
4.2. Evaluation Metrics
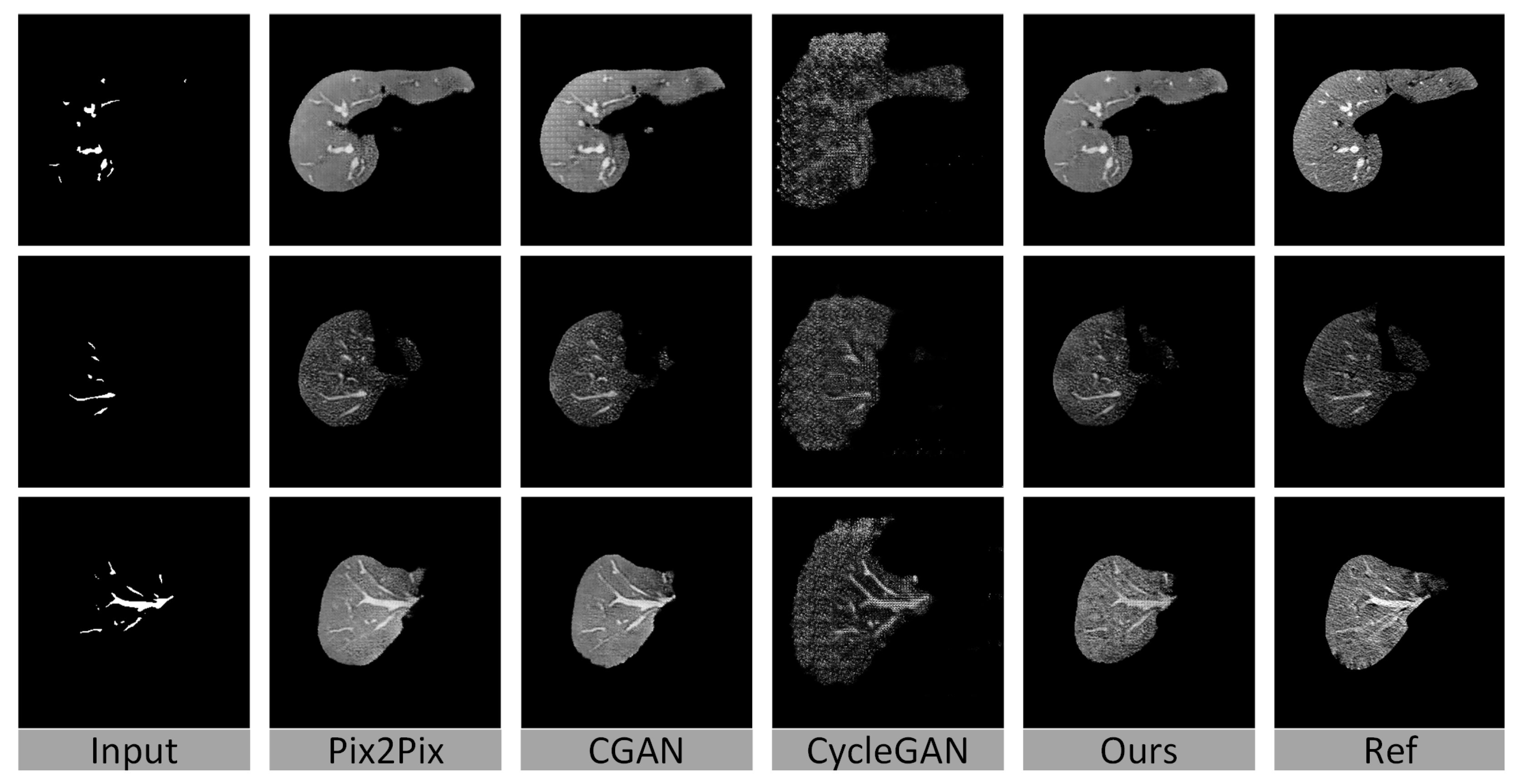
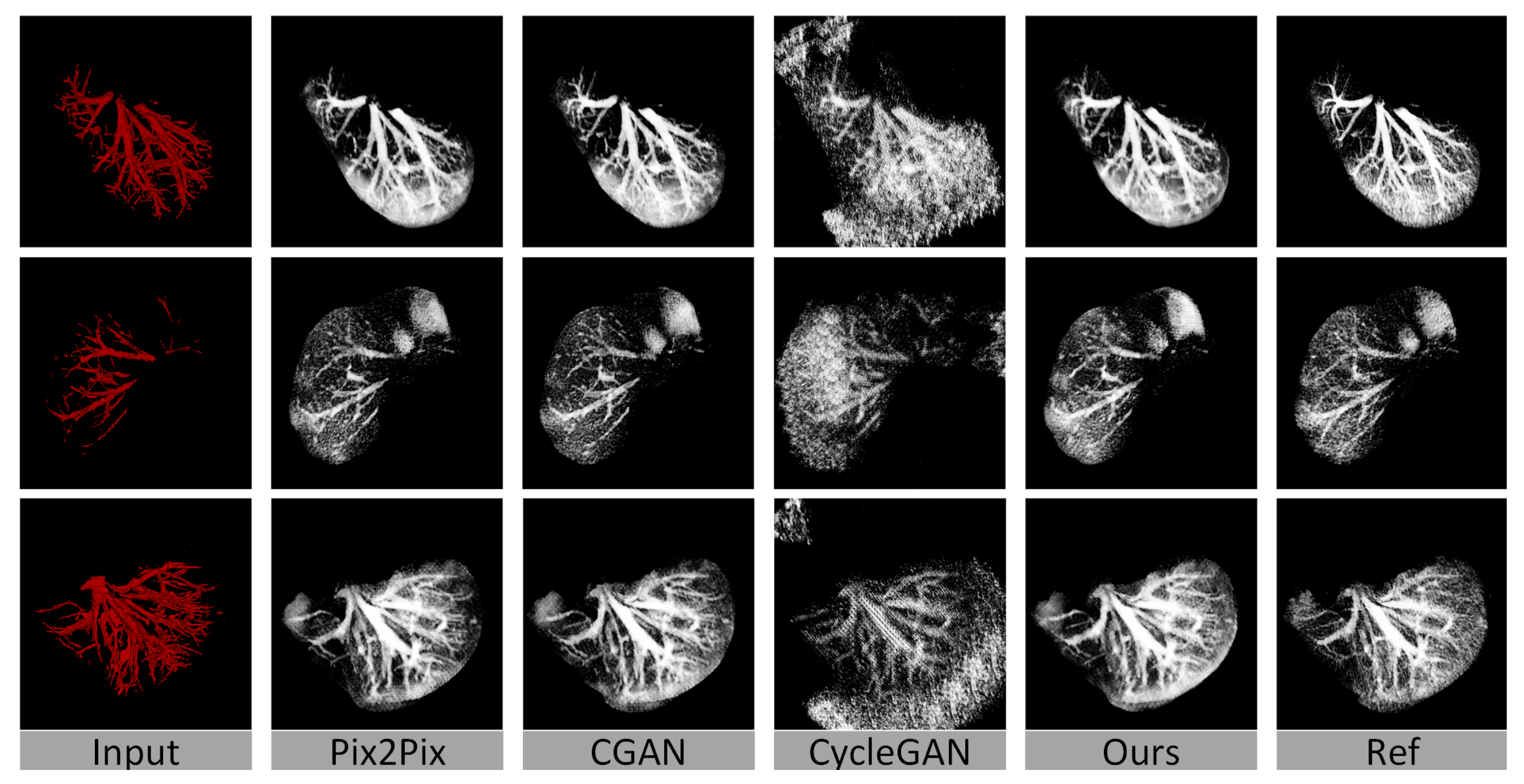
4.3. Baseline Methods
4.4. Quantitative Evaluation
4.5. Visualization of the Results
5. Discussion
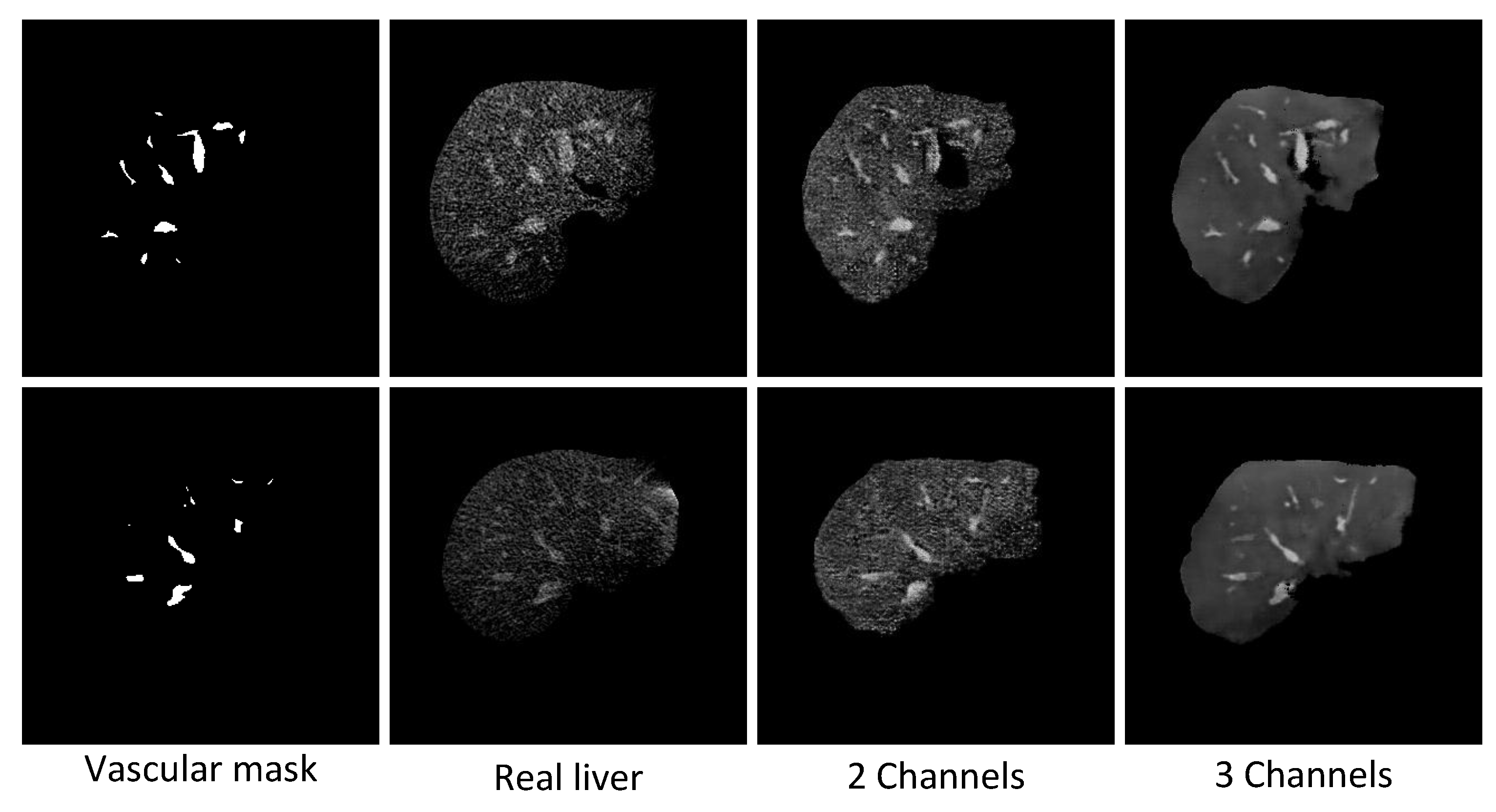
5.1. Discussion of the Discriminator Inputs
5.2. Advantages of the Multi-Task Generator
5.2.1. Discussion of the Differences in Texture Details
5.2.2. Discussion of the Effect of Lesion Synthesis
5.3. Limitations of Our Method
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huettl, F.; Saalfeld, P.; Hansen, C.; Preim, B.; Poplawski, A.; Kneist, W.; Lang, H.; Huber, T. Virtual reality and 3D printing improve preoperative visualization of 3D liver reconstructions—Results from a preclinical comparison of presentation modalities and user’s preference. Ann. Transl. Med. 2021, 9, 1074. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Lu, Y.; Li, K.; Pu, B.; Tan, Y.; Zhu, N. A YOLOX-based Deep Instance Segmentation Neural Network for Cardiac Anatomical Structures in Fetal Ultrasound Images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 1–12. [Google Scholar] [CrossRef]
- Wu, X.; Tan, G.; Pu, B.; Duan, M.; Cai, W. DH-GAC: Deep hierarchical context fusion network with modified geodesic active contour for multiple neurofibromatosis segmentation. Neural Comput. Appl. 2022, 1–16. [Google Scholar] [CrossRef]
- Pu, B.; Lu, Y.; Chen, J.; Li, S.; Zhu, N.; Wei, W.; Li, K. Mobileunet-fpn: A semantic segmentation model for fetal ultrasound four-chamber segmentation in edge computing environments. IEEE J. Biomed. Health Inform. 2022, 26, 5540–5550. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Yuan, L.; Wen, X.; Xu, H.; Wang, J. End-To-End Retina Image Synthesis Based on CGAN Using Class Feature Loss and Improved Retinal Detail Loss. IEEE Access 2022, 10, 83125–83137. [Google Scholar] [CrossRef]
- Mendes, J.; Pereira, T.; Silva, F.; Frade, J.; Morgado, J.; Freitas, C.; Negrão, E.; de Lima, B.F.; da Silva, M.C.; Madureira, A.J.; et al. Lung CT image synthesis using GANs. Expert Syst. Appl. 2023, 215, 119350. [Google Scholar] [CrossRef]
- Costa, P.; Galdran, A.; Meyer, M.I.; Niemeijer, M.; Abràmoff, M.; Mendonça, A.M.; Campilho, A. End-to-end adversarial retinal image synthesis. IEEE Trans. Med. Imaging 2017, 37, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Jabbarpour, A.; Mahdavi, S.R.; Sadr, A.V.; Esmaili, G.; Shiri, I.; Zaidi, H. Unsupervised pseudo CT generation using heterogenous multicentric CT/MR images and CycleGAN: Dosimetric assessment for 3D conformal radiotherapy. Comput. Biol. Med. 2022, 143, 105277. [Google Scholar] [CrossRef] [PubMed]
- Skandarani, Y.; Jodoin, P.M.; Lalande, A. Gans for medical image synthesis: An empirical study. J. Imaging 2023, 9, 69. [Google Scholar] [CrossRef]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.H.; Wei, Z.; Heidari, A.A.; Zheng, N.; Li, Z.; Chen, H.; Hu, H.; Zhou, Q.; Guan, Q. Generative adversarial networks in medical image augmentation: A review. Comput. Biol. Med. 2022, 144, 105382. [Google Scholar] [CrossRef] [PubMed]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Yu, B.; Zhou, L.; Wang, L.; Shi, Y.; Fripp, J.; Bourgeat, P. Ea-GANs: Edge-Aware Generative Adversarial Networks for Cross-Modality MR Image Synthesis. IEEE Trans. Med Imaging 2019, 38, 1750–1762. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Karras, T.; Laine, S.; Aila, T. A style-based generator architecture for generative adversarial networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 4401–4410. [Google Scholar]
- He, R.; Xu, S.; Liu, Y.; Li, Q.; Liu, Y.; Zhao, N.; Yuan, Y.; Zhang, H. Three-Dimensional Liver Image Segmentation Using Generative Adversarial Networks Based on Feature Restoration. Front. Med. 2021, 8, 794969. [Google Scholar] [CrossRef]
- Ho, J.; Jain, A.; Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 2020, 33, 6840–6851. [Google Scholar]
- Nichol, A.Q.; Dhariwal, P. Improved denoising diffusion probabilistic models. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 18 July 2021; pp. 8162–8171. [Google Scholar]
- Ramesh, A.; Dhariwal, P.; Nichol, A.; Chu, C.; Chen, M. Hierarchical text-conditional image generation with clip latents. arXiv 2022, arXiv:2204.06125. [Google Scholar]
- Shin, H.C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In Proceedings of the Simulation and Synthesis in Medical Imaging: Third International Workshop, SASHIMI 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; pp. 1–11. [Google Scholar]
- Han, C.; Hayashi, H.; Rundo, L.; Araki, R.; Shimoda, W.; Muramatsu, S.; Furukawa, Y.; Mauri, G.; Nakayama, H. GAN-based synthetic brain MR image generation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 734–738. [Google Scholar]
- Oliveira, D.A.B. Implanting synthetic lesions for improving liver lesion segmentation in CT exams. arXiv 2020, arXiv:2008.04690. [Google Scholar]
- Jiang, Y.; Chen, H.; Loew, M.; Ko, H. COVID-19 CT image synthesis with a conditional generative adversarial network. IEEE J. Biomed. Health Inform. 2020, 25, 441–452. [Google Scholar] [CrossRef]
- Ying, X.; Guo, H.; Ma, K.; Wu, J.; Weng, Z.; Zheng, Y. X2CT-GAN: Reconstructing CT from biplanar X-rays with generative adversarial networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 10619–10628. [Google Scholar]
- Hering, A.; Hansen, L.; Mok, T.C.; Chung, A.C.; Siebert, H.; Häger, S.; Lange, A.; Kuckertz, S.; Heldmann, S.; Shao, W.; et al. Learn2Reg: Comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning. IEEE Trans. Med. Imaging 2022, 42, 697–712. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q. A survey on multi-task learning. IEEE Trans. Knowl. Data Eng. 2021, 34, 5586–5609. [Google Scholar] [CrossRef]
- Pu, B.; Li, K.; Li, S.; Zhu, N. Automatic fetal ultrasound standard plane recognition based on deep learning and IIoT. IEEE Trans. Ind. Inform. 2021, 17, 7771–7780. [Google Scholar] [CrossRef]
- Zhao, L.; Li, K.; Pu, B.; Chen, J.; Li, S.; Liao, X. An ultrasound standard plane detection model of fetal head based on multi-task learning and hybrid knowledge graph. Future Gener. Comput. Syst. 2022, 135, 234–243. [Google Scholar] [CrossRef]
- Wang, G.; Gong, E.; Banerjee, S.; Martin, D.; Tong, E.; Choi, J.; Chen, H.; Wintermark, M.; Pauly, J.M.; Zaharchuk, G. Synthesize High-Quality Multi-Contrast Magnetic Resonance Imaging From Multi-Echo Acquisition Using Multi-Task Deep Generative Model. IEEE Trans. Med. Imaging 2020, 39, 3089–3099. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zheng, F.; Cong, R.; Huang, W.; Scott, M.R.; Shao, L. MCMT-GAN: Multi-Task Coherent Modality Transferable GAN for 3D Brain Image Synthesis. IEEE Trans. Image Process. 2020, 29, 8187–8198. [Google Scholar] [CrossRef] [PubMed]
- Vandenhende, S.; Georgoulis, S.; Van Gansbeke, W.; Proesmans, M.; Dai, D.; Van Gool, L. Multi-task learning for dense prediction tasks: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3614–3633. [Google Scholar] [CrossRef]
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.; Moreau, J.; Osswald, A.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient Specific Anatomical and Medical Image Database; Tech. Rep; IRCAD: Strasbourg, France, 2010; Volume 1. [Google Scholar]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Zhao, L. Liver Vessel Segmentation. 2022. Available online: https://doi.org/10.21227/rwys-mk84 (accessed on 30 May 2023).
- Yan, Q.; Wang, B.; Zhang, W.; Luo, C.; Xu, W.; Xu, Z.; Zhang, Y.; Shi, Q.; Zhang, L.; You, Z. Attention-guided deep neural network with multi-scale feature fusion for liver vessel segmentation. IEEE J. Biomed. Health Inform. 2020, 25, 2629–2642. [Google Scholar] [CrossRef]
- Su, J.; Liu, Z.; Zhang, J.; Sheng, V.S.; Song, Y.; Zhu, Y.; Liu, Y. DV-Net: Accurate liver vessel segmentation via dense connection model with D-BCE loss function. Knowl.-Based Syst. 2021, 232, 107471. [Google Scholar] [CrossRef]
- Gao, Z.; Zong, Q.; Wang, Y.; Yan, Y.; Wang, Y.; Zhu, N.; Zhang, J.; Wang, Y.; Zhao, L. Laplacian salience-gated feature pyramid network for accurate liver vessel segmentation. IEEE Trans. Med. Imaging 2023. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Caruna, R. Multitask learning: A knowledge-based source of inductive bias1. In Proceedings of the ICML’93: Proceedings of the Tenth International Conference on International Conference on Machine Learning, Amherst, MA, USA, 27–29 July 1993; pp. 41–48. [Google Scholar]
- Chen, Z.; Badrinarayanan, V.; Lee, C.Y.; Rabinovich, A. Gradnorm: Gradient normalization for adaptive loss balancing in deep multitask networks. In Proceedings of the International Conference on Machine Learning, Macau, China, 26–28 February 2018; pp. 794–803. [Google Scholar]
- Désidéri, J.A. Multiple-gradient descent algorithm (MGDA) for multiobjective optimization. Comptes Rendus Math. 2012, 350, 313–318. [Google Scholar] [CrossRef]
- Sener, O.; Koltun, V. Multi-task learning as multi-objective optimization. Adv. Neural Inf. Process. Syst. 2018, 31. [Google Scholar]
- Karush, W. Minima of Functions of Several Variables with Inequalities as Side Constraints. Master’s Thesis, Department of Mathematics, University of Chicago, Chicago, IL, USA, 1939. [Google Scholar]
- Bertsekas, D.P. Nonlinear programming. J. Oper. Res. Soc. 1997, 48, 334. [Google Scholar] [CrossRef]
- Lin, X.; Zhen, H.L.; Li, Z.; Zhang, Q.F.; Kwong, S. Pareto multi-task learning. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Demir, U.; Unal, G. Patch-based image inpainting with generative adversarial networks. arXiv 2018, arXiv:1803.07422. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. Gans trained by a two time-scale update rule converge to a local nash equilibrium. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Bińkowski, M.; Sutherland, D.J.; Arbel, M.; Gretton, A. Demystifying mmd gans. arXiv 2018, arXiv:1801.01401. [Google Scholar]
- Zhang, R.; Isola, P.; Efros, A.A.; Shechtman, E.; Wang, O. The unreasonable effectiveness of deep features as a perceptual metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 586–595. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Obukhov, A.; Seitzer, M.; Wu, P.W.; Zhydenko, S.; Kyl, J.; Lin, E.Y.J. High-Fidelity Performance Metrics for Generative Models in PyTorch; Version: 0.3.0; Zenodo: Geneva, Switzerland, 2020. [Google Scholar] [CrossRef]







| Input Size | 320 × 320 × 64 | 320 × 320 × 48 | 320 × 320 × 32 |
| Generator Encoder | 160 × 160 × 32 | 160 × 160 × 24 | 160 × 160 × 32 |
| 80 × 80 × 16 | 80 × 80 × 12 | 80 × 80 × 16 | |
| 40 × 40 × 8 | 40 × 40 × 6 | 40 × 40 × 8 | |
| 20 × 20 × 4 | 20 × 20 × 3 | 20 × 20 × 4 | |
| 10 × 10 × 2 | 10 × 10 × 1 | 10 × 10 × 2 | |
| Bottleneck | 5 × 5 × 1 | 5 × 5 × 1 | 5 × 5 × 1 |
| Input Size | 320 × 320 × 64 | 320 × 320 × 48 | 320 × 320 × 32 |
| Discriminator | 160 × 160 × 32 | 160 × 160 × 24 | 160 × 160 × 16 |
| 80 × 80 × 16 | 80 × 80 × 12 | 80 × 80 × 8 | |
| 40 × 40 × 8 | 40 × 40 × 6 | 40 × 40 × 4 | |
| 20 × 20 × 4 | 20 × 20 × 3 | 20 × 20 × 2 | |
| Output Size | 10 × 10 × 4 | 10 × 10 × 3 | 10 × 10 × 2 |
| Segmentation Layers | 64 s | 48 s | 32 s |
| Number of liver sequence segments | 1621 | 2323 | 3579 |
| Number | Model/Evaluation Metrics | FID (dim = 2048) | KID | LPIPS | DSC |
|---|---|---|---|---|---|
| 1 | CGAN_64s | 101.459 | 0.1172 | 0.180 | 0.865 |
| 2 | Pix2Pix_64s | 85.045 | 0.097 | 0.174 * | 0.867 |
| 3 | CycleGAN_64s | 245.226 | 0.307 | 0.449 | 0.599 |
| 4 | MTv2l_64s (Ours) | 79.295 * | 0.086 * | 0.177 | 0.872 * |
| 5 | CGAN_48s | 98.485 | 0.1168 | 0.186 | 0.863 |
| 6 | Pix2Pix_48s | 111.586 | 0.136 | 0.192 | 0.856 |
| 7 | CycleGAN_48s | 195.270 | 0.234 | 0.422 | 0.614 |
| 8 | MTv2l_48s (Ours) | 61.900 * | 0.062 * | 0.181 * | 0.865 * |
| 9 | CGAN_32s | 109.561 | 0.129 | 0.194 | 0.855 |
| 10 | Pix2Pix_32s | 107.912 | 0.124 | 0.186 * | 0.833 |
| 11 | CycleGAN_32s | 297.366 | 0.344 | 0.515 | 0.464 |
| 12 | MTv2l_32s (Ours) | 85.992 * | 0.094 * | 0.187 | 0.858 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Q.; Zhao, L. End-to-End 3D Liver CT Image Synthesis from Vasculature Using a Multi-Task Conditional Generative Adversarial Network. Appl. Sci. 2023, 13, 6784. https://doi.org/10.3390/app13116784
Xiao Q, Zhao L. End-to-End 3D Liver CT Image Synthesis from Vasculature Using a Multi-Task Conditional Generative Adversarial Network. Applied Sciences. 2023; 13(11):6784. https://doi.org/10.3390/app13116784
Chicago/Turabian StyleXiao, Qianmu, and Liang Zhao. 2023. "End-to-End 3D Liver CT Image Synthesis from Vasculature Using a Multi-Task Conditional Generative Adversarial Network" Applied Sciences 13, no. 11: 6784. https://doi.org/10.3390/app13116784
APA StyleXiao, Q., & Zhao, L. (2023). End-to-End 3D Liver CT Image Synthesis from Vasculature Using a Multi-Task Conditional Generative Adversarial Network. Applied Sciences, 13(11), 6784. https://doi.org/10.3390/app13116784




